Abstract
HIV-1 resists neutralization by most antibodies. Two somatically related human antibodies, PG9 and PG16, however, each neutralize 70 to 80% of circulating HIV-1 isolates. Here we present the structure of the antigen-binding fragment of PG16 in monoclinic and orthorhombic lattices at 2.4 and 4.0 Å, respectively, and use a combination of structural analysis, paratope dissection, and neutralization assessment to determine the functional relevance of three unusual PG9/PG16 features: N-linked glycosylation, extensive affinity maturation, and a heavy chain-third complementarity-determining region (CDR H3) that is one of the longest observed in human antibodies. Glycosylation extended off the side of the light chain variable domain and was not required for neutralization. The CDR H3 formed an axe-shaped subdomain, which comprised 42% of the CDR surface, with the axe head looming ∼20 Å above the other combining loops. Comprehensive sets of chimeric swaps between PG9 and PG16 of light chain, heavy chain, and CDR H3 were employed to decipher structure-function relationships. Chimeric swaps generally complemented functionally, with differences in PG9/PG16 neutralization related primarily to residue differences in CDR H3. Meanwhile, chimeric reversions to genomic V genes showed isolate-dependent effects, with affinity maturation playing a significant role in augmenting neutralization breadth (P = 0.036) and potency (P < 0.0001). The structural and functional details of extraordinary CDR H3 and extensive affinity maturation provide insights into the neutralization mechanism of and the elicitation pathway for broadly neutralizing antibodies like PG9 and PG16.
To create antibodies capable of effectively neutralizing human immunodeficiency virus type 1 (HIV-1), the adaptive humoral response is driven to exceptional lengths (reviewed in reference 8). Indeed, the response often fails, and sera from individuals infected with HIV-1 typically display limited neutralization breadth (59). After several years of infection, however, antibodies capable of neutralizing diverse viral strains develop in 15 to 25% of infected individuals (3, 16, 32, 33, 49, 53). Details of the adaptive changes that allow for effective recognition are of direct vaccine relevance, and clues from rare neutralizing antibodies have been eagerly sought.
Two broadly neutralizing antibodies, PG9 and PG16, were recently identified with single cell-sequencing techniques after direct microneutralization assessment of secreted antibody from individually plated, stimulated B cells (58). These antibodies are somatically related and appear to be derived from the same recombination of heavy and light chains. They both recognize a site on HIV-1 gp120 composed of elements from the second and third variable regions (V2 and V3). Despite the vaunted diversity of the HIV-1 gp120 envelope and the even higher sequence variability in the V2 and V3 regions (26), neutralization assays indicate that the recognized epitope is conserved in 70 to 80% of circulating viral isolates (58).
To investigate the molecular features of PG9 and PG16 that account for their neutralization effectiveness, we prepared antigen-binding fragments (Fabs) of each antibody and screened for crystallization. We were able to obtain a number of crystals, and those of PG16 proved suitable for structural analysis. Determination of the PG16 structure visualized several unusual features, and structure-function analysis indicated that two features, extensive affinity maturation and an exceptionally long heavy chain-third complementarity-determining region (CDR H3), were critical to its neutralization effectiveness. Barriers to eliciting these two features provide a likely explanation for the rarity of antibodies like PG9 and PG16; understanding and overcoming such barriers may form the basis for an effective HIV-1 vaccine.
MATERIALS AND METHODS
Production of PG9 and PG16.
The sequences of PG9 and PG16 light and heavy chains were synthesized by Geneart and cloned into the pVRC8400 expression vector. To express PG9 and PG16 antibodies, 250 μg of light chain plasmid and 250 μg of heavy chain plasmid were mixed with 1 ml of 293fectin for 20 min and added to 1 liter of 293 FreeStyle or HEK293S GnTI− cells (47). Cells were incubated in FreeStyle 293 expression medium (Invitrogen, Carlsbad, CA) for suspension culture at 8% CO2, 37.0°C, and 125 rpm. Five days after transfection, supernatants were harvested and passed over a protein A column. Bound antibody was eluted using IgG elution buffer (Pierce) and dialyzed against phosphate-buffered saline (PBS).
Production and deglycosylation of Fab.
Immunoglobulins were proteolyzed to Fab by Lys-C for PG9 and Ficin for PG16 followed by passage over a protein A affinity column (Pierce) using previously described methodology (27). The flowthrough was collected and further purified by gel filtration chromatography (Superdex 200; GE Healthcare) to obtain glycosylated Fab. To produce deglycosylated Fab, full-length antibodies were produced in GnTI− cells and processed to Fab as described above and then treated with His-tagged endoglycosidase H. Deglycosylated Fab was passed through a Ni2+ column, and the flowthrough was collected and further purified by gel filtration. The protein was used immediately or flash-frozen and stored at −80°C.
Crystallization of PG9 and PG16.
Fab fragments of PG9 and PG16 (both glycosylated and deglycosylated) were screened for crystallization using 576 conditions derived from Crystal Screen (Hampton Research), Precipitant Synergy Screen (Emerald BioSystems), and Wizard Screen (Emerald BioSystems), where each of the conditions in the commercially specified screen was modified with 3 to 4 levels of precipitant to increase screen diversity. Vapor-diffusion sitting droplet crystallization trials were set up robotically (Cartesian Honeybee robot; 0.1 μl of protein plus 0.1 μl of crystallization solution). Droplets were allowed to incubate at 20°C and observed daily during the first week after setup and biweekly thereafter. Crystallization hits obtained by robotic screening were optimized manually using vapor-diffusion hanging droplets (0.5 to 1 μl of protein plus 0.5 to 1 μl of crystallization solution).
For PG9, robotic crystallization of the glycosylated Fab identified a single hit (20% polyethylene glycol 3350 [PEG 3350], 20% isopropanol, 100 mM Tris-HCl, pH 8.5) whereas no hits were found with the deglycosylated Fab (see Fig. S1A in the supplemental material).
For the glycosylated PG16 Fab, four hits were obtained by robotic screening and further optimized. One condition (1.96 M ammonium sulfate, 0.2 M NaCl, 0.1 M acetate, pH 4.5) gave small crystals (0.05 × 0.01 × ∼0.005 mm). These crystals were optimized further by use of the Additive Screen (Hampton Research) and by microseeding (55). To produce crystals suitable for structural analysis, 0.5 μl of glycosylated PG16 Fab was combined with 0.5 μl of reservoir solution (1.37 M ammonium sulfate, 1.4 M NaCl, 0.07 M acetate, pH 4.5, with 3% 1,6-diaminohexane) in vapor-diffusion hanging droplets over 0.5-ml reservoirs. After 24 h, crystal seeds (prepared from small clusters of crystals using the Seed Bead kit [Hampton Research] following the manufacturer's suggested directions) were introduced with a hair into the drop. These orthorhombic crystals grew to a maximum size of 0.20 × 0.01 × 0.02 mm at 20°C (see Fig. S1B in the supplemental material).
For the deglycosylated PG16, five hits were obtained with the robot and further optimized. One condition [1.1 M sodium dihydrogen phosphate, 0.74 M potassium dihydrogen phosphate, 0.2 M lithium sulfate monohydrate, 0.1 M 3-(cyclohexylamino)-1-propanesulfonic acid (CAPS), pH 10.5] gave well-formed and reproducible crystals. Increasing the concentration of the Fab and using microseeding helped to obtain single crystals (0.5 × 0.3 × 0.1 mm) (see Fig. S1C in the supplemental material).
Data collection, structure solution, and refinement (monoclinic form).
Crystals of deglycosylated PG16 Fab were transferred to a cryoprotectant solution containing 15% 2R3R-butanediol, 1 M potassium phosphate, 1.5 M sodium phosphate, 0.2 M lithium sulfate, and 0.1 M CAPS, pH 10.5, for 1 to 2 min; mounted in a cryoloop (Hampton Research); and flash-cooled in a liquid nitrogen cryostat (100 K). X-ray data were collected at a wavelength of 1.00 Å with a bending magnet beam-line (BM-22; Southeast Region Collaborative Access team [SER-CAT]) at the Advanced Photon Source and processed and reduced with HKL2000 (42). The monoclinic crystals were indexed as primitive monoclinic lattice, and solvent considerations suggested four complexes per asymmetric unit. The crystal structure was solved by molecular replacement in space group P21 using Phaser (35, 36) as implemented in CCP4i (11) and Protein Data Bank (PDB) identification (ID) code 1NL0 (20) as the search model. Refinement was carried out with PHENIX (1), and model building was performed in COOT (18). Validation of the structure was performed using the program MolProbity (10) and the PDB validation server (http://deposit.pdb.org/validate/). X-ray crystallographic data collection and refinement statistics are shown in Table S1 in the supplemental material.
Data collection, structure solution, definition of CDR H3 structure, and refinement (orthorhombic form).
Crystals of the glycosylated PG16 Fab were transferred to a cryoprotectant solution containing 15% xylitol, 15% ethylene glycol, 1.96 M ammonium sulfate, 0.2 M NaCl, 0.1 M acetate, pH 4.5, and 3% 1,6-diaminohexane for 1 to 2 min; mounted in a cryoloop (Hampton Research); and flash-cooled in a liquid nitrogen cryostat (100 K). X-ray data were collected at a wavelength of 1.00 Å with the third-generation undulator beam-line (ID-22; SER-CAT) at the Advanced Photon Source and processed and reduced with HKL2000 (42). Data from the orthorhombic crystals were indexed in a C-centered orthorhombic lattice, and solvent considerations suggested three complexes per asymmetric unit. The crystal structure was solved by molecular replacement in space group C222 using Phaser (35, 36) as implemented in CCP4i (11) and PDB ID code 1NL0 (20) as the search model. Due to the low resolution of the data, the Rfree values converged initially to ∼40% (4). Once the high-resolution structure of the deglycosylated form was available (monoclinic structure described here; PDB code 3LRS), it was used as the search model for molecular replacement. Translation-libration-screw motion (TLS) and rigid body refinement in PHENIX (1) yielded an Rfree value of ∼33% with electron density for the CDR H3 clearly visible in one Fab molecule and somewhat visible in a second molecule but not clear in the third molecule. Each of the four independent Fab molecules from the refined monoclinic structure was independently superimposed with each of the three Fab molecules in the orthorhombic lattice, and the combined model with lowest Rfree was used for phasing. Considerations for low-resolution refinement as described by DeLaBarre and Brunger (13) were followed. EM-IMO (65), a real-space refinement program for building and refining protein models within cryo-electron density maps, was used to construct the initial backbone trace of the CDR H3, which was improved by manual model building with O (21) and XtalView/Xfit (37). Validation of the structure was performed using the program MolProbity (10) and the PDB validation server (http://deposit.pdb.org/validate/). X-ray crystallographic data collection and refinement statistics are shown in Table S1 in the supplemental material.
Structural homology of PG16 CDR H3.
To search for structural homologs of the CDR H3, we submitted its structure to Dali (19). No homologs of significant structural similarity were identified. We next constructed a database of proteins using the PISCES server (http://dunbrack.fccc.edu/PISCES.php) with the following criteria: sequence identity of ≤90%, resolution of ≤3.0 Å, and R value of ≤1.0. The resulting database contains 16,938 proteins. We then structurally aligned the CDR H3 onto each of the proteins in the database using TMalign (62) and processed the output using the following criteria: the entire CDR H3 (28 amino acids) being aligned onto the protein, the number of gaps in the CDR H3 sequence being ≤3, and the root mean square deviation (RMSD) of aligned residues being ≤4.0 Å. Note that although stringent alignment criteria had been applied to the CDR H3 structure, the proteins to be searched were not constrained, as such constraints may eliminate potential structural homologs.
Homology modeling of PG9 and of PG9/PG16 chimeras.
Alignments between PG9 and PG16 sequences were generated using ClustalW2 (28). A structural model of PG9 (heavy or light) was constructed using Nest, a homology modeling program based on rigid-body optimization (45). The compatibleness of PG16/PG9 swaps was evaluated using two residue-based scoring functions: one is a table function derived from the van der Waals energy function in the OPLS (optimized potentials for liquid simulations) united-atom force field and softened using a free energy perturbation formula (56), and the other is a normalized DFIRE statistical potential (63). The van der Waals function was mainly used to detect residue clashes on the interface, while DFIRE was used to check for other unfavorable residue interactions. The two scoring functions have been used in the model quality assessment server http://wiki.c2b2.columbia.edu/honiglab_public/index.php/Software:Model_Quality_Assessment.
Creation of PG9/PG16 chimeras and genomic revertants.
Variants of PG9 and PG16 antibodies were synthesized as follows: PG9 and PG16 light chain V-gene regions were reverted to the genomic precursor gene IGLV2-14*01 as determined using the IMGT website (http://imgt.cines.fr/). The V-gene reverted light chain sequences were synthesized by Geneart and cloned into the pVRC8400 vector. PG9 and PG16 heavy chain V-gene regions were reverted to germ line IGHV3-33*05. The sequence alignments of the putative V-gene germ line predecessors of PG9 and PG16 and the affinity-matured antibodies can be seen in Fig. S2 in the supplemental material. Finally the CDR H3 sequences of PG9 and PG16 were swapped to obtain a PG16 heavy chain with the PG9 CDR H3 and PG9 heavy chain with the PG16 CDR H3. The modified heavy chain sequences of PG9 and PG16 were synthesized by Geneart and cloned into the pVRC8400 vector. All swap antibodies were expressed and purified as described under “Production of PG9 and PG16.”
Virus stocks and neutralization assays.
HIV-1 Env pseudoviruses were prepared by transfecting 293T cells (6 × 106 cells in 50 ml growth medium in a T-175 culture flask) with 10 μg of rev/env expression plasmid and 30 μg of an env-deficient HIV-1 backbone vector (pSG3ΔEnvelope), using Fugene 6 transfection reagent (Invitrogen). Pseudovirus-containing culture supernatants were harvested 2 days after transfection, filtered (0.45 μm), and stored at −80°C or in the vapor phase of liquid nitrogen. Neutralization was measured using HIV-1 Env pseudoviruses to infect TZM-bl cells as described previously (30, 31). Briefly, 40 μl of pseudovirus was incubated for 30 min at 37°C with 10 μl of serially diluted test antibody in duplicate wells of a 96-well flat-bottomed culture plate. To keep assay conditions constant, sham medium was used in place of antibody in control wells. The pseudovirus input was set at a multiplicity of infection of approximately 0.01, which generally results in 100,000 to 400,000 relative light units (RLU) in a luciferase assay (Bright Glo; Promega, Madison, WI). Neutralization curves were fit by nonlinear regression using a 5-parameter hill slope equation as previously described (52). The 50% inhibitory concentrations (IC50) were reported as the antibody concentrations required to inhibit viral entry by 50% (see Tables S2, S3, and S4 in the supplemental material). Note that for statistical analyses, an arbitrary value of twice the maximum concentration used was reported for IC50 of chimeric antibodies that did not inhibit viral entry by 50%.
Analysis of neutralization data.
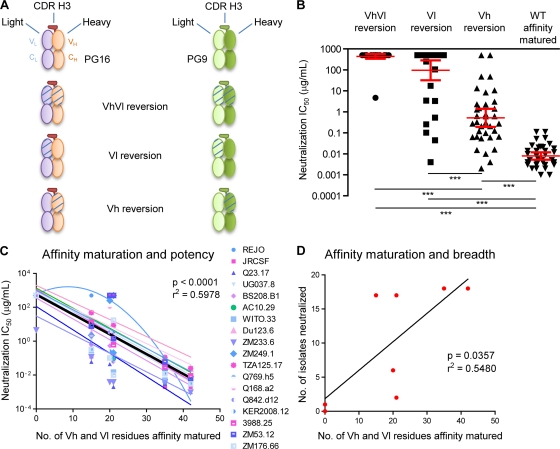
The following statistical analyses were performed using the GraphPad Prism software package (version 5.00 for Windows; GraphPad). The median IC50 values of eight chimeric antibodies (PG9 and PG16 wild type [WT], PG9 and PG16 Vh reverted, PG9 and PG16 Vl reverted, and PG9 and PG16 VhVl reverted) on 18 viral isolates were compared using a nonparametric Kruskal-Wallis test (see Fig. 6B). Linear and quadratic regression methods were used to model the effect of affinity maturation on potency for all 18 isolates (see Fig. 6C). Neutralization of 17 isolates (all except the REJO isolate) was best modeled using linear regression. Analysis of covariance (ANCOVA) indicated that these 17 isolates could be accurately modeled with one pooled linear regression line (Y-int = 2.751, slope = −0.1171, F = 199.2, P < 0.0001, and R2 = 0.5978). Linear regression was also used to model the relationship between affinity maturation and breadth (Y-int = 1.840, slope = 0.4174, F = 7.275, P = 0.0357, and R2 = 0.5480) (see Fig. 6D).
FIG. 6.
V-gene reverted chimeras and dissection of functional relevance. Resolution-enhancing methods are particularly useful when analyzing diffuse effects such as somatic hypermutation, which alters ∼50 residues between PG9 and PG16. (A) Schematic V-gene reverted chimeras of PG16 and PG9 Fabs are shown: both heavy and light chain V genes reverted to germ line precursor, only heavy V gene reverted, or only light chain V gene reverted (see Fig. S2 in the supplemental material). PG16 light chain is in light blue, PG16 heavy chain is in tan, and PG16 CDR H3 is in red; PG9 light chain is in light green, PG9 heavy chain is in green, and PG9 CDR H3 is bordered in dark green. V-gene reversion to germ line is shown by hatching. (B) IC50 are reported for each category of antibodies: V-gene heavy and light chain reverted to germ line (VhVl), Vl reverted, Vh reverted, and affinity-matured antibodies, i.e., wild-type PG9 and PG16. There were significant differences in IC50 among the four groups according to a Kruskal-Wallis (KW) test (KW = 112, P < 0.0001) or a one-way analysis of variance (F = 129.3, P < 0.0001). Dunn's multiple comparison post hoc tests indicated that differences between the groups were highly significant (P < 0.001, indicated by a triple asterisk on the graph). (C) Correlation between affinity maturation and potency. The IC50 (potency) values were plotted as a function of the number of residues that are affinity matured, i.e., 15 and 21 residues for the Vl of PG9 and PG16, respectively, and 20 residues for the Vh of both PG9 and PG16. The Vh and Vl reverted chimeras contain no affinity maturation in the V gene, and the PG9 and PG16 have 35 and 41 residues affinity matured in the V gene, respectively. Analysis indicated that one isolate, REJO, could not be fitted to a linear regression (see Materials and Methods). The rest of the data could be fitted by linear regression, and statistical analysis using Prism (version 5.00 for Windows; GraphPad) indicated that it was valid to model the entire data set (minus REJO) with a single linear regression with Y-int = 2.751, slope = −0.1171, F = 199.2, P < 0.0001, and R2 = 0.5978. Note that if the REJO isolate data set is included, the linear regression values are similar (Y-int = 2.831, slope = −0.1184, F = 209.5, P < 0.0001, and R2 = 0.5960). (D) Correlation between affinity maturation and breadth. The numbers of isolates that are neutralized at 50% using chimeric antibodies (at a concentration up to 50 μg/ml) are reported as a function of the number of residues that are V gene affinity matured. A linear regression model was used and showed a correlation between affinity maturation and breadth (Y-int = 1.840, slope = 0.4174, F = 7.275, P = 0.0357, and R2 = 0.5480). IC50 are reported in Table S3, and neutralization curves are shown in Fig. S6.
Structural alignments and figures.
Structural superpositions were carried out with the program “superpose” as implemented in CCP4i (11). Figures were prepared using the program PyMOL (14).
Protein structure accession numbers.
Coordinates and structure factors for the crystal structures of PG16 in monoclinic and orthorhombic lattices have been deposited with the Protein Data Bank under PDB accession codes 3LRS and 3MME, respectively.
RESULTS
Structure of PG16 in monoclinic and orthorhombic crystal lattices.
To obtain a structural understanding of the PG9 and PG16 antibodies, we proteolytically processed antibody to Fab, purified it, and screened it for crystallization. Initial robotic screens produced microcrystals of PG9 and PG16. Optimization of the PG16 crystallization by additive screening and microseeding produced rectangular needles with dimensions of up to 0.20 × 0.01 × 0.02 mm (see Fig. S1B in the supplemental material). Diffraction extended to 4.0 Å, and structure solution identified three Fabs per asymmetric unit in a C222 lattice. Two of the Fabs appeared to be reasonably ordered, including the complete complementarity-determining region, but a lack of diffraction beyond 4 Å hindered the refinement required for atomic-level definition of the CDR H3. Optimization of the PG9 crystallization condition did not yield crystals suitable for diffraction.
To obtain improved diffraction, we enzymatically deglycosylated Fabs to remove a single light chain N-linked sugar and screened for crystallization. Robotic crystallizations of deglycosylated PG9 produced no crystals; five conditions with crystals of deglycosylated PG16, however, were found (see Fig. S1C in the supplemental material). Optimization enabled the collection of diffraction data to 2.4 Å, and structure solution identified four Fabs per asymmetric unit in a P21 lattice. Refinement resulted in an R value of 21.1% (Rfree = 26.0%) (Table S1).
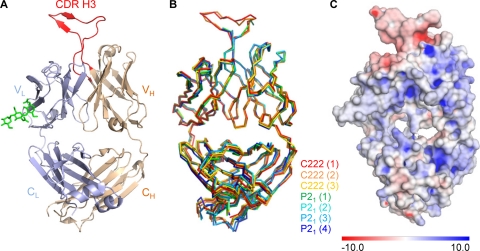
All four Fabs showed strong similarity with pairwise superposition yielding Cα RMSDs ranging from 0.16 to 0.33 Å for variable domain and 0.21 to 0.98 Å for the entire Fab. Electron density for residues 3 to 209 of the light chain and residues 1 to 214 of the heavy chain could be seen, but residues 100A to 100J of the heavy chain CDR H3 were disordered in all four molecules in the asymmetric unit (Fig. 1B).
FIG. 1.
Crystal structure of the antigen-binding fragment (Fab) of antibody PG16. Anti-HIV-1 antibodies that effectively neutralize HIV-1 often have unusual structural characteristics. With PG16, an extraordinary CDR H3 forms a separate subdomain, which towers over the combining-region surface. (A) Structure of antibody PG16, with complete combining region. A Cα-ribbon representation shows the light chain in blue, the heavy chain in tan, and its CDR H3 in red. The variable domains comprise the top half of the image, and the constant regions comprise the bottom. The single N-linked glycan (shown in green stick representation) was sufficiently well ordered at its protein-proximal base to define the placement of three sugars. The displayed Fab molecule is molecule 1 from the C222 lattice as shown in panel B. The CDR H3 of PG16 dominates the combining site and comprises 42% of the total surface area of the complementarity-determining regions. (B) Superposition of asymmetric unit components. PG16 crystallized in orthorhombic (C222) and monoclinic (P21) lattices, with 3 and 4 molecules per asymmetric unit, respectively. Shown are the Cα-backbone traces for all seven of the crystallized forms of antibody PG16, after superposition of variable domain-framework regions. The number for each molecule in each lattice is indicated in parentheses. (C) PG16 Fab electrostatic surface potentials. The PG16 electrostatic potentials are displayed on the molecular surface of PG16, with blue showing positively charged surfaces and red showing negatively charged surfaces. The scale is from −10 to +10 kT/e.
To visualize the entire combining region, we used the 2.4-Å refined monoclinic PG16 structure to bootstrap the refinement of the 4.0-Å orthorhombic crystals. Each of the four independent Fabs from the monoclinic lattice was superimposed onto the three independent Fabs in the orthorhombic lattice and rigid body refined. Then the combined model with lowest Rfree (4) was used for phasing. Inspection of the resultant electron density found the combining region in two of the molecules reasonably well ordered. The CDR H3 for one of these molecules proved to be sufficiently well defined to allow the entire CDR H3 to be traced (Fig. 1A; see also Fig. 3B). Highly restrained refinement (RMSD on bonds of 0.003 Å) led to an R value of 25.6% (Rfree, 31.7%) (see Table S1 in the supplemental material). The electrostatics of the Fab are shown in Fig. 1C and indicate that the Fab is mainly positively charged whereas the CDR H3 is negatively charged.
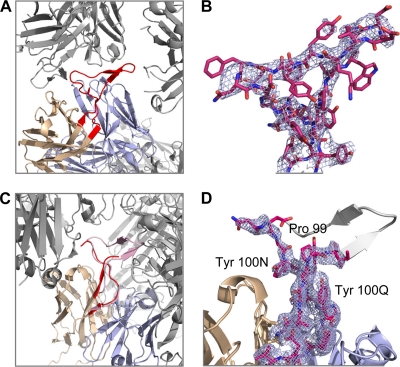
FIG. 3.
CDR H3 flexibility and tyrosine clasp. The head of the CDR H3 appears to have the ability to deform substantially in response to different environments, perhaps enabling it to maneuver into a recessed epitope on HIV-1. In contrast, the handle of the CDR H3 appears invariant, fixed by Tyr100N and Tyr100Q, which embrace and support the CDR H3 N terminus. (A) Ordered CDR H3 as seen in the C222 lattice. Neighboring molecules are colored gray whereas the CDR H3 is highlighted in red. (B) The CDR H3 of PG16 Fab from the C222 lattice is shown in stick representation, and the 2Fo-Fc map obtained after rigid body and TLS refinement in PHENIX is shown in light blue at 1σ. (C) Disordered CDR H3 (red) as seen in the P21 lattice. The ordered CDR H3 (pink) from the C222 lattice has been superimposed to highlight potential clashes within the crystal lattice. (D) A close-up of the CDR H3 (in red sticks) from the P21 lattice is shown to highlight the tyrosine clasp. The missing portion of the CDR H3 loop is modeled in gray. 2Fo-Fc electron density surrounding the CDR H3 loop is also shown in light blue at 1σ.
CDR H3 of PG16 forms a separate subdomain.
The CDR H3 region of PG16 consists of 28 residues (Kabat numbering) (23), one of the longest CDR H3 regions observed in a human antibody. Overall the structure resembles an axe, extending above the antibody-variable domains. The handle of the axe is formed by the N-terminal residues (AGGP99) and the C-terminal residues (YYNY100Q) of the CDR H3; residues AGG98 extend from strand F of the variable domain and hydrogen bond in antiparallel fashion to YNY100Q, which extends into strand G of the variable domain (Fig. 2B). In addition to the hydrogen bonding, the N-terminal amino acids are held in place by the side chains of Tyr100N and Tyr100Q, which grip Gly98 and Pro99 in a “tyrosine clasp” (Fig. 3D), a structural motif which is conserved in all seven of the crystallized forms of PG16.
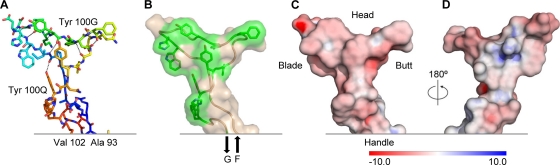
FIG. 2.
Structural and chemical properties of the PG16 CDR H3. One of the longest human CDR H3s ever observed, the PG16 CDR H3 is anionic and secured by a number of hydrogen bonds but lacks the hydrophobic core associated with more stable configurations. (A) Stick representation with rainbow coloring of carbon atoms from blue to red, Ala93 to Val102, respectively. Hydrogen bonds are depicted as dotted lines. Backbone hydrogen bonds include NGly98-OTyr100O, OGly98-NTyr100O, NIle100-OTyr100G, OIle100-NTyr100G, NTrp100A-OVal100E, OHis100B-NVal100E, NAsp100I-OAla100M, and OAsp100I-NAsp100L residue numbering follows the Kabat format (23). (B) Transparent molecular surface over a backbone ribbon with the side chains of aromatic residues shown as sticks and colored green. The CDR H3 extends from strand F and returns into strand G of the Fab. (C) Molecular surface colored according to electrostatic potentials from −10 to +10 kT/e, red to blue, respectively, with analogy to the axe vocabulary indicated. (D) 180° rotation of panel C about the y axis.
Continuing from Pro99 at the heel of the handle, the CDR H3 takes a sharp turn to form the head of the axe (Fig. 2A). Three residues (IWH100B) outline the bottom of the head with a β-strand, which—after a two-residue turn (DD100D) that forms the blade of the axe—forms four hydrogen bonds to the returning strand (VKY100G). This strand continues for three more residues (YDF100J) to outline the top of the head, before turning (N100K) at the butt of the head and returning (NGY100N) to the heel of the handle. Despite extensive H bonding, the head of the axe is ordered in only two of the seven crystallized lattices, perhaps influenced by lattice packing, which in the monoclinic crystal form does not leave enough room for a fully ordered head (Fig. 3A and C). Overall the axe is anionic (Fig. 2C and D), and many of its residues are aromatic (Fig. 2B).
Structural homology of PG16 CDR H3.
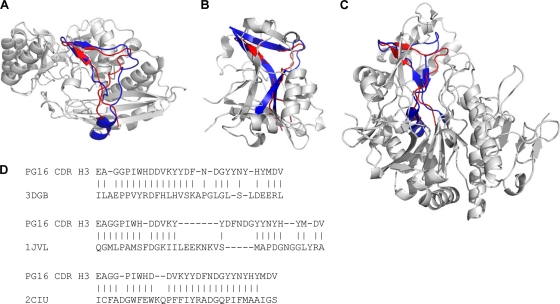
Six PDB entries were found to contain structural homologs of the CDR H3. The observed structural similarity could be categorized into three families (Fig. 4). One family consisted of three homologs, two from the enolase superfamily and one with unknown function (PDB IDs 2NQL, 3DGB [48], and 2OZ8). The resemblance of blade and butt of the CDR H3 axe motif could be recognized, while the handle region, which is made up by a twisted β sheet in the CDR H3, was replaced by a loop and a helix in all three proteins (Fig. 4A). Although the structural resemblance in the blade region was high, due to the structural mismatch of the handle region, the RMSD of aligned residues ranged from 3.47 to 3.71 Å.
FIG. 4.
Structural homologs of PG16 CDR H3. Analysis of structural homologs allows insight into the function of the PG16 subdomain in other contexts. Six structural homologs were identified from a database of 16,938 proteins. Based on the structural similarity, they could be categorized into three families, and a single example of each family is shown. Regions of homology to PG16 are highlighted in blue, and the CDR H3 of PG16 is shown in red. (A) One family consisted of three proteins, two from the enolase superfamily and one with unknown function (PDB IDs 2NQL, 3DGB, and 2OZ8). 3DGB is depicted here. The region of structural homology to the CDR H3 was packed on the protein surface, and the RMSD of aligned residues ranged from 3.47 to 3.71 Å. (B) In a second family, the CDR H3 was matched to a buried region sandwiched by secondary structure elements on both sides in two pyrophosphorylase proteins (PDB IDs 1JV1 and 2YQC), with an RMSD of 2.16 to 2.26 Å. 1JV1 is shown as the example of the second family. (C) The third family had only one protein, 2ICU, in which the CDR H3 of PG16 matched to a core segment surrounded by other structural components with an RMSD of 3.0 Å. None of the structural homologs protrudes from the protein body as does the CDR H3 of PG16. (D) Sequence alignments of the PG16 CDR H3 with the three structural homologs shown in panels A to C. The residues that have been aligned between the CDR H3 of PG16 and the structural homolog to calculate RMSD are shown with vertical lines.
In a second family, the CDR H3 was matched to a buried region in two pyrophosphorylase proteins (PDB IDs 1JV1 [44] and 2YQC [34]), with an RMSD of 2.16 to 2.26 Å (Fig. 4B). The blade and twisted handle regions were observed to fit better than the butt region, where an insertion can be found in the two proteins. As opposed to being packed on the protein surface, the structural homolog of the CDR H3 in this second family was sandwiched by secondary structure elements on both sides.
The third family of structural homology had only one member (PDB ID 2ICU), which showed a different structural pattern from that of the other two families. The CDR H3 matched to a core segment surrounded by other structural components of the protein. This segment contains more β-sheet content and showed moderate similarity to the CDR H3 with an RMSD of 3.0 Å (Fig. 4C). Our results suggest that the axe motif is rare in existing protein structures and that, in the few cases where it is found, it is utilized as a structural building block rather than as an independent domain. Thus, in PG16, the CDR H3 protrudes from the antibody and appears to fold as a semi-independent subdomain, whereas all observed structural homologs were integral parts of other domains. Sequence alignments showed that the CDR H3 contains more aromatic residues than do any of the structural homologs (Fig. 4D); these aromatic residues, however, were spread throughout the CDR H3 and did not assemble into a core (Fig. 2B).
Unusual features of PG9/PG16 from sequence analysis.
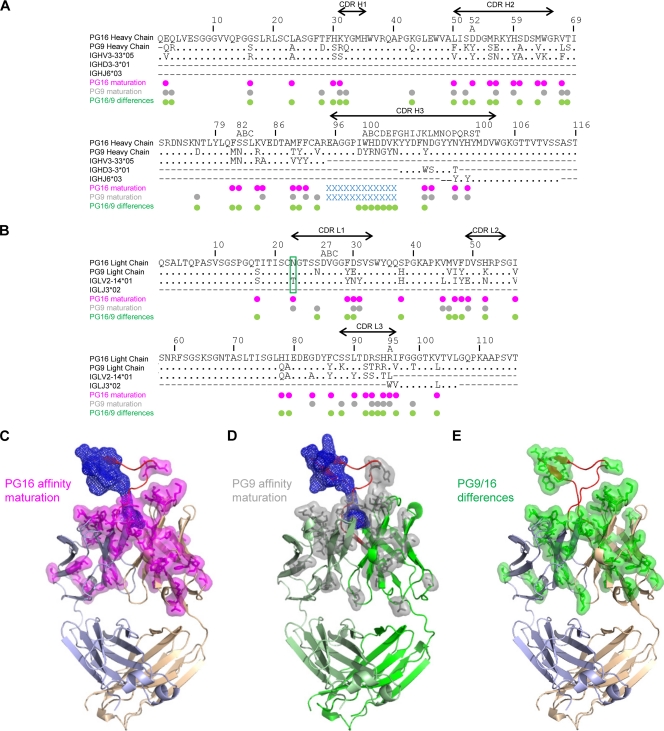
In addition to the extraordinary CDR H3, the sequences of PG9 and PG16 showed two other unusual features, N-linked glycosylation and extensive affinity maturation, as 20.4% of the Vh gene was altered for both PG9 and PG16 and 15.1 and 21.2% of the Vl gene were altered for PG9 and PG16, respectively (Fig. 5A and B). In total, 17.5% of the PG9 and PG16 heavy chains (including V-D-J) and 14.5% and 20.9% of the PG9 and PG16 light chains (V-J), respectively, are affinity matured. The IMGT website (29) and JOINSOLVER (54) were used to determine germ line genes. For the D and Jl genes, the genomic precursors were ambiguous as the two servers did not rank the same precursor. Nonetheless, IGHD3-3*01 and IGLJ3*02 were designated the common precursor as proposed by JOINSOLVER and IMGT, respectively. Despite similarity in antibody phenotypes, significant differences were observed between the variable domain sequences of PG9 and PG16 (21.9% divergent) (Fig. 5A and B). These differences were mapped onto the structures (Fig. 5E).
FIG. 5.
Affinity maturation and sequence differences between PG9 and PG16. Somatic hypermutation alters residues throughout the variable domains of both PG9 and PG16. (A) Sequence alignments of PG16 and PG9 heavy chain with genomic precursor gene (Vh, D, and Jh) using ClustalW2 (28). (B) Sequence alignments of PG16 and PG9 light chain with genomic precursor gene (Vl and Jl). Dots indicate identical residues, and a dash indicates that there is no corresponding sequence for that region. PG16 and PG9 affinity maturation from germ line is shown in magenta and gray dots, respectively. The blue crosses (“x”) indicate the residues that are not included in the Vh or D gene. The site of glycosylation is boxed in green on the light chain. PG9 and PG16 differences are shown with green dots. The sequences of the CDRs are indicated by lines with arrowheads. (C) Structural mapping of PG16 affinity maturation changes. Cα-ribbon representation of PG16 Fab with somatic mutations shown in magenta (stick and transparent surface representation). The CDR H3 residues not included in the Vh or D alignment are shown in blue mesh. The CDR H3 is shown in red, the PG16 light chain in light blue, and the PG16 heavy chain in tan. (D) Structural mapping of PG9 affinity maturation changes. Cα-ribbon representation of PG9 Fab homology model with somatic mutations shown in gray (stick and transparent surface representation). The CDR H3 residues not included in the Vh or Dh alignment are shown in blue mesh. The CDR H3 is shown in red, the PG9 light chain in light green, and the PG9 heavy chain in green. (E) Structural mapping of PG9 and PG16 sequence differences. Cα-ribbon representation of the PG16 Fab structure with CDR H3, light chain, and heavy chain colored as in panel C. Residues where PG9 differs from PG16 are displayed in green stick and transparent surface representation.
N-linked glycosylation.
N-linked glycosylation is not present in the genomic V domains (Vh, Vk, and Vl) and is a product of somatic hypermutation. The structure of PG16 revealed that its single N-linked glycan extended off the side of the light chain variable domain (see Fig. S3A in the supplemental material). While the location of the N-linked site was substantially separated from the expected location of the antibody paratope, complex N-linked sugars of the type likely to be present at this site are quite large and could conceivably affect antibody recognition. Electron density, moreover, was observed for only the three protein-proximal sugar residues, suggesting considerable glycan mobility.
To determine the influence of the N-linked glycan on neutralization, we used endoglycosidase H to remove most of the glycan, leaving only the protein-proximal N-acetylglucosamine. The resultant deglycosylated PG9 and PG16 were tested in both Fab and IgG formats and showed slight isolate-dependent alterations in neutralization. Overall, the presence of the N-linked glycan was not required for neutralization (see Fig. S3B, S4, and S5 and Table S2 in the supplemental material). (Parenthetically, we note that Fab and IgG formats for PG9 and PG16 had comparable neutralization potencies [Fig. S3B, S4, and S5 and Table S2]. Related observations have been made elsewhere for CD4-binding-site antibodies [9] and for select broadly neutralizing ones [24]).
Domain swapping in structure-function analysis.
Structure-function analysis by hypothesis-driven mutational dissection is a well-established paradigm. With the PG9 and PG16 antibodies, however, two features of interest, affinity maturation and paratope definition, did not appear particularly suited to such a hypothesis-based approach. Affinity maturation affects about a quarter of the residues throughout the variable domains of both PG9 and PG16 (Fig. 5C and D), making it difficult to focus on any particular change in altering neutralization; moreover, the solved structure contained only free antibody, not the antibody-epitope complex, making it difficult to focus on a particular site of interaction. We therefore chose to use a resolution-enhancing approach as opposed to a hypothesis-driven approach (39). We created coarse screens of functional relevance by utilizing chimeric antibodies to take advantage of both the functional similarity and amino acid divergence of the somatically related PG9 and PG16 antibodies. Because the expression of heavy and light chains already involved separate plasmids, chimeras between heavy and light chain variants were made by merely transfecting combinations of different heavy and light chain plasmids during transient-transfection expression. As the CDR H3 comprised almost half of the surface area of the combining region, we chose to segregate its contribution from the rest of the heavy chain and the light chain. Here we describe structure-function analysis of the resolution-enhancing chimeric antibody constructs.
V-gene genomic reversion and neutralization assessment.
To parse the contribution of affinity maturation to neutralization, we reverted the Vl portion of the light chain, altering 15 and 21 residues for PG9 and PG16, respectively, and the Vh portion of the heavy chain, changing 20 residues for both PG9 and PG16. Chimeric antibodies of all six possible combinations (Fig. 6A) were expressed and tested for neutralization along with wild-type PG9 and PG16 against 18 HIV-1 isolates and two non-HIV-1 virus controls (Fig. 6B; see also Fig. S6 and Table S3 in the supplemental material).
Overall, isolate-specific results were obtained, indicating that the chimeric V-gene reversions interacted with the HIV-1 Env in different ways. Heavy chain V-gene reversion resulted in chimeric antibodies that were still broad and potent neutralizers, whereas light chain V-gene reversion produced chimeric antibodies that were less able to neutralize. Reversions of both heavy and light V genes were generally inactive, although the fully V-gene-reverted PG9 showed an IC50 of ∼5 μg/ml against the clade C isolate ZM233.6 (see Table S3 in the supplemental material). Since expression of the V-gene reverted chimeric antibodies was similar to that of the mature antibodies and they neutralized at least one isolate, we believe that the chimeric antibodies were able to form functional IgG. Despite substantial isolate-specific variation, affinity maturation of PG9 and PG16 correlated with antibody neutralization breadth (P = 0.037) and potency (P < 0.0001) (Fig. 6C and D).
Dissection of PG9/PG16 functional differences with antibody chimeras.
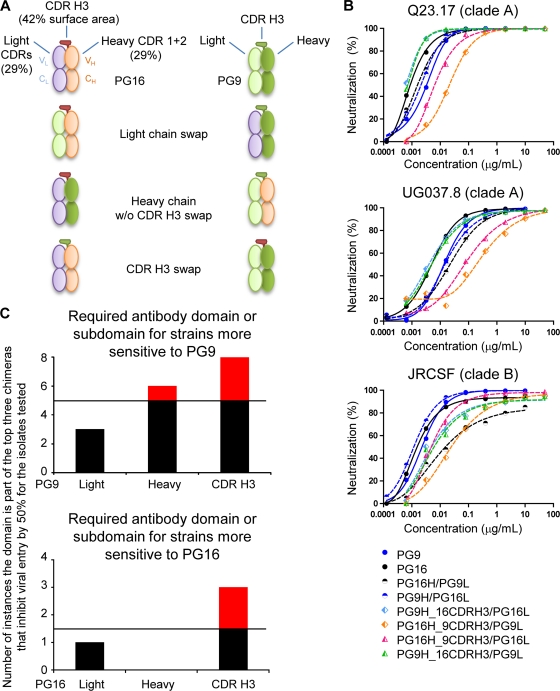
Most viruses are neutralized by PG9 and PG16 with less than 10-fold difference in antibody IC50; some viruses, however, show greater discrimination, being substantially more sensitive to PG9 or to PG16 (58). To decipher the source of functional differences between PG9 and PG16, swaps of light chain, heavy chain, and CDR H3 were assessed for functional competence, and a full complement of chimeric PG9/PG16 antibodies was tested for neutralization against a panel of HIV-1 isolates (Fig. 7; see also Table S4 in the supplemental material).
FIG. 7.
PG9 and PG16 swaps and neutralization assessment. (A) PG16- and PG9-swap chimeras were made as shown: PG9 and PG16 light chains were swapped, PG9 and PG16 heavy chains without the CDR H3 changed were swapped, and PG9 and PG16 CDR H3 were swapped. PG16 light chain is in light blue, heavy chain is in tan, and CDR H3 is in red; PG9 light chain is in light green, heavy chain is in green, and CDR H3 is bordered in dark green. (B) Neutralization curves are shown for three isolates that are similarly sensitive to PG9 and PG16. Little difference in neutralization potency was observed using the swapped chimeras, indicating that the chimeras were functional and properly folded. (C) To dissect the contribution of the different domains or subdomains to the neutralization capacity of the PG9 and PG16 antibodies, the presence of each (light, heavy, or CDR H3) in chimeric antibodies capable of viral neutralization was enumerated. With six different chimeras and two variables (a domain or subdomain being either from PG9 or from PG16), if a particular domain were the sole source of neutralization, it would be present in the top three of the neutralizing chimeras. If a particular domain or subdomain played no role in neutralization, it would be randomly assorted in the neutralizing chimeras. And if a particular domain or subdomain interfered with neutralization, it would be present in the bottom three of the neutralizing chimeras. Five viral strains were tested that showed substantially enhanced sensitivity to PG9 over PG16, and five viral strains were tested that showed substantially enhanced sensitivity to PG16 (see Fig. S7 and S8 in the supplemental material). However, many of the chimeric antibodies could not reach a 50% neutralization threshold at 50 μg/ml against any of the isolates; indeed, there were only 10 instances in which chimeras reached this 50% threshold and were in the top three of the six chimeras with isolates sensitive to PG9, and there were only three instances in which chimeras reached this 50% threshold with isolates sensitive to PG16. Thus, the “domain” or subdomain that renders isolates more sensitive to one antibody than to the other should appear in the top half of the number of instances that the chimeras neutralized, i.e., above five instances for isolates sensitive to PG9 and above 1.5 instances for isolates sensitive to PG16. These appearances are highlighted in red. IC50 are reported in Table S4, and neutralization curves are shown in Fig. S7 and S8.
The PG9/PG16 chimeras were first screened for activity against three pseudoviruses of high (and similar) sensitivities to neutralization by PG9 and PG16. All of these chimeras were functional, and IC50 were within ∼100-fold of each other (Fig. 7B), indicating functional complementation between heavy-light and CDR H3 regions of PG9 and PG16. We next tested pseudoviruses that were preferentially sensitive to either PG9 or PG16 (see Fig. S7 and S8 and Table S4 in the supplemental material) to decipher which parts of the antibody contributed to functional enhancement of each of the antibodies relative to the other. For PG16-sensitive isolates, the PG16 CDR H3 appeared to be the primary contributor; for PG9-sensitive isolates, the PG9 CDR H3 was a dominant factor, although the PG9 heavy chain also had some contribution (Fig. 7C).
Identification of a common paratope.
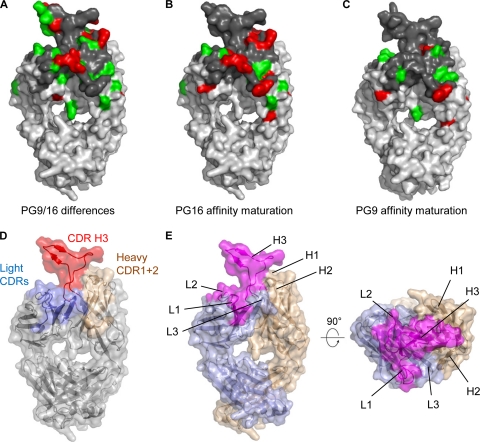
Neutralization by PG9 correlates strongly with that of PG16 (58), indicating that these antibodies recognize a common HIV-1 epitope. This suggests that a common surface on PG9 and PG16 might be involved in recognition of HIV-1. Substantial differences in sequence are found between PG9 and PG16. Mapping of these differences onto the structure of PG16 showed that they were located throughout the variable region (Fig. 5E). In light of the number and ubiquity of the differences between PG9 and PG16, it is remarkable that their virus neutralization properties are so similar. That is, 34% of the combining region differs between the two antibodies—leaving no completely conserved surface of sufficient size for antigen binding (Fig. 8A). We therefore asked whether such differences might constrain the potential location of common surface used by PG9 and PG16 to recognize HIV-1. In particular, we asked whether an area of the size of a typical paratope (e.g., roughly 20 × 20 Å) could be found within the combining surface of PG16 that contained conservative amino acid substitutions with PG9 (Fig. 8A to C). Because the CDR H3 was the source of functional enhancements, nonconservative substitutions there were allowed. With the rest of the combining surface, nonconservative changes in the heavy chain and the light chain CDR L3 were observed. A potential patch, which was composed of elements from the CDR L1 and L2 and the CDR H3, appeared to be a likely site of HIV-1 Env interactions (Fig. 8E; see also Fig. S9 in the supplemental material).
FIG. 8.
Identification of a potential site on PG16 for recognition of gp120. A combination of structure analysis and resolution-enhancing chimeras permits the boundaries of the likely paratope of PG16 to be identified. (A) Conservative (green) and nonconservative (red) substitutions between PG9 and PG16 Fabs are highlighted on the PG16 surface. Conservative and nonconservative substitutions were defined based on the GONNET PAM 250 matrix (28). The combining surface including identical residues is highlighted in dark gray. (B) Conservative and nonconservative substitutions between PG16 and its genomic precursor are highlighted on the PG16 surface, using the same color scheme as that in panel A. (C) Conservative and nonconservative substitutions between PG9 and its genomic precursor are highlighted on a PG9 surface model, using the same color scheme as that in panel A. (D) The PG16 structure is shown in Cα-ribbon representation covered with a transparent surface. CDRs for the heavy and light chains are colored in tan and light blue, respectively, while the CDR H3 loop is shown in red. (E) Front and top views of the PG16 structure highlighting a potential paratope including CDR L1, CDR L2, and CDR H3 regions mapped out in magenta. The highlighted paratope is defined by a conserved surface between PG9 and PG16 which includes identical and conservative substitutions and allowed residues based on affinity maturation and functional differences. Heavy and light chains are depicted in tan and light blue, respectively.
DISCUSSION
In the past, successful vaccines have often been achieved by mimicking naturally elicited neutralizing responses, and one potential template for an effective HIV-1 vaccine is represented by the PG9 and PG16 antibodies. Our structural and functional analysis of PG9 and PG16 antibodies suggests that the two most critical elements to mimic are the CDR H3 and the extensive affinity maturation.
Antibodies with long CDR H3s.
Analysis of antibodies elicited by viral antigens shows that long CDR H3 regions are often present (12), with an average length of 16.5 residues. Indeed, many of the broadly neutralizing monoclonal antibodies found to neutralize HIV-1 effectively have longer CDR H3s. These include the CD4 binding site antibody b12 (CDR H3 of 18 residues) (7) and the gp41-reactive antibodies 2F5 (CDR H3 of 22 residues) (46) and 4E10 (CDR H3 of 18 residues) (5). In the case of b12, the CDR H3 reaches toward a site of glycosylation (64), whereas with the gp41-reactive antibodies, the CDR H3 performs an additional function, involving hydrophobic interactions with membrane (2, 22, 40, 41, 51). It may be that the CDR H3 of the PG9/PG16 antibodies exhibits both of these activities (reaching toward the site of glycosylation and performing additional activity). The HIV-1 epitope reached by the PG9/PG16 CDR H3 is glycosylated (58), and the CDR H3 forms a separate subdomain that is critical for neutralization. It will be interesting to see the exact role played by the CDR H3 (see Fig. S9 in the supplemental material): does it achieve neutralization by thrusting its anionic surface into a recessed cationic crevice on gp120?
Affinity maturation.
HIV-1 Env-reactive antibodies often show significant degrees of affinity maturation (50). It has been unclear, however, whether such extensive maturation is a consequence of long-term antigen exposure or required for neutralization. Here, we show that affinity maturation has direct functional implications, since it correlates with increased neutralization breadth and potency. Whether the extensive maturation necessary for neutralization requires several years of antigen exposure to elicit PG9- and PG16-like antibodies needs to be determined; this nonetheless appears to be the case with HIV-1-infected individuals whose sera exhibit neutralization properties similar to PG9- and PG16-like antibodies (38, 57). Reversion of the V gene shows that not all of the antibody maturation is required for neutralization of select isolates. It remains to be seen whether the long CDR H3 is a product of affinity maturation (growing like a pedunculated tumor from the combining site), or if a rare natural process allows it to spring fully formed from the combining loops as has been shown previously in other cases (25).
Domain swapping to enhance functional resolution.
Before embarking on a hypothesis-driven approach to map the functional regions of the PG9 and PG16 antibodies, we chose to use simple, coarse screens of functional domains by creating chimeric antibodies. A complete set of eight chimeric revertants required synthesis of only four plasmids (Fig. 6A), while a complete set of eight PG9/PG16 chimeras swapping CDR H3s, light chains, and heavy chains (without CDR H3) required synthesis of only two (Fig. 7A), with all other antibodies created by mixing of heavy and light chain plasmids during transient transfection. The use of complete sets of chimeric swaps, moreover, created sufficient diversity to allow correlative-approaches of data analysis to be used (Fig. 6C and D and 7C). Whether hypothesis-driven or resolution-enhancing approaches are most effective ultimately depends on the questions asked and the answers desired (39); nonetheless, our results suggest that resolution-enhancing approaches may have advantages in some instances.
Eliciting additional PG9- and PG16-like antibodies.
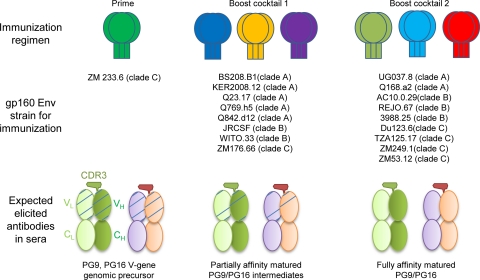
Independent of the precise mechanism by which the CDR H3 of PG9/PG16 is formed, it seems reasonable that at a minimum, elicitation of additional PG9- and PG16-like antibodies requires affinity between an immunogen and progenitor PG9- and PG16-like antibodies. In the case of other broadly neutralizing antibodies such as b12, 2G12, and 2F5, the ablating of neutralization observed upon reverting these antibodies to their genomic precursors suggests that such affinity may not be present in the initial rearrangement (60). Identifying antibodies that are intermediates in the maturation pathway and their use in creating novel vaccine immunogens has also been proposed in the case of the CD4 binding site antibody b12 (15, 61). Our analysis of V-gene chimeric revertants shows that a fully V-gene reverted PG9 as well as affinity-matured intermediates with Vh or Vl reverted was able to neutralize select HIV-1 isolates. These results suggest that one potential strategy for obtaining PG9- and PG16-like antibodies is to immunize successively with variants of HIV-1, as outlined by their neutralization susceptibility to the chimeric set of putative PG9 and PG16 germ line antibodies (e.g., starting with the ones that can be neutralized by V-gene genomic precursor antibodies) (Fig. 9).
FIG. 9.
Immunization scheme to elicit PG9- and PG16-like antibodies. The use of chimeric antibodies with V-gene germ line reversions permitted HIV-1 isolates that contained V-gene-sensitive epitopes to be identified, and such identification may prove to be useful in elaborating a vaccine strategy. Because the PG9 and PG16 antibodies recognize a quaternary epitope, appropriate immunogens may be those where the HIV-1 envelope is expressed in genetic format (DNA, adenovirus) or particle format (virus-like particles). One immunization scheme consistent with this strategy would be to immunize with a quaternary-epitope-retaining format of isolate ZM233.6 gp160 Env (shown in green with gp120 trimer represented in oval and trimer of gp41 in rectangle) as it can be neutralized by V-gene reverted PG9 (5 μg/ml) (see Table S3 in the supplemental material). Such an immunogen may have the ability to elicit V-gene genomic precursor (schematic of V-gene genomic precursor VhVl as shown in Fig. 6). Then Env isolates that are neutralized by Vh or Vl reverted chimeric PG9 or PG16 antibodies would be used as a first boost (boost cocktail 1). Partially affinity-matured antibodies may be elicited (partial maturation is shown with less hatching). Another boost (boost cocktail 2) with Env isolates that require complete PG9 and PG16 affinity maturation to be neutralized may lead to the elicitation of more fully affinity-matured antibodies. The success of such a scheme would depend in part on the precursor frequency of long and anionic CDR H3s capable of functioning like those of PG9 and PG16. Indeed, such a scheme coupled to sequence characterization of elicited antibodies for CDR H3s characteristic of PG9/PG16 may prove to be one way to assess this frequency. An understanding of the ability of the clade C isolate ZM233.6 to be recognized by V-gene reverted PG9—or at least the identification of additional isolates with similar neutralization sensitivities—may also prove to be useful in eliciting quaternary-specific antibodies like PG9 and PG16.
It has been suggested that one route to a vaccine is to determine the structure of a broadly neutralizing antibody with its epitope, modify that epitope to be immunogenic, and immunize with the modified epitope to elicit antibodies similar to the original template broadly neutralizing antibody (6, 17). Our results suggest the possibility for a variation of this process, eliminating the structural analysis of antibody-epitope interactions and potentially the epitope modification. Instead, the structure and sequence of the target antibody are analyzed for clues to elicitation barriers, putative germ line predecessor antibodies are produced, and immunogens are sequentially used, which bind with high affinity to each stage of the antibody transition: from initial V-D-J/V-J recombinant to fully affinity-matured antibody. Such a modified process might have advantages, for example, if it is technically difficult to determine the structure of the antibody-epitope complex, as may be the case with the quaternary-specific PG9 and PG16 antibodies.
ADDENDUM
The structure of antibody PG16 was recently reported by Pejchal, Wilson, and colleagues (43). Their structure determination was based on crystals that differed from those reported here. Nonetheless, many of the biological findings are similar, including the structure of the CDR H3 subdomain and its functional importance in HIV-1 neutralization. One notable difference is that their crystals permitted a 2.5-Å view of a fully ordered CDR H3, which allowed for the identification of a tyrosine-sulfated residue in PG16. This posttranslational modification is fully compatible with the analysis reported here. Overall, the two studies are highly complementary: the structures of PG16 from all crystalline lattices are similar; they utilized hypothesis-based mutagenesis to decipher function of PG9 and PG16, while we used a resolution-enhancing chimeric approach.
Supplementary Material
Acknowledgments
We thank D. R. Burton, N. Longo, R. Pejchal, J. Skinner, L. M. Walker, and I. A. Wilson for discussions; members of the Structural Biology Section and Structural Bioinformatics Core, Vaccine Research Center, for comments on the manuscript; and J. Stuckey for assistance with figures.
M.P., J.S.M., A.C., X.W., J.R.M., and P.D.K. designed research; M.P., J.S.M., S.D.S., X.W., Y.Y., T.Z., and J.Z. performed research; S.P. contributed new reagents; M.P., J.S.M., A.C., S.D.S., X.W., J.Z., J.R.M., and P.D.K. analyzed the data; and M.P., J.S.M., X.W., J.Z., and P.D.K. wrote the first draft of the paper, on which all authors commented.
Support for this work was provided by the International AIDS Vaccine Initiative (IAVI) and by the Intramural Research Program of the NIH. Use of sector 22 (Southeast Region Collaborative Access team) at the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract number W-31-109-Eng-38.
Footnotes
Published ahead of print on 10 June 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Adams, P. D., K. Gopal, R. W. Grosse-Kunstleve, L. W. Hung, T. R. Ioerger, A. J. McCoy, N. W. Moriarty, R. K. Pai, R. J. Read, T. D. Romo, J. C. Sacchettini, N. K. Sauter, L. C. Storoni, and T. C. Terwilliger. 2004. Recent developments in the PHENIX software for automated crystallographic structure determination. J. Synchrotron Radiat. 11:53-55. [DOI] [PubMed] [Google Scholar]
- 2.Alam, S. M., M. Morelli, S. M. Dennison, H. X. Liao, R. Zhang, S. M. Xia, S. Rits-Volloch, L. Sun, S. C. Harrison, B. F. Haynes, and B. Chen. 2009. Role of HIV membrane in neutralization by two broadly neutralizing antibodies. Proc. Natl. Acad. Sci. U. S. A. 106:20234-20239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Binley, J. M., E. A. Lybarger, E. T. Crooks, M. S. Seaman, E. Gray, K. L. Davis, J. M. Decker, D. Wycuff, L. Harris, N. Hawkins, B. Wood, C. Nathe, D. Richman, G. D. Tomaras, F. Bibollet-Ruche, J. E. Robinson, L. Morris, G. M. Shaw, D. C. Montefiori, and J. R. Mascola. 2008. Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J. Virol. 82:11651-11668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brunger, A. T. 1992. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355:472-475. [DOI] [PubMed] [Google Scholar]
- 5.Buchacher, A., R. Predl, K. Strutzenberger, W. Steinfellner, A. Trkola, M. Purtscher, G. Gruber, C. Tauer, F. Steindl, A. Jungbauer, et al. 1994. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res. Hum. Retroviruses 10:359-369. [DOI] [PubMed] [Google Scholar]
- 6.Burton, D. R. 2002. Antibodies, viruses and vaccines. Nat. Rev. Immunol. 2:706-713. [DOI] [PubMed] [Google Scholar]
- 7.Burton, D. R., J. Pyati, R. Koduri, S. J. Sharp, G. B. Thornton, P. W. Parren, L. S. Sawyer, R. M. Hendry, N. Dunlop, P. L. Nara, et al. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024-1027. [DOI] [PubMed] [Google Scholar]
- 8.Burton, D. R., R. L. Stanfield, and I. A. Wilson. 2005. Antibody vs. HIV in a clash of evolutionary titans. Proc. Natl. Acad. Sci. U. S. A. 102:14943-14948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, L., Y. D. Kwon, T. Zhou, X. Wu, S. O'Dell, L. Cavacini, A. J. Hessell, M. Pancera, M. Tang, L. Xu, Z. Y. Yang, M. Y. Zhang, J. Arthos, D. R. Burton, D. S. Dimitrov, G. J. Nabel, M. R. Posner, J. Sodroski, R. Wyatt, J. R. Mascola, and P. D. Kwong. 2009. Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science 326:1123-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, V. B., W. B. Arendall III, J. J. Headd, D. A. Keedy, R. M. Immormino, G. J. Kapral, L. W. Murray, J. S. Richardson, and D. C. Richardson. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66:12-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collaborative Computational Project No. 4 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760-763. [DOI] [PubMed] [Google Scholar]
- 12.Collis, A. V., A. P. Brouwer, and A. C. Martin. 2003. Analysis of the antigen combining site: correlations between length and sequence composition of the hypervariable loops and the nature of the antigen. J. Mol. Biol. 325:337-354. [DOI] [PubMed] [Google Scholar]
- 13.DeLaBarre, B., and A. T. Brunger. 2006. Considerations for the refinement of low-resolution crystal structures. Acta Crystallogr. D Biol. Crystallogr. 62:923-932. [DOI] [PubMed] [Google Scholar]
- 14.DeLano, W. L. 2002. The PyMOL Molecular Graphics System. DeLano Scientific, San Carlos, CA.
- 15.Dimitrov, D. S. 2010. Therapeutic antibodies, vaccines and antibodyomes. mAbs 2:347-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doria-Rose, N. A., R. M. Klein, M. G. Daniels, S. O'Dell, M. Nason, A. Lapedes, T. Bhattacharya, S. A. Migueles, R. T. Wyatt, B. T. Korber, J. R. Mascola, and M. Connors. 2010. Breadth of human immunodeficiency virus-specific neutralizing activity in sera: clustering analysis and association with clinical variables. J. Virol. 84:1631-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Douek, D. C., P. D. Kwong, and G. J. Nabel. 2006. The rational design of an AIDS vaccine. Cell 124:677-681. [DOI] [PubMed] [Google Scholar]
- 18.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 19.Holm, L., and C. Sander. 1995. Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 20:478-480. [DOI] [PubMed] [Google Scholar]
- 20.Huang, M., B. C. Furie, and B. Furie. 2004. Crystal structure of the calcium-stabilized human factor IX Gla domain bound to a conformation-specific anti-factor IX antibody. J. Biol. Chem. 279:14338-14346. [DOI] [PubMed] [Google Scholar]
- 21.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47(Pt. 2):110-119. [DOI] [PubMed] [Google Scholar]
- 22.Julien, J. P., N. Huarte, R. Maeso, S. G. Taneva, A. Cunningham, J. L. Nieva, and E. F. Pai. 2010. Ablation of the complementarity-determining region H3 apex of the anti-HIV-1 broadly neutralizing antibody 2F5 abrogates neutralizing capacity without affecting core epitope binding. J. Virol. 84:4136-4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kabat, E. A., T. T. Wu, H. M. Perry, K. S. Gottesman, and C. Foeller. 1991. Sequences of proteins of immunological interest, 5th ed. National Institutes of Health, U.S. Department of Health and Human Services, Bethesda, MD.
- 24.Klein, J. S., P. N. Gnanapragasam, R. P. Galimidi, C. P. Foglesong, A. P. West, Jr., and P. J. Bjorkman. 2009. Examination of the contributions of size and avidity to the neutralization mechanisms of the anti-HIV antibodies b12 and 4E10. Proc. Natl. Acad. Sci. U. S. A. 106:7385-7390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobrin, C., M. Bendandi, and L. Kwak. 2001. Novel secondary Ig VH gene rearrangement and in-frame Ig heavy chain complementarity-determining region III insertion/deletion variants in de novo follicular lymphoma. J. Immunol. 166:2235-2243. [DOI] [PubMed] [Google Scholar]
- 26.Korber, B., B. Gaschen, K. Yusim, R. Thakallapally, C. Kesmir, and V. Detours. 2001. Evolutionary and immunological implications of contemporary HIV-1 variation. Br. Med. Bull. 58:19-42. [DOI] [PubMed] [Google Scholar]
- 27.Kwong, P. D., R. Wyatt, E. Desjardins, J. Robinson, J. S. Culp, B. D. Hellmig, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1999. Probability analysis of variational crystallization and its application to gp120, the exterior envelope glycoprotein of type 1 human immunodeficiency virus (HIV-1). J. Biol. Chem. 274:4115-4123. [DOI] [PubMed] [Google Scholar]
- 28.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 29.Lefranc, M. P., V. Giudicelli, C. Ginestoux, J. Jabado-Michaloud, G. Folch, F. Bellahcene, Y. Wu, E. Gemrot, X. Brochet, J. Lane, L. Regnier, F. Ehrenmann, G. Lefranc, and P. Duroux. 2009. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 37:D1006-D1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, M., F. Gao, J. R. Mascola, L. Stamatatos, V. R. Polonis, M. Koutsoukos, G. Voss, P. Goepfert, P. Gilbert, K. M. Greene, M. Bilska, D. L. Kothe, J. F. Salazar-Gonzalez, X. Wei, J. M. Decker, B. H. Hahn, and D. C. Montefiori. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79:10108-10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, M., J. F. Salazar-Gonzalez, C. A. Derdeyn, L. Morris, C. Williamson, J. E. Robinson, J. M. Decker, Y. Li, M. G. Salazar, V. R. Polonis, K. Mlisana, S. A. Karim, K. Hong, K. M. Greene, M. Bilska, J. Zhou, S. Allen, E. Chomba, J. Mulenga, C. Vwalika, F. Gao, M. Zhang, B. T. Korber, E. Hunter, B. H. Hahn, and D. C. Montefiori. 2006. Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in southern Africa. J. Virol. 80:11776-11790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, Y., S. A. Migueles, B. Welcher, K. Svehla, A. Phogat, M. K. Louder, X. Wu, G. M. Shaw, M. Connors, R. T. Wyatt, and J. R. Mascola. 2007. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat. Med. 13:1032-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, Y., K. Svehla, M. K. Louder, D. Wycuff, S. Phogat, M. Tang, S. A. Migueles, X. Wu, A. Phogat, G. M. Shaw, M. Connors, J. Hoxie, J. R. Mascola, and R. Wyatt. 2009. Analysis of neutralization specificities in polyclonal sera derived from human immunodeficiency virus type 1-infected individuals. J. Virol. 83:1045-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maruyama, D., Y. Nishitani, T. Nonaka, A. Kita, T. A. Fukami, T. Mio, H. Yamada-Okabe, T. Yamada-Okabe, and K. Miki. 2007. Crystal structure of uridine-diphospho-N-acetylglucosamine pyrophosphorylase from Candida albicans and catalytic reaction mechanism. J. Biol. Chem. 282:17221-17230. [DOI] [PubMed] [Google Scholar]
- 35.McCoy, A. J. 2007. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 63:32-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCoy, A. J., R. W. Grosse-Kunstleve, P. D. Adams, M. D. Winn, L. C. Storoni, and R. J. Read. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40:658-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McRee, D. E. 1999. XtalView/Xfit—a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125:156-165. [DOI] [PubMed] [Google Scholar]
- 38.Morris, L., E. Gray, P. Moore, M. Madiga, K. Mlisana, and S. A. Karim. 2010. How does neutralization breadth develop in HIV infection? Abstr. 013, p. 65. 2010 Keystone HIV Vaccine Symp. Banff, Canada.
- 39.Nabel, G. J. 2009. Philosophy of science. The coordinates of truth. Science 326:53-54. [DOI] [PubMed] [Google Scholar]
- 40.Ofek, G., K. McKee, Y. Yang, Z. Y. Yang, J. Skinner, F. J. Guenaga, R. Wyatt, M. B. Zwick, G. J. Nabel, J. R. Mascola, and P. D. Kwong. 2010. Relationship between antibody 2F5 neutralization of HIV-1 and hydrophobicity of its heavy chain third complementarity-determining region. J. Virol. 84:2955-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ofek, G., M. Tang, A. Sambor, H. Katinger, J. R. Mascola, R. Wyatt, and P. D. Kwong. 2004. Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J. Virol. 78:10724-10737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 43.Pejchal, R., L. M. Walker, R. L. Stanfield, S. K. Phogat, W. C. Koff, P. Poignard, D. R. Burton, and I. A. Wilson. 2 June 2010, posting date. Structure and function of broadly reactive antibody PG16 reveal an H3 subdomain that mediates potent neutralization of HIV-1. Proc. Natl. Acad. Sci. U. S. A. [Epub ahead of print.] [DOI] [PMC free article] [PubMed]
- 44.Peneff, C., P. Ferrari, V. Charrier, Y. Taburet, C. Monnier, V. Zamboni, J. Winter, M. Harnois, F. Fassy, and Y. Bourne. 2001. Crystal structures of two human pyrophosphorylase isoforms in complexes with UDPGlc(Gal)NAc: role of the alternatively spliced insert in the enzyme oligomeric assembly and active site architecture. EMBO J. 20:6191-6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Petrey, D., Z. Xiang, C. L. Tang, L. Xie, M. Gimpelev, T. Mitros, C. S. Soto, S. Goldsmith-Fischman, A. Kernytsky, A. Schlessinger, I. Y. Koh, E. Alexov, and B. Honig. 2003. Using multiple structure alignments, fast model building, and energetic analysis in fold recognition and homology modeling. Proteins 53(Suppl. 6):430-435. [DOI] [PubMed] [Google Scholar]
- 46.Purtscher, M., A. Trkola, G. Gruber, A. Buchacher, R. Predl, F. Steindl, C. Tauer, R. Berger, N. Barrett, A. Jungbauer, et al. 1994. A broadly neutralizing human monoclonal antibody against gp41 of human immunodeficiency virus type 1. AIDS Res. Hum. Retroviruses 10:1651-1658. [DOI] [PubMed] [Google Scholar]
- 47.Reeves, P. J., N. Callewaert, R. Contreras, and H. G. Khorana. 2002. Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc. Natl. Acad. Sci. U. S. A. 99:13419-13424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sakai, A., A. A. Fedorov, E. V. Fedorov, A. M. Schnoes, M. E. Glasner, S. Brown, M. E. Rutter, K. Bain, S. Chang, T. Gheyi, J. M. Sauder, S. K. Burley, P. C. Babbitt, S. C. Almo, and J. A. Gerlt. 2009. Evolution of enzymatic activities in the enolase superfamily: stereochemically distinct mechanisms in two families of cis, cis-muconate lactonizing enzymes. Biochemistry 48:1445-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sather, D. N., J. Armann, L. K. Ching, A. Mavrantoni, G. Sellhorn, Z. Caldwell, X. Yu, B. Wood, S. Self, S. Kalams, and L. Stamatatos. 2009. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J. Virol. 83:757-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scheid, J. F., H. Mouquet, N. Feldhahn, M. S. Seaman, K. Velinzon, J. Pietzsch, R. G. Ott, R. M. Anthony, H. Zebroski, A. Hurley, A. Phogat, B. Chakrabarti, Y. Li, M. Connors, F. Pereyra, B. D. Walker, H. Wardemann, D. Ho, R. T. Wyatt, J. R. Mascola, J. V. Ravetch, and M. C. Nussenzweig. 2009. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 458:636-640. [DOI] [PubMed] [Google Scholar]
- 51.Scherer, E. M., D. P. Leaman, M. B. Zwick, A. J. McMichael, and D. R. Burton. 2010. Aromatic residues at the edge of the antibody combining site facilitate viral glycoprotein recognition through membrane interactions. Proc. Natl. Acad. Sci. U. S. A. 107:1529-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seaman, M. S., H. Janes, N. Hawkins, L. E. Grandpre, C. Devoy, A. Giri, R. T. Coffey, L. Harris, B. Wood, M. G. Daniels, T. Bhattacharya, A. Lapedes, V. R. Polonis, F. E. McCutchan, P. B. Gilbert, S. G. Self, B. T. Korber, D. C. Montefiori, and J. R. Mascola. 2010. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J. Virol. 84:1439-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simek, M. D., W. Rida, F. H. Priddy, P. Pung, E. Carrow, D. S. Laufer, J. K. Lehrman, M. Boaz, T. Tarragona-Fiol, G. Miiro, J. Birungi, A. Pozniak, D. A. McPhee, O. Manigart, E. Karita, A. Inwoley, W. Jaoko, J. Dehovitz, L. G. Bekker, P. Pitisuttithum, R. Paris, L. M. Walker, P. Poignard, T. Wrin, P. E. Fast, D. R. Burton, and W. C. Koff. 2009. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J. Virol. 83:7337-7348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Souto-Carneiro, M. M., N. S. Longo, D. E. Russ, H. W. Sun, and P. E. Lipsky. 2004. Characterization of the human Ig heavy chain antigen binding complementarity determining region 3 using a newly developed software algorithm, JOINSOLVER. J. Immunol. 172:6790-6802. [DOI] [PubMed] [Google Scholar]
- 55.Stura, E. A., and I. A. Wilson. 1990. Analytical and production seeding techniques. Methods Companion Methods Enzymol. 1:38-39. [Google Scholar]
- 56.Taylor, R. D., P. J. Jewsbury, and J. W. Essex. 2003. FDS: flexible ligand and receptor docking with a continuum solvent model and soft-core energy function. J. Comput. Chem. 24:1637-1656. [DOI] [PubMed] [Google Scholar]
- 57.van Loggerenberg, F., K. Mlisana, C. Williamson, S. C. Auld, L. Morris, C. M. Gray, Q. Abdool Karim, A. Grobler, N. Barnabas, I. Iriogbe, and S. S. Abdool Karim. 2008. Establishing a cohort at high risk of HIV infection in South Africa: challenges and experiences of the CAPRISA 002 acute infection study. PLoS One 3:e1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Walker, L. M., S. K. Phogat, P. Y. Chan-Hui, D. Wagner, P. Phung, J. L. Goss, T. Wrin, M. D. Simek, S. Fling, J. L. Mitcham, J. K. Lehrman, F. H. Priddy, O. A. Olsen, S. M. Frey, P. W. Hammond, S. Kaminsky, T. Zamb, M. Moyle, W. C. Koff, P. Poignard, and D. R. Burton. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weiss, R. A., P. R. Clapham, R. Cheingsong-Popov, A. G. Dalgleish, C. A. Carne, I. V. Weller, and R. S. Tedder. 1985. Neutralization of human T-lymphotropic virus type III by sera of AIDS and AIDS-risk patients. Nature 316:69-72. [DOI] [PubMed] [Google Scholar]
- 60.Xiao, X., W. Chen, Y. Feng, Z. Zhu, P. Prabakaran, Y. Wang, M. Y. Zhang, N. S. Longo, and D. S. Dimitrov. 2009. Germline-like predecessors of broadly neutralizing antibodies lack measurable binding to HIV-1 envelope glycoproteins: implications for evasion of immune responses and design of vaccine immunogens. Biochem. Biophys. Res. Commun. 390:404-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xiao, X., W. Chen, Y. Feng, and S. D. Dimitrov. 2009. Maturation pathways of cross-reactive HIV-1 neutralizing antibodies. Viruses 1:802-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang, Y., and J. Skolnick. 2005. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 33:2302-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou, H. Y., and Y. Q. Zhou. 2002. Distance-scaled, finite ideal-gas reference state improves structure-derived potentials of mean force for structure selection and stability prediction. Protein Sci. 11:2714-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou, T., L. Xu, B. Dey, A. J. Hessell, D. Van Ryk, S. H. Xiang, X. Yang, M. Y. Zhang, M. B. Zwick, J. Arthos, D. R. Burton, D. S. Dimitrov, J. Sodroski, R. Wyatt, G. J. Nabel, and P. D. Kwong. 2007. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445:732-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu, J., L. Cheng, Q. Fang, Z. H. Zhou, and B. Honig. 2010. Building and refining protein models within cryo-electron microscopy density maps based on homology modeling and multiscale structure refinement. J. Mol. Biol. 397:835-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.