Abstract
Homologs of the pseudorabies virus (PrV) essential large tegument protein pUL36 are conserved throughout the Herpesviridae. pUL36 functions during transport of the nucleocapsid to and docking at the nuclear pore as well as during virion formation after nuclear egress in the cytoplasm. Deletion analyses revealed several nonessential regions within the 3,084-amino-acid PrV pUL36 (S. Böttcher, B. G. Klupp, H. Granzow, W. Fuchs, K. Michael, and T. C. Mettenleiter, J. Virol. 80:9910-9915, 2006; S. Böttcher, H. Granzow, C. Maresch, B. Möhl, B. G. Klupp, and T. C. Mettenleiter, J. Virol. 81:13403-13411, 2007), while the C-terminal 62 amino acids are essential for virus replication (K. Coller, J. Lee, A. Ueda, and G. Smith, J. Virol. 81:11790-11797, 2007). To identify additional functional domains, we performed random mutagenesis of PrV pUL36 by transposon-mediated insertion of a 15-bp linker. By this approach, 26 pUL36 insertion mutants were selected and tested in transient transfection assays for their ability to complement one-step growth and/or viral spread of a PrV UL36 null mutant. Ten insertion mutants in the N-terminal half and 10 in the C terminus complemented both, whereas six insertion mutants clustering in the center of the protein did not complement in either assay. Interestingly, several insertions within conserved parts yielded positive complementation, including those located within the essential C-terminal 62 amino acids. For 15 mutants that mediated productive replication, stable virus recombinants were isolated and further characterized by plaque assay, in vitro growth analysis, and electron microscopy. Except for three mutant viruses, most insertion mutants replicated like wild-type PrV. Two insertion mutants, at amino acids (aa) 597 and 689, were impaired in one-step growth and viral spread and exhibited a defect in virion maturation in the cytoplasm. In contrast, one functional insertion (aa 1800) in a region which otherwise yielded only nonfunctional insertion mutants was impaired in viral spread but not in one-step growth without a distinctive ultrastructural phenotype. In summary, these studies extend and refine previous analyses of PrV pUL36 and demonstrate the different sensitivities of different regions of the protein to functional loss by insertion.
The herpesvirus particle is composed of four structural elements. The DNA genome-containing core is enclosed in an icosahedral capsid, which, in turn, is embedded in a proteinaceous layer termed the tegument and enveloped by a cell-derived membrane containing viral glycoproteins (35). The tegument of the Alphaherpesvirinae contains more than 15 different viral and several cellular proteins and can be structurally and functionally separated into at least two layers: a capsid-proximal “inner” part and an envelope-associated “outer” part (reviewed in references 34 and 35). The largest tegument proteins in all herpesviruses analyzed so far are homologs of herpes simplex virus type 1 (HSV-1) pUL36, which are essential for viral replication. pUL36, its interaction partner, pUL37, and the pUS3 kinase are part of the inner tegument and remain associated with nucleocapsids during their transport along microtubules to the nuclear pore (2, 3, 19, 31). In contrast, other tegument proteins like pUL46, pUL47, and pUL49 rapidly diffuse in the cytoplasm after fusion of the virion envelope with the plasma membrane. Proteolytic cleavage of HSV-1 pUL36 after docking of the nucleocapsid to the nuclear pore appears to be required for release of viral DNA into the nucleus (22). Besides these roles early in infection, pUL36 also functions during later stages of replication in virion maturation. After assembly in the nucleus, nucleocapsids are translocated to the cytoplasm by budding at the inner nuclear membrane and fusion with the outer nuclear membrane (34). Although functional nuclear localization motifs have been described for pseudorabies virus (PrV) and HSV-1 pUL36 (1, 37), in PrV-infected cells, pUL36 was never detected in the nucleus but was added to nascent virions early after nuclear egress (18, 27, 31, 37). It has been suggested that pUL36 interacts either directly (9, 32, 42, 44) or indirectly via capsid-associated pUL25 (10) with the capsid shell starting the tegumentation process in the cytosol.
In PrV, pUL36 is the only tegument protein which has been shown to be truly essential. It consists of 3,084 amino acids (aa), resulting in a molecular mass of more than 300 kDa (27). Deletion of pUL36 in HSV-1 and PrV abolished viral replication. Ultrastructurally, similar phenotypes with nonenveloped nucleocapsids present in the cytoplasm and the lack of extracellular particles indicated a defect in virion maturation in the cytoplasm (13, 16). Several functional domains have been identified in pUL36. The interaction domain of pUL36 with pUL37 (5, 16, 20, 27, 36, 42) could be located in the N-terminal part of PrV and HSV-1 pUL36 (16, 36) (Fig. 1). Deletion of the pUL37 binding site in PrV pUL36 (PrV-UL36BSF) resulted in a similar phenotype to deletion of pUL37 with an impairment of secondary envelopment in the cytoplasm (16, 26). Unlike in PrV, pUL37 is essential for replication in HSV-1 (14, 30).
FIG. 1.
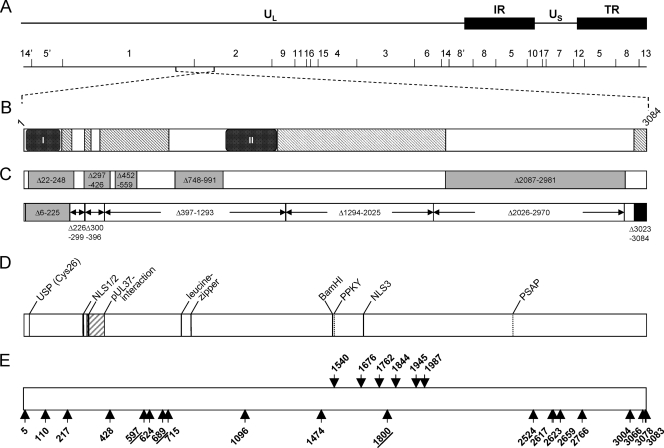
Schematic overview of PrV pUL36 and corresponding insertion mutants. (A) Diagram of the PrV genome with the unique long (UL) and unique short (US) regions as well as repeat regions (internal repeat, IR; terminal repeat, TR). The positions of BamHI restriction sites are indicated, and restriction fragments are numbered according to their size. (B) Schematic diagram of the UL36 open reading frame with conserved regions. Pfam analysis (4; http://www.sanger.ac.uk/Software/Pfam/) delineated two highly conserved PfamA domains within pUL36 homologs of herpesviruses of all three herpesvirus subfamilies [box I, Herpes_teg_N PrV (p)UL36, aa 11 to 178] and of alphaherpesviruses [box II, Herpes_UL36 PrV (p)UL36, aa 1000 to 1251] as well as PfamB domains (hatched rectangles) (6) (C) Known essential and nonessential regions in PrV pUL36. Nonessential regions are shown in gray, with the positions of the amino acids deleted in the corresponding constructs (6, 8). Deletions tested by Lee et al. (28) are shown below, marked by arrows. The essential C terminus is shown in black. Besides the N-terminal deletion Δ6-225, none of the truncated proteins was functional. (D) Predicted or identified motifs in pUL36: USP (Cys26), active-site cysteine of the deubiquitinating activity (24); pUL37 interaction domain (16, 27); NLS, nuclear localization signal (37); leucine zipper (27); and late domain motifs PPKY and PSAP (6). (E) Locations of linker insertions in pUL36 are indicated by arrows and the position of the amino acid immediately preceding the insertion. Insertions shown by arrows pointing upwards yielded functional proteins, while arrows pointing downwards indicate nonfunctional mutants. Insertions resulting in proteins which were impaired but not fully deficient in complementation are underlined. For orientation, the BamHI site separating BamHI fragments 1 and 2 is indicated.
A second functional domain in the N terminus of pUL36 comprises a ubiquitin-specific cysteine protease (USP) activity which could be identified in all three herpesvirus subfamilies (24, 40, 41). Interestingly, the USP activity is not essential for virus replication in cell culture (7, 21, 25, 43). However, it is relevant for oncogenicity of Marek′s disease virus (MDV) (21) and for virion maturation and neuroinvasion of PrV (7, 8, 29).
Several other regions in PrV pUL36 were deleted without abolishing virus replication (6, 8, 28). While deletion of nearly 1/3 of the protein in the C-terminal part (aa 2087 to 2981) had only a slight effect, deletion of a region containing two leucine zipper motifs impaired virus replication and spread more strongly (8). The highly conserved C-terminal 62 amino acids, except for the extreme C-terminal 6 amino acids, are essential for virus replication (6, 28). Due to the size of the protein, a more detailed mutagenesis analysis has, however, not yet been undertaken.
Therefore, the aim of our study was to construct random insertion mutants of PrV pUL36 using transposon-mediated insertion mutagenesis resulting in a 5-amino-acid linker insertion. Mutant proteins were analyzed functionally in transient transfection assays for complementation, and stable recombinants were isolated and further characterized.
MATERIALS AND METHODS
Viruses and cells.
PrV strain Kaplan (PrV-Ka) (23) was used as the parental wild-type strain. Viruses were propagated in rabbit kidney (RK13) or porcine kidney (PSEK) cells in minimum essential medium supplemented with 10% or 5% fetal calf serum, respectively. PrV-ΔUL36F, which lacks almost the complete UL36 coding region, was grown in RK13-UL36ΔFse cells, which express a fully functional pUL36 lacking amino acids 2087 to 2795 (6).
Plasmids and mutagenesis.
Random UL36 insertion mutants were generated using the GPS-LS linker-scanning system (New England Biolabs) according to the manufacturer's protocol. The in vitro transposase reaction was carried out on pcDNA-UL36 (6, 27), and linker-containing insertion mutants were selected with kanamycin. Insertions within the UL36 coding region could be identified by a size shift after digestion of the plasmid DNA with KpnI, separating the insert and vector fragment. The insertion site could be further localized by sequencing with GPS-specific primers N (5′-ACTTTATTGTCATAGTTTAGATCTATTTTG-3′) and S (5′-ATAATCCTTAAAAACTCCATTTCCACCCCT-3′). The kanamycin resistance cassette within the transposon was removed by cleavage with PmeI, leaving an insertion of 15 bp, consisting of 10 bp from the transprimer, including the PmeI cleavage site, and a 5-bp duplication of the target DNA. UL36-specific primers were then used to determine the newly generated codons at the linker insertion site (Table 1).
TABLE 1.
Positions and deduced amino acid sequences in pUL36 of the characterized GPS insertion mutants
| aa position | aa insertion |
|---|---|
| 5 | CLNNA |
| 110 | MFKHQ |
| 217 | CLNMD |
| 428 | FKHMV |
| 597 | CLNTK |
| 624 | CLNID |
| 689 | CLNIT |
| 715 | VFKHM |
| 1096 | VFKHY |
| 1474 | CLNTG |
| 1540 | FKHTL |
| 1676 | CLNTR |
| 1762 | CLNMD |
| 1800 | CLNNW |
| 1844 | LFKHP |
| 1945 | MFKHL |
| 1987 | CLNSL |
| 2524 | CLNRE |
| 2617 | CLNFS |
| 2623 | MFKHR |
| 2659 | CLNTQ |
| 2766 | VFLQA |
| 3004 | VFKQE |
| 3066 | VFKHK |
| 3078 | VFKHQ |
| 3083 | V stopa |
PrV-UL36GPS[3083] contains a premature stop codon eliminating the penultimate amino acid of pUL36.
Transient complementation assay and isolation of recombinant viruses.
Viral DNA of PrV-ΔUL36F was cotransfected with the pcDNA3 constructs expressing the UL36 linker insertion mutants into RK13 cells by calcium-phosphate coprecipitation (17). Cells were observed for cytopathic effects (CPE), and harvested after 2 to 3 days. Transfection progeny were titrated on complementing cells and compared to cells transfected with pcDNA3 as a negative control or pcDNA-UL36 as a positive control. For isolation of stable recombinants, transfection progeny were subjected to three rounds of plaque purification, and viral DNA of one isolate of each positive transfection was further analyzed by restriction enzyme digestion with BamHI and PmeI. To verify and precisely localize the linker insertions in the isolated recombinants, the corresponding region was amplified by PCR and sequenced.
In vitro replication studies.
For plaque size analysis, RK13 and RK13-UL36ΔFse cells in six-well culture dishes were infected with 500 PFU per well under plaque assay conditions. Cells were fixed with 5% formaldehyde 2 days postinfection (p.i.) and stained with crystal violet. For each virus, diameters of 50 plaques were analyzed microscopically in three independent experiments for RK13 cells and once for RK13-UL36 ΔFse cells and values were calculated relative to the plaque size of wild-type PrV-Ka, which was set at 100%.
For assay of one-step replication kinetics, RK13 cells were infected with PrV-Ka, PrV-ΔUL36F, and PrV-UL36GPS linker insertion mutants at a multiplicity of infection (MOI) of 5 and incubated on ice for 1 h. Thereafter, the inoculum was replaced by prewarmed medium and cells were further incubated for 1 h at 37°C. Extracellular virus was inactivated by low-pH treatment (33), and cells were scraped into the medium immediately (0 h) and at the indicated times after temperature shift and frozen at −70°C. Progeny virus titers were determined by plaque assays on RK13 cells, and the mean titers of three parallel experiments were calculated.
Electron microscopy.
RK13 cells were infected with PrV-Ka or the corresponding linker insertion mutants at an MOI of 1 and incubated for 14 h at 37°C. Fixation and embedding were done as described previously (19), and ultrathin sections were examined with a Tecnai 12 electron microscope (Philips, Eindhoven, Netherlands).
Metabolic labeling and immunoprecipitation.
PSEK cells were infected with PrV-Ka, PrV-ΔUL36F, or the PrV-UL36GPS mutants 597, 689, and 1800 at an MOI of 5 and labeled with 100 μCi Met-35S label (Hartmann Analytic, Braunschweig, Germany) from 3 h onwards. Cells were harvested after approximately 16 h, and lysates were incubated with monospecific sera against pUL36 (37) and pUL37 (26). The precipitated proteins were separated on an SDS-6% polyacrylamide gel under reducing conditions. The dried gels were exposed to an image plate and examined in an image analyzer (FLA-3000; Fuji).
RESULTS
Random transposon-mediated mutagenesis of PrV UL36.
Targeted deletion analyses revealed several nonessential and essential regions in PrV pUL36, as schematically depicted in Fig. 1C (6-8, 28). To identify additional functional domains, we used random mutagenesis by transposon-mediated insertion of 15-bp linkers, resulting in 48 insertion mutants within the UL36 open reading frame. Twelve of the 48 mutants contained stop codons, and one revealed a frameshift. These mutants were not examined any further. For detailed analysis, we selected 26 insertion mutants which were designated as pcDNA-UL36GPS followed by the number corresponding to the amino acid behind which the linker was inserted. An overview of the analyzed mutants is shown in Fig. 1E. Nine mutants were not further investigated since the insertions clustered in regions which had previously already been identified as nonessential.
Functionality of the UL36GPS mutants.
The UL36GPS mutants were tested in transfection assays for their ability to complement the replication defect of PrV-ΔUL36F, which is unable to produce viral progeny or plaques (16). To this end, RK13 cells were cotransfected with viral DNA of PrV-ΔUL36F and either pcDNA3 or pcDNA-UL36 as negative and positive controls, respectively, or with pcDNA3 expressing the mutated UL36. Twenty of the 26 insertion mutants (UL36GPS 5, 110, 217, 428, 597, 624, 689, 715, 1096, 1474, 1800, 2524, 2617, 2623, 2659, 2766, 3004, 3066, 3078, and 3083) complemented the replication defect of PrV-ΔUL36F (Fig. 1E, arrows pointing upwards). Interestingly, 6 of the 26 UL36GPS mutants (1540, 1676, 1762, 1844, 1945, and 1987), which did not complement PrV-ΔUL36F, clustered in the central part of pUL36 (Fig. 1E, arrows pointing downwards). This region contains an area of high conservation between the UL36 homologs of alphaherpesviruses (shown as gray hatched boxes in Fig. 1B) and is closely located next to a region which could be deleted without loss of function (PrV-UL36Δ2087-2981) (8). In congruence with our present findings, extension of this deletion toward the N terminus yielded a nonfunctional protein (PrV-UL36Δ1581-3012) (8). Surprisingly, two insertions in this region were tolerated (UL36GPS 1474 and 1800).
Isolation and characterization of PrV-UL36GPS mutant viruses.
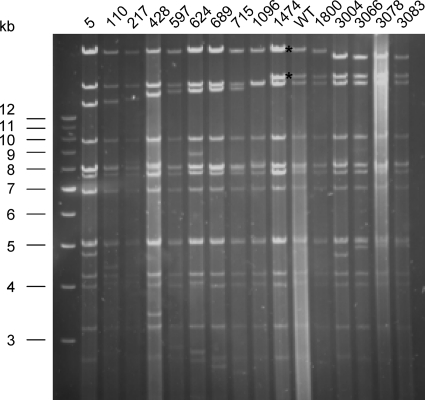
UL36 insertion mutants which in the transient assay resulted in the formation of infectious progeny were recombined into the genome of PrV-ΔUL36F. Stable recombinants expressing the complementing UL36GPS mutant proteins were isolated, and insertion of the linker was verified after digestion with BamHI and PmeI by agarose gel electrophoresis (Fig. 2). The decrease in size of BamHI fragments 1 and 2 of viral genomic DNA, each of which contains part of the UL36 open reading frame (Fig. 1A and E), correlates with the location of the linker insertion. Moreover, the exact insertion site was verified by sequencing of the corresponding regions (data not shown).
FIG. 2.
Analysis of insertion mutants using agarose gel electrophoresis. PrV UL36 is located on the largest BamHI fragments 1 and 2 (see Fig. 1). To verify insertion of the transposon linker sequences in the virus recombinants, viral DNA was digested with BamHI and PmeI. The recognition site of the latter is unique in the PrV genome and had been introduced by the linker. BamHI fragments 1 and 2 of wild-type (WT) PrV-Ka are indicated by asterisks. In viral DNAs of the PrV-UL36GPS mutants, a shift in size of either BamHI fragment 1 or 2 is observed. Sizes of the marker fragments (1-kb ladder; Invitrogen) are given on the left.
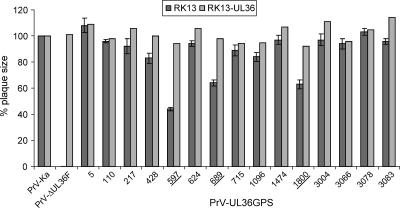
To investigate the effects of the insertions on virus replication, plaque sizes and one-step growth kinetics were determined. While the UL36 deletion mutant PrV-ΔUL36F was completely unable to spread in noncomplementing RK13 cells, most of the UL36GPS insertion mutants showed similar or only slightly decreased plaque diameters compared to PrV-Ka (Fig. 3). In contrast, plaque formation of PrV-UL36GPS[597] was decreased to 44%, and PrV-UL36GPS[689] and PrV-UL36GPS[1800] reached only 64% and 63% of PrV-Ka diameters, respectively, indicating an impairment in cell-to-cell spread (Fig. 3). Wild-type-like plaque sizes were found for all viruses on complementing pUL36-expressing cells, indicating that the observed defects were due to the insertions in pUL36 (Fig. 3). Surprisingly, insertion mutants in the conserved and essential C-terminal domain (behind amino acids 3066, 3078, and 3083) did not affect plaque formation.
FIG. 3.
Determination of plaque size. For analysis of plaque diameters, nontransgenic and pUL36-expressing RK13 cells were infected under plaque assay conditions. Cells were fixed 2 days p.i., and plaque diameters were measured microscopically. For each virus, 50 plaques were measured in three independent experiments on RK13 cells and once on complementing cells. Values were calculated compared to wild-type PrV-Ka, which was set at 100%. Standard deviations are indicated where appropriate. UL36GPS insertion mutants which show a significant decrease in plaque diameter are underlined.
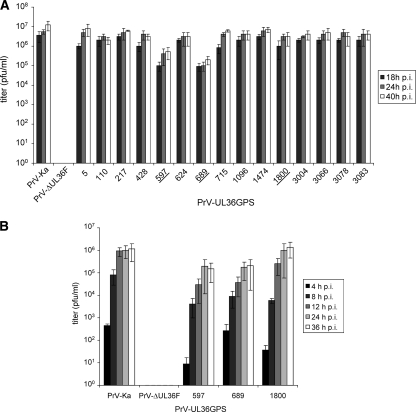
In one-step growth analyses, most insertion mutants also showed only slightly decreased titers compared to PrV-Ka (Fig. 4A). Correlating with the plaque analysis, mutants PrV-UL36GPS[597] and PrV-UL36GPS[689] replicated to approximately 10- to 50-fold-reduced titers (Fig. 4A and B). In contrast, PrV-UL36GPS[1800], which formed smaller plaques, showed only a slight reduction in virus titer (Fig. 4B). The C-terminal insertion mutants PrV-UL36GPS[3004], PrV-UL36GPS[3066], PrV-UL36GPS[3078], and PrV-UL36GPS[3083] replicated to titers comparable to those of PrV-Ka (Fig. 4A).
FIG. 4.
One-step growth kinetics. (A) RK13 cells were infected with PrV-Ka, PrV-ΔUL36F, or PrV-UL36GPS insertion mutants at an MOI of 5, harvested 18, 24, and 40 h p.i., and titrated. (B) Infectious progeny of RK13 cells infected by PrV-Ka, PrV-ΔUL36F, PrV-UL36GPS[597], PrV-UL36GPS[689], or PrV-UL36GPS[1800] were determined 4, 8, 12, 24, and 36 h p.i. Shown are mean values with standard deviations from three independent experiments.
Ultrastructural analysis.
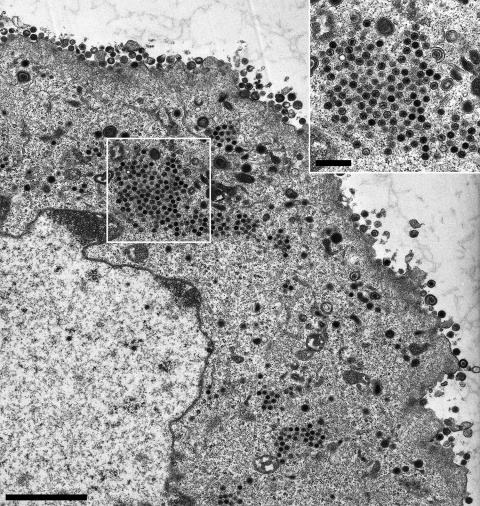
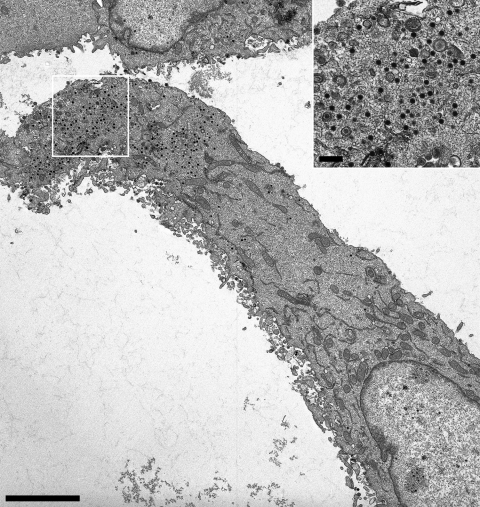
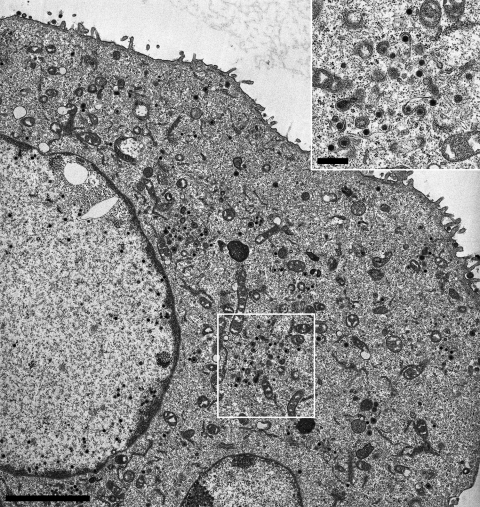
To further investigate the effects of the insertions in pUL36 on virion formation, ultrastructural analyses were performed. RK13 cells were infected with the UL36GPS insertion mutants 5, 597, 689, 715, 1096, 1474, 1800, and 3004 at an MOI of 1 and analyzed by electron microscopy 14 h after infection. Most of the insertion mutants tested showed no detectable defect in virion morphogenesis. In cells infected with PrV-UL36GPS[597] or PrV-UL36GPS[689], which formed small plaques and replicated with reduced titers (Fig. 3 and 4), a defect in secondary envelopment concomitant with accumulations of nucleocapsids in the cytoplasm was evident (Fig. 5 and 6). In contrast, PrV-UL36GPS[1800], with an insertion in the central region of pUL36, which formed small plaques but was unimpaired in one-step growth, showed no detectable defect in virion formation (Fig. 7).
FIG. 5.
Ultrastructural analysis of PrV-UL36GPS[597]-infected RK13 cells. RK13 cells were infected with PrV-UL36GPS[597] at an MOI of 1 and processed for electron microscopy after 14 h. Shown is an overview of a PrV-UL36GPS[597]-infected cell. Although stages of secondary envelopment in the cytoplasm and extracellular virions are detectable, accumulations of nucleocapsids are obvious, which are shown enlarged in the inset. Size bars: main panel, 3 μm; inset, 500 nm.
FIG. 6.
Ultrastructural analysis of PrV-UL36GPS[689]-infected RK13 cells. RK13 cells were infected with PrV-UL36GPS[689] at an MOI of 1 and processed for electron microscopy after 14 h. Shown is an overview of a PrV-UL36GPS[689]-infected cell. Intracytoplasmic nucleocapsids are enlarged in the inset. Bars: main panel, 4 μm; inset, 500 nm.
FIG. 7.
Ultrastructural analysis of PrV-UL36GPS[1800]-infected cells. RK13 cells were fixed 14 h postinfection with PrV-UL36GPS[1800] at an MOI of 1, and ultrathin sections were analyzed by electron microscopy. Shown is an overview of a PrV-UL36GPS[1800]-infected cell, in which cytoplasmic steps of virions morphogenesis were apparently unimpaired. An enlarged view showing nucleocapsids undergoing secondary envelopment is shown in the inset. Bars: main panel, 2 μm; inset, 500 nm.
Complex formation with pUL37.
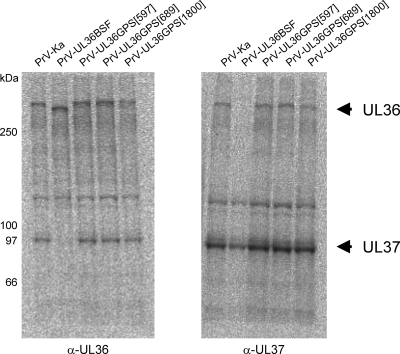
Since absence of the pUL36 complex partner pUL37 also resulted in defects in secondary envelopment and intracytoplasmic accumulation of capsids (26), we tested insertion mutants 597, 689, and 1800 for their ability to interact with pUL37 by radioimmunocoprecipitation using monospecific anti-pUL37 (26) and anti-pUL36 (37) antisera. As shown in Fig. 8, all insertion mutants tested, including PrV-UL36GPS[597] and PrV-UL36GPS[689], showed coprecipitation of both complex partners using either antiserum, in contrast to mutant PrV-UL36BSF which lacks the UL37 interaction domain (16). Thus, the defect in virion morphogenesis observed in cells infected by PrV-UL36GPS[597] or PrV-UL36GPS[689] is not attributable to a lack of interaction with pUL37.
FIG. 8.
Radioimmunocoprecipitation of pUL36 and pUL37. Cells infected with the indicated virus mutants were radiolabeled with Met-35S, and proteins were precipitated with monospecific antisera against pUL36 (α-UL36) or pUL37 (α-UL37). Protein bands corresponding to pUL36 and pUL37 are marked by arrows. Locations of marker proteins are indicated on the left.
DISCUSSION
Homologs of the HSV-1 large tegument protein pUL36 are the largest herpesvirus proteins and are essential for virus replication in all viruses studied so far (13, 16). They play important roles during entry as well as virion morphogenesis, indicating multiple functions. Previously, we and others have identified several functional regions within this large protein by introduction of sizeable deletions as well as mutation of predicted or known motifs (6-8, 10, 16, 28, 37). In this study, we used random linker insertion mutagenesis of PrV pUL36 for identification of further functional regions. The salient findings of this study are as follows: (i) linker insertions in the amino-terminal half of the protein did not impair pUL36 function; (ii) linker insertions in the conserved and essential C-terminal 62 amino acids were also tolerated; (iii) loss-of-function insertions clustered in a region from aa 1540 to 1987 in the center of the protein; (iv) most linker insertion mutants which retain function of pUL36 do not lead to detectable impairment of in vitro replication; (v) linker insertions at aa 597 and 689 resulted in impairment of one-step growth, decreased plaque sizes, and a defect in virion formation; and (vi) one linker insertion in the central essential region at aa 1800 resulted in impaired plaque sizes but no defects in one-step growth and virion formation.
In this study, we characterized a set of 26 linker insertion mutants of pUL36 for their ability to complement the one-step growth and plaque formation defects of the null mutant PrV-ΔUL36F. While cotransfection of the majority of mutated UL36 plasmids with viral DNA of the UL36 deletion mutant resulted in infectious progeny, no complementation was found in repeated assays for 6 linker insertion mutants, which all clustered in the central region of the protein ranging from amino acids 1540 to 1987. Thus, our study identified a central region in pUL36 which is highly sensitive to loss of function by linker insertion. This correlates well with our previous findings that a large deletion in the C-terminal part ranging from aa 2087 to 2981, which was functionally tolerated, could not be extended to either side without loss of function (8).
The insertion after amino acid position 1540 is very close to a predicted late domain motif, “PPKY” (aa 1528 to 1531) (6). Late domain motifs, which are necessary for recruitment of endosomal sorting complexes (ESCRT), were originally described as important for budding of RNA viruses (12, 15, 38). Recently, it was proposed that the ATPase Vps4 and ESCRT III complexes play a role in secondary envelopment of HSV-1 nucleocapsids (11, 39). It remains to be tested whether this predicted late domain motif is functional and, if so, whether insertion in this position has an effect on its interaction with components of the ESCRT machinery.
The linker insertion at position 1676 immediately precedes the predicted third nuclear localization signal, NLS3 (aa 1679 to 1682) (6). However, NLS3 was not functional when tested after fusion to green fluorescent protein (GFP), in contrast to NLS1 and NLS2, located in the N terminus of pUL36 (37). Thus, the predicted NLS3 may execute a different function.
Surprisingly, a single linker insertion mutant, PrV-UL36GPS[1800], located within this region proved to be functional and conferred replication competence to PrV-ΔUL36F with only slightly reduced titers but a small-plaque phenotype. Ultrastructural analysis did not show any gross impairment of virion morphogenesis, and mature virus particles were detectable on the cell surface. A second insertion located after aa 1474, preceding the cluster of nonfunctional mutants, showed no impairment in in vitro virus replication, delineating this important domain between amino acids 1474 and 2087. This region shows high sequence conservation between alphaherpesvirus UL36 homologs (Fig. 1B). The nonfunctional UL36GPS insertions 1844 and 1987 are located adjacent to highly conserved motifs “VLARMCIP” and “NPIENACL,” whose functional relevance remains unclear at present.
Interestingly, pUL36 with insertions within the 62 C-terminal amino acids which have been shown by deletion analysis to be essential for pUL36 function (6, 28), was found to complement PrV-ΔUL36F, and no defect in replication in cell culture of the resulting virus mutants could be observed in either plaque size or one-step growth kinetics. Unimpaired replication of PrV-UL36GPS[3083], encoding an additional valine and a premature stop, and PrV-UL36GPS[3078] is congruent with earlier data showing the nonessential nature of the last 6 amino acids (28). PrV-UL36GPS[3066] carries an insertion prior to the last 18 amino acids. An alignment of the amino acid sequences of the corresponding part in alphaherpesvirus homologs is shown in Fig. 9. Linker insertion after amino acid position 3066 is just behind a highly conserved region, indicating that its important function is probably encoded upstream of the insertion site. In consequence, a truncation mutant up to aa 3066 might also be functional, in contrast to the deletion up to aa 3054 (28).
FIG. 9.
Amino acid comparison of the conserved C terminus of pUL36. Amino acid sequences for several alphaherpesvirus pUL36 homologs are shown for the conserved C-terminal part of pUL36. Indicated by arrows are the locations of the functional linker insertions. BHV-1, bovine herpesvirus 1; EHV-1, equine herpesvirus 1; VZV, varicella-zoster virus. Analysis was performed with SYSTERS (http://systers.molgen.mpg.de/).
Functional complementation by insertion mutants in the N terminus (positions 110 and 217) or in the C-terminus (aa 2524, 2617, 2623, 2659, and 2766) is congruent with the nonessential nature of these regions, as shown for the targeted deletion mutants PrV-UL36Δ22-248 and PrV-UL36Δ2087-2981 (8), which affected virus replication in cell culture only marginally. The unimpaired replication competence of PrV-UL36GPS[5] indicates that the complete N terminus probably up to NLS1 might be dispensable for virus replication in cell culture, although a PrV-UL36 mutant lacking aa 6 to 225 showed a substantial decrease in one-step growth (28). Mutant PrV-UL36BSF, which carries a deletion of the pUL37 interaction domain (aa 297 to 426) (16) and an unintended second in-frame deletion (aa 452 to 559) delineates an additional nonessential region, with the PrV-UL36GPS[428] insertion located between these two deletions.
An interesting conserved region is located between the pUL37 interaction domain and the predicted leucine zipper motifs. Targeted deletion of the leucine zipper motifs (aa 748 to 991) impaired viral replication, resulting in approximately 100-fold-reduced titers and a small-plaque phenotype (8). While viral titers and plaque sizes of PrV-UL36GPS[624] and PrV-UL36GPS[715] were similar to those of PrV-Ka, 10- to 50-fold-reduced titers were found, and plaque sizes were significantly reduced for PrV-UL36GPS[597] and PrV-UL36GPS[689] to 44% and 64%, respectively. In line with these observations, ultrastructural analyses indicated accumulations of cytoplasmic nucleocapsids, which are characteristic of impaired secondary envelopment, in cells infected with either virus mutant. Intracytoplasmic nucleocapsids were observed in clusters in PrV-UL36GPS[597]-infected cells, similar to the situation found in cells infected with PrV-ΔUL37 or PrV-UL36BSF (16, 26). In contrast, in cells infected with PrV-UL36[689] the phenotype was more reminiscent of deletion of the full-length pUL36, which resulted in unenveloped nucleocapsids dispersed throughout the cytoplasm (16).
pUL36 interacts with pUL37 to form a physical complex (27). In the absence of pUL37, nucleocapsids accumulate in the cytoplasm due to an impairment in secondary envelopment. Thus, the defects in virion morphogenesis in PrV-UL36GPS[597]- and PrV-UL36GPS[689]-infected cells could be due to an impairment of complex formation with pUL37. However, in radioimmunoprecipitation analyses, both proteins were coprecipitated, indicating that interaction between pUL36 and pUL37 is not affected by the insertions. Thus, similar defects in virion formation can occur after either deletion of pUL37 or insertion at sites distant from the pUL37 interaction domain. It is conceivable that mutations at both sites distort the higher-order structure of pUL36 such as to impair its function.
In summary, with this random mutagenesis approach we further identified and delineated functional domains in pUL36 of PrV. The region located in the central part of the protein comprising amino acids 1540 to 1987 appears to be critical for virus replication, although no specific function has so far been assigned to this region of the protein. In contrast, major parts of the N and C termini are apparently nonessential. We are currently selecting cell lines expressing the noncomplementing UL36GPS mutant proteins to study at which step virus replication is blocked. Furthermore, glutathione S-transferase (GST) pulldowns and/or yeast two-hybrid studies with fusion proteins containing this central region should reveal putative viral or cellular interaction partners specific for this domain.
Acknowledgments
This study was supported by the Deutsche Forschungsgemeinschaft (SPP1175, Me 854/8-2).
We thank Mandy Jörn, Petra Meyer, and Diana Werner for expert technical assistance.
Footnotes
Published ahead of print on 9 June 2010.
REFERENCES
- 1.Abaitua, F., and P. O'Hare. 2008. Identification of a highly conserved, functional nuclear localization signal within the N-terminal region of herpes simplex virus type 1 VP1-2 tegument protein. J. Virol. 82:5234-5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antinone, S. E., and G. A. Smith. 2010. Retrograde axon transport of herpes simplex virus and pseudorabies virus: a live-cell comparative analysis. J. Virol. 84:1504-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antinone, S. E., G. T. Shubeita, K. E. Coller, J. I. Lee, S. Haverlock-Moyns, S. P. Gross, and G. A. Smith. 2006. The herpesvirus capsid surface protein, VP26, and the majority of the tegument proteins are dispensable for capsid transport toward the nucleus. J. Virol. 80:5494-5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, S. Griffiths-Jones, A. Khanna, M. Marshall, S. Moxon, E. L. Sonnhammer, D. J. Studholme, C. Yeats, and S. R. Eddy. 2004. The Pfam protein families database. Nucleic Acids Res. 32:D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bechtel, J. T., and T. Shenk. 2002. Human cytomegalovirus UL47 tegument protein functions after entry and before immediate-early gene expression. J. Virol. 76:1043-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Böttcher, S., B. G. Klupp, H. Granzow, W. Fuchs, K. Michael, and T. C. Mettenleiter. 2006. Identification of a 709-amino-acid internal nonessential region within the essential conserved tegument protein (p)UL36 of pseudorabies virus. J. Virol. 80:9910-9915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Böttcher, S., C. Maresch, H. Granzow, B. G. Klupp, J. P. Teifke, and T. C. Mettenleiter. 2008. Mutagenesis of the active-site cysteine in the ubiquitin-specific protease contained in large tegument protein pUL36 of pseudorabies virus impairs viral replication in vitro and neuroinvasion in vivo. J. Virol. 82:6009-6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Böttcher, S., H. Granzow, C. Maresch, B. S. Möhl, B. G. Klupp, and T. C. Mettenleiter. 2007. Identification of functional domains within the essential large tegument protein pUL36 of pseudorabies virus. J. Virol. 81:13403-13411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, D. H., H. Jiang, M. Lee, F. Liu, and Z. H. Zhou. 1999. Three-dimensional visualization of tegument/capsid interactions in the intact human cytomegalovirus. Virology 260:10-16. [DOI] [PubMed] [Google Scholar]
- 10.Coller, K. E., J. I. Lee, A. Ueda, and G. A. Smith. 2007. The capsid and tegument of the alphaherpesviruses are linked by an interaction between the UL25 and VP1/2 proteins. J. Virol. 81:11790-11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crump, C. M., C. Yates, and T. Minson. 2007. Herpes simplex virus type 1 cytoplasmic envelopment requires functional Vps4. J. Virol. 81:7380-7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Demirov, D. G., and E. O. Freed. 2004. Retrovirus budding. Virus Res. 106:87-102. [DOI] [PubMed] [Google Scholar]
- 13.Desai, P. 2000. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J. Virol. 74:11608-11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desai, P., G. L. Sexton, J. M. McCaffery, and S. Person. 2001. A null mutation in the gene encoding the herpes simplex virus type 1 UL37 polypeptide abrogates virus maturation. J. Virol. 75:10259-10271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freed, E. O. 2002. Viral late domains. J. Virol. 76:4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuchs, W., B. G. Klupp, H. Granzow, and T. C. Mettenleiter. 2004. Essential function of the pseudorabies virus UL36 gene product is independent of its interaction with the UL37 protein. J. Virol. 78:11879-11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graham, F. L., and A. J. Van der Eb. 1973. New technique for assay of infectivity of human adenovirus 5 DNA. Virology 52:456-467. [DOI] [PubMed] [Google Scholar]
- 18.Granzow, H., B. G. Klupp, and T. C. Mettenleiter. 2004. The pseudorabies virus US3 protein is a component of primary and of mature virions. J. Virol. 78:1314-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Granzow, H., B. G. Klupp, and T. C. Mettenleiter. 2005. Entry of pseudorabies virus: an immunogold-labeling study. J. Virol. 79:3200-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harmon, M. E., and W. Gibson. 1996. High molecular weight virion protein of human cytomegalovirus forms complex with product of adjacent open reading frame, abstr. W35-4, p. 144. Proc. 15th Annu. Meet. Am. Soc. Virol.
- 21.Jarosinski, K., L. Kattenhorn, B. Kaufer, H. Ploegh, and N. Osterrieder. 2007. A herpesvirus ubiquitin-specific protease is critical for efficient T cell lymphoma formation. Proc. Natl. Acad. Sci. U. S. A. 104:20025-20030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jovasevic, V., L. Liang, and B. Roizman. 2008. Proteolytic cleavage of VP1-2 is required for release of herpes simplex virus 1 DNA into the nucleus. J. Virol. 82:3311-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaplan, A. S., and A. E. Vatter. 1959. A comparison of herpes simplex and pseudorabies viruses. Virology 7:394-407. [DOI] [PubMed] [Google Scholar]
- 24.Kattenhorn, L. M., G. A. Korbel, B. M. Kessler, E. Spooner, and H. L. Ploegh. 2005. A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that is conserved across the family Herpesviridae. Mol. Cell 19:547-557. [DOI] [PubMed] [Google Scholar]
- 25.Kim, E. T., S. E. Oh, Y.O. Lee, W. Gibson, and J. H. Ahn. 2009. Cleavage specificity of the UL48 deubiquitinating protease activity of human cytomegalovirus and the growth of an active-site mutant virus in cultured cells. J. Virol. 83:12046-12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klupp, B. G., H. Granzow, E. Mundt, and T. C. Mettenleiter. 2001. Pseudorabies virus UL37 gene product is involved in secondary envelopment. J. Virol. 75:8927-8936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klupp, B. G., W. Fuchs, H. Granzow, R. Nixdorf, and T. C. Mettenleiter. 2002. Pseudorabies virus UL36 tegument protein physically interacts with the UL37 protein. J. Virol. 76:3065-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, J. I., G. W. Luxton, and G. A. Smith. 2006. Identification of an essential domain in the herpesvirus VP1/2 tegument protein: the carboxy terminus directs incorporation into capsid assemblons. J. Virol. 80:12086-12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, J. I., P. J. Sollars, S. B. Baver, G. E. Pickard, M. Leelawong, and G. A. Smith. 2009. A herpesvirus encoded deubiquitinase is a novel neuroinvasive determinant. PLoS Pathog. 5:e1000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leege, T., H. Granzow, W. Fuchs, B. G. Klupp, and T. C. Mettenleiter. 2009. Phenotypic similarities and differences between UL37-deleted pseudorabies virus and herpes simplex virus type 1. J. Gen. Virol. 90:1560-1568. [DOI] [PubMed] [Google Scholar]
- 31.Luxton, G. W., S. Haverlock, K. E. Coller, S. E. Antinone, A. Pincetic, and G. A. Smith. 2005. Targeting of herpesvirus capsid transport in axons is coupled to association with specific sets of tegument proteins. Proc. Natl. Acad. Sci. U. S. A. 102:5832-5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McNabb, D. S., and R. J. Courtney. 1992. Characterization of the large tegument protein (ICP1/2) of herpes-simplex virus type-1. Virology 190:221-232. [DOI] [PubMed] [Google Scholar]
- 33.Mettenleiter, T. C. 1989. Glycoprotein gIII deletion mutants of pseudorabies virus are impaired in virus entry. Virology 171:623-625. [DOI] [PubMed] [Google Scholar]
- 34.Mettenleiter, T. C., B. G. Klupp, and H. Granzow. 2006. Herpesvirus assembly: a tale of two membranes. Curr. Opin. Microbiol. 9:423-429. [DOI] [PubMed] [Google Scholar]
- 35.Mettenleiter, T. C., B. G. Klupp, and H. Granzow. 2009. Herpesvirus assembly: an update. Virus Res. 143:222-234. [DOI] [PubMed] [Google Scholar]
- 36.Mijatov, B., A. L. Cunningham, and R. J. Diefenbach. 2007. Residues F593 and E596 of HSV-1 tegument protein pUL36 (VP1/2) mediate binding of tegument protein pUL37. Virology 368:26-31. [DOI] [PubMed] [Google Scholar]
- 37.Möhl, B. S., S. Böttcher, H. Granzow, J. Kuhn, B. G. Klupp, and T. C. Mettenleiter. 2009. Intracellular localization of the pseudorabies virus large tegument protein pUL36. J. Virol. 83:9641-9651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morita, E., and W. I. Sundquist. 2004. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 20:395-425. [DOI] [PubMed] [Google Scholar]
- 39.Pawliczek, T., and C. M. Crump. 2009. Herpes simplex virus type 1 production requires a functional ESCRT-III complex but is independent of TSG101 and ALIX expression. J. Virol. 83:11254-11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schlieker, C., G. A. Korbel, L. M. Kattenhorn, and H. L. Ploegh. 2005. A deubiquitinating activity is conserved in the large tegument protein of the Herpesviridae. J. Virol. 79:15582-15585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schlieker, C., W. A. Weihofen, E. Frijns, L. M. Kattenhorn, R. Gaudet, and H. L. Ploegh. 2007. Structure of a herpesvirus-encoded cysteine protease reveals a unique class of deubiquitinating enzymes. Mol. Cell 9:677-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vittone, V., E. Diefenbach, D. Triffett, M. W. Douglas, A. L. Cunningham, and R. J. Diefenbach. 2005. Determination of interactions between tegument proteins of herpes simplex virus type 1. J. Virol. 79:9566-9571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, J., A. N. Loveland, L. M. Kattenhorn, H. L. Ploegh, and W. Gibson. 2006. High-molecular-weight protein (pUL48) of human cytomegalovirus is a competent deubiquitinating protease: mutant viruses altered in its active site cysteine or histidine are viable. J. Virol. 80:6003-6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou, Z. H., D. H. Chen, J. Jakana, F. J. Rixon, and W. Chiu. 1999. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J. Virol. 73:3210-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]