Abstract
The deadly paramyxovirus Nipah virus (NiV) contains a fusion glycoprotein (F) with canonical structural and functional features common to its class. Receptor binding to the NiV attachment glycoprotein (G) triggers F to undergo a two-phase conformational cascade: the first phase progresses from a metastable prefusion state to a prehairpin intermediate (PHI), while the second phase is marked by transition from the PHI to the six-helix-bundle hairpin. The PHI can be captured with peptides that mimic F's heptad repeat regions, and here we utilized a NiV heptad repeat peptide to quantify PHI formation and the half-lives (t1/2) of the first and second fusion cascade phases. We found that ephrinB2 receptor binding to G triggered ∼2-fold more F than that triggered by ephrinB3, consistent with the increased rate and extent of fusion observed with ephrinB2- versus ephrinB3-expressing cells. In addition, for a series of hyper- and hypofusogenic F mutants, we quantified F-triggering capacities and measured the kinetics of their fusion cascade phases. Hyper- and hypofusogenicity can each be manifested through distinct stages of the fusion cascade, giving rise to vastly different half-lives for the first (t1/2, 1.9 to 7.5 min) or second (t1/2, 1.5 to 15.6 min) phase. While three mutants had a shorter first phase and a longer second phase than the wild-type protein, one mutant had the opposite phenotype. Thus, our results reveal multiple critical parameters that govern the paramyxovirus fusion cascade, and our assays should help efforts to elucidate other class I membrane fusion processes.
Nipah (NiV) and Hendra (HeV) viruses are emerging members of the new Paramyxoviridae genus Henipavirus (12, 19). The Paramyxoviridae family comprises important viral pathogens, such as measles, mumps, human parainfluenza, respiratory syncytial, and Newcastle disease viruses and the henipaviruses (HNV), and NiV is its deadliest known member (4, 5). NiV has a broad host range and causes respiratory and neurological symptoms that often lead to encephalitis and a mortality rate of up to 75% in humans (21, 47). It can also spread efficiently and cause morbidity in economically important livestock (21). NiV is a biosafety level 4 (BSL4) pathogen and is considered a select agent with bio- and agro-terrorism potential. Both animal-to-human and human-to-human transmissions have been documented (4, 5), underscoring the need for research and treatment development. Since microvascular endothelial cell-cell fusion (syncytium formation) is a pathognomonic hallmark of NiV infection (50), understanding virus-cell and cell-cell membrane fusion should assist in the development of therapeutics to target this aspect of NiV pathobiology.
Paramyxovirus membrane fusion requires the coordinated action of the attachment (G, HN, or H) and fusion (F) glycoproteins, and numerous canonical structural and functional features of G/HN/H and F proteins are conserved among paramyxoviruses (20, 23, 46, 48). G/HN/H proteins have a receptor-binding globular domain formed by a six-bladed beta-propeller connected to its transmembrane anchor via a flexible stalk domain (10, 51). For NiV and HeV, both ephrinB2 (B2) and ephrinB3 (B3) can be used as cell receptors (8, 33, 34), although B2 appears to be the higher-affinity receptor (34). B2 or B3 receptors bind to and activate G, which in turn triggers a conformation cascade in F that leads to membrane fusion (1). HNV F proteins are trimeric class I fusion proteins with structural/functional features common to their class (23, 52). HNV F proteins are synthesized as precursors that are cleaved and hence activated into a metastable conformation, poised for enabling membrane fusion. Cleavage generates a new N terminus that contains a hydrophobic fusion peptide (48). For NiV and HeV, the precursor (F0) reaches the plasma membrane uncleaved, but endocytosis exposes F0 to cathepsin L in the endosomes, cleaving F0 to generate mature disulfide-linked F1 and F2 subunits that are trafficked back to the cell surface (14, 31). The structures of the retroviral Moloney murine leukemia virus p15E, lentiviral human immunodeficiency virus type 1 (HIV-1) gp41, Ebola virus GP2, influenza virus HA, and paramyxovirus SV5 and NiV-F fusion proteins all share similar trimeric coiled-coil core structures (6, 11, 17, 27, 53) and, in general, similar membrane fusion mechanisms (22, 23, 48).
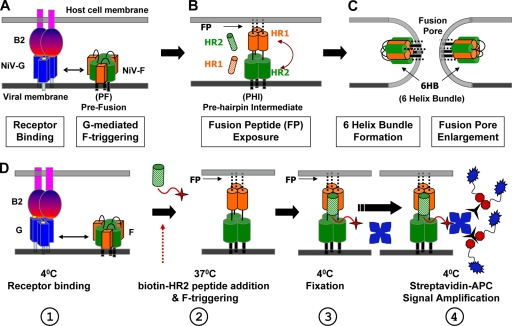
Receptor binding to paramyxoviral G/HN/H triggers a conformational cascade in F, leading to membrane fusion (Fig. 1). Although the determinants for F triggering on G/HN/H have not been defined clearly, evidence suggests that the stalk domain (7, 13, 24, 28, 29) and, at least for NiV, a region at the base of the globular domain of G (1) are involved in F triggering. Additionally, recent evidence indicates an interaction between the stalk region of the measles virus H protein and the globular domain of the cognate F protein (35). Once triggered, F progresses through a prehairpin intermediate (PHI) (Fig. 1A and B). In the PHI conformation, the fusion peptide is harpooned into the host cell membrane, and the N- and C-terminal heptad repeat domains (HR1 and HR2, respectively) are exposed. The HR domains then coalesce into the postfusion six-helix-bundle (6HB) hairpin conformation. In the 6HB, the transmembrane and fusion peptide domains are juxtaposed, bringing viral and target cell membranes together and driving membrane fusion (Fig. 1C) (30, 48). Much evidence suggests that 6HB formation is coincident with membrane merger and that synthetic HR1 and HR2 peptides only bind to and inhibit fusion intermediates (e.g., PHI) prior to 6HB formation (9, 30, 37, 43, 48). Additionally, HR1 peptides can inhibit an earlier fusion intermediate than that inhibited by HR2 peptides (43), and HR2 peptides are invariably more potent inhibitors of fusion than HR1 peptides. HR2 peptides trap the PHI by binding to the radial interstices formed by the trimeric HR1 core, inhibiting 6HB formation and membrane fusion (22, 23, 48). Altogether, there is much evidence to support the fusion cascade shown in Fig. 1 and the use of HR2 peptides to physically capture fusion intermediates (9, 30, 43, 48).
FIG. 1.
Nipah virus fusion cascade. The schematic shows the NiV fusion cascade broken down into three major stages. (A) EphrinB2 or ephrinB3 binding to NiV-G triggers the metastable NiV-F protein through allosteric mechanisms that are still being elucidated. (B) After F is triggered, it forms the PHI, in which a fusion peptide is harpooned into the host cell membrane. The PHI can be captured by peptides that mimic the NiV-F HR1 (orange-striped cylinder) or HR2 (green-striped cylinder) region and bind the F HR2 or HR1 region, respectively. (C) The HR1 and HR2 regions in the PHI coalesce to form the 6HB conformation, bringing the viral and cell membranes together and facilitating virus-host membrane fusion and viral entry. The viral membrane can be replaced by a cell membrane expressing the F and G glycoproteins in cell-cell fusion, resulting in syncytium formation. We term the transitions from A to B and from B to C phases I and II, respectively, of the fusion cascade. (D) Schematic representation of the F-triggering assay, showing its four main steps: (1) receptor binding at 4°C, (2) biotinylated HR2 peptide addition and induction of F triggering at 37°C, (3) fixation at 4°C with paraformaldehyde, and (4) signal amplification at 4°C. In the “time-of-addition” and “time-of-stopping” experiments, step 2 was modified as indicated in the text. The HR2 peptide (green hatched column) is shown with its N-terminal biotin modification (red star). Blue stars, streptavidin-APC; black, three-pronged symbols, activator; blue symbols with red octagons, enhancer.
We previously developed a fluorescence-activated cell sorting (FACS)-based NiV-F-triggering assay by measuring the amount of HR2 peptide binding to F/G-expressing cells triggered by cell surface ephrinB2 (1). In this study, we further optimized our assay for robust quantification of HR2 peptide binding and used this assay to monitor the differential degree of F triggering induced by B2 or B3. In addition, through “time-of-addition” and “time-of-stopping” experiments (described below), we show that this HR2 binding assay can measure the half-lives of various fusion intermediates, i.e., the transition times from the prefusion (PF) state to PHI and from PHI to 6HB. Using a panel of hyper- and hypofusogenic mutants, we show that hyper- and hypofusogenicity can each be manifested through distinct effects on the half-lives of these fusion intermediates and/or the absolute amounts of F triggering. Thus, we elucidated the impacts of different mutations on individual steps of the fusion cascade. Since HR2 peptides can generally capture the PHI of class I fusion proteins, our assays should help efforts to understand fusion processes mediated by other class I fusion proteins.
MATERIALS AND METHODS
Expression plasmids.
Codon-optimized NiV-G and NiV-F genes, tagged at their C termini with hemagglutinin (HA) and AU1 tags, respectively, were utilized. Their constructs were previously described (25), and all mutant F protein constructs utilized were previously published (2, 3).
Cell culture.
CHOpgsA745 (CHO) cells were cultured in minimal essential medium alpha with 10% fetal bovine serum (FBS). PK13 and 293T cells were cultured in Dulbecco's modified Eagle's medium with 10% FBS. We obtained 293T and CHOpgsA745 cells from the ATCC and PK13 (porcine fibroblast) cells from Irvin Chen at UCLA. CHOpgsA745 cells stably expressing ephrinB2 (CHOB2) or ephrinB3 (CHOB3) were described previously and were cultured in CHOpgsA745 medium supplemented with 1 mg/ml G418 (34).
Quantification of syncytium formation.
NiV-G and wild-type or mutant NiV-F expression plasmids (1:1 ratio; 1 μg in total) were transfected into CHOB2 or CHOB3 cells growing in 12-well plates at 80% confluence. At 18 h posttransfection, cells were fixed in 2% paraformaldehyde and cell-cell fusion was quantified by counting the number of nuclei within syncytia per field (magnification, ×200; 5 fields were counted per well). Syncytia were defined as 4 or more nuclei visualized within a common cell membrane (3, 25).
Quantification of NiV-F, NiV-G, and receptor cell surface expression levels by flow cytometry.
Production of anti-NiV-F or -G antisera from genetically immunized rabbits was previously described (33). Anti-F and anti-G antisera 834 and 806, respectively (3), were used at a 1:1,000 dilution for flow cytometry on NiV-F/G-transfected cells. Bound antibody was detected with phycoerythrin (PE)-conjugated goat anti-rabbit antibodies (Caltag, Burlingame, CA). For detection of cell surface B2 or B3, soluble EphB3 conjugated to human Fc (R&D Systems, Minneapolis, MN) was used at a 1:1,000 dilution. B3 was detected with PE-conjugated anti-human antibodies (Caltag). Background values were obtained by binding equal concentrations of antibodies or EphB3-Fc and secondary reagents to untransfected CHOpgsA745 cells and were subtracted from the cognate binding level to CHOB2 or CHOB3 cells.
Real-time fusion kinetics.
The fusion kinetics of cells expressing wild-type (WT) NiV-F and NiV-G with cells expressing either ephrinB2 (CHOB2), ephrinB3 (CHOB3), or neither (CHO) was determined in a β-lactamase reporter cell-cell fusion assay, as previously described (2, 3, 26, 42). For enhanced sensitivity, a catalytically enhanced and codon-optimized β-lactamase gene was used (16, 49). Fusion-nonpermissive PK13 effector cells were cotransfected with β-lactamase, NiV-G, and NiV-F by use of BioT transfection reagent (Bioland Scientific, Cerritos, CA). These were then mixed with CHO, CHO-B2, or CHO-B3 cells labeled with the CCF2-AM dye. Effector and target cells were incubated for 30 min at 4°C and then moved immediately to 37°C. Cell-cell fusion was detected by analyzing the shift from green to blue fluorescence at 37°C, indicating β-lactamase cleavage of CCF2-AM. Fluorescence was quantified every 3 min with a Synergy 2 Multimode microplate reader (BioTek Instruments, Winooski, VT). The results are expressed as the ratio of blue to green fluorescence obtained with NiV-G- and NiV-F-transfected effectors minus the background blue-to-green fluorescence ratios obtained with effector cells lacking NiV-F.
Triggering of NiV-F.
F-triggering assays were performed as previously described (1), but with the optimized protocols and signal amplification methods described here. CHO cells were transfected with F, G, and green fluorescent protein (GFP) expression plasmids, at a 13:6:1 ratio. At 18 h posttransfection, the transfected cells and untransfected CHO (negative control), CHOB2, or CHOB3 cells were mixed at a 1:1 ratio and incubated for 90 min at 4°C or 37°C in the presence of excess (500 μM) biotinylated HR2 peptide (biotin-KVDISSQISSMNQSLQQSKDYIKEAQRLLDTVNPSL). Where indicated, the HR2 peptide was added at various times during the 37°C incubation. Subsequently, the cells were moved back to 4°C, washed with wash buffer (1% FBS in phosphate-buffered saline [PBS]), fixed in 2% paraformaldehyde for 15 min, and washed again twice with wash buffer. The biotinylated HR2 peptide bound to F was detected using allophycocyanin (APC)-conjugated streptavidin and was quantified by flow cytometric analysis. To enhance our signals and signal/noise ratios, an APC amplification kit was used (Miltenyi Biotec, Auburn, CA), and we focused our analysis on F-expressing cells by gating on the brightest GFP-expressing cells and analyzing for APC fluorescence within the GFP++ gate.
RESULTS
Optimization of NiV-F-triggering assay.
The paramyxovirus attachment protein triggers the fusion protein to initiate the fusion cascade, but the parameters that govern the rate and extent of the fusion cascade that leads to membrane fusion remain to be elucidated (22, 46, 48). An F-triggering assay that can quantify the rate and extent of the fusion conformational cascade induced by receptor variants or envelope protein mutations would provide valuable insights into the paramyxovirus entry process. We previously developed a flow cytometry-based NiV-F-triggering assay to measure the effects of NiV-G fusion mutations on F triggering (1). Essentially, detection of binding of a biotinylated HR2 peptide that trapped the PHI conformation on the surfaces of NiV-F/G-expressing cells, triggered with receptor-containing CHOB2 cells, served as a surrogate marker for F triggering and fusion peptide exposure (Fig. 1D).
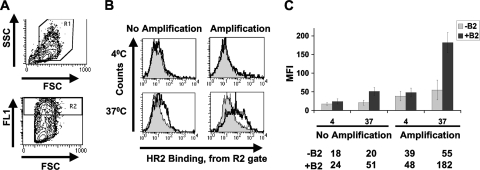
However, in order to efficiently utilize this technique to quantify the effects of receptor subtypes or F mutations on F triggering, as well as to dissect the half-lives of the fusion cascade steps affected by hyper- or hypofusogenic F fusion mutants, we improved the robustness of the assay by increasing the signal/noise ratio and the absolute HR2 peptide binding signals. The optimized methods utilized, including the use of signal amplification, are described in Materials and Methods. Figure 2 A shows our gating strategy on live (R1; top panel) and high-GFP-expressing (R2; bottom panel) cells. We reasoned that HR2 binding would be most prominent on the highest-level F/G-expressing cells. A comparison of representative histograms (gated on R1 and R2 cells) resulting from experiments with and without the utilization of an APC signal amplification kit is shown in Fig. 2B. Note that increased HR2 binding was apparent only under fusion-permissive conditions, such as when CHOB2 cells (solid line, unfilled histogram), but not CHO cells (filled histogram), were used to trigger the fusion cascade at 37°C. At 4°C, there was no difference in HR2 peptide binding, regardless of whether CHO or CHOB2 cells were used, underscoring the specificity of HR2 peptide binding and consistent with current fusion models where fusion peptide exposure is energy dependent (1, 43, 48).
FIG. 2.
Optimization of NiV-F-triggering assay. (A) NiV-F/G- and GFP-cotransfected CHO cells were gated for live cells (R1 gate; top panel) expressing high levels of GFP (well-transfected cells) in the high-FL1 gate (R2; bottom panel). (B) To increase the magnitudes and signal/noise ratios of the biotin-HR2 peptide binding signals, the streptavidin-APC signals were increased with an anti-APC signal amplification kit. Representative histograms of APC signals from the R2 gate (A) are shown for samples incubated at 4°C versus 37°C, with and without amplification. Filled histograms represent triggering with the CHO cell negative control, while the open histograms represent the positive signals obtained using CHOB2 cells as target cells. (C) Data from panel B are shown as MFI averages ± standard deviations (SD) for 3 independent experiments.
Figure 2C summarizes the mean fluorescence intensities (MFI) from the histograms in Fig. 2B, measured over multiple experiments. The average signal/noise ratios improved when amplification was used, and importantly, the overall average HR2 binding signal also increased (51 MFI units with no amplification versus 182 MFI units with amplification), providing a larger dynamic range to discern the amount of HR2 binding (ΔMFI values of ∼127 and ∼31 with and without amplification, respectively).
EphrinB2 receptor engagement triggers NiV-F to a greater extent, and produces faster fusion kinetics, than ephrinB3.
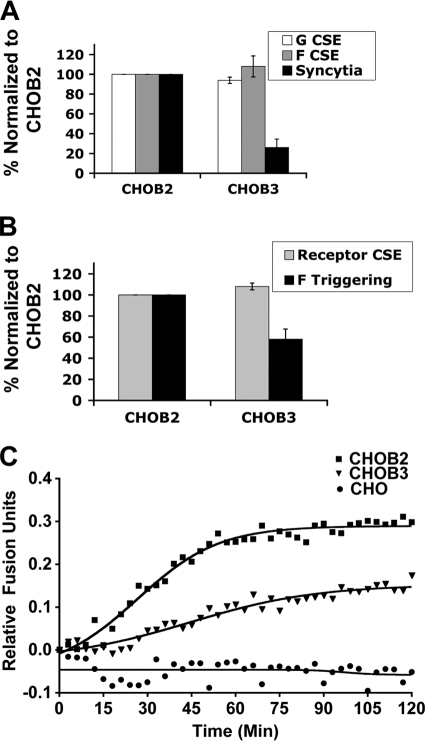
The B2 receptor binds NiV-G with a higher affinity than that of the B3 receptor (34) and induces more syncytia when infected with live NiV (34). However, whether the levels of receptor binding and syncytium formation correspond to levels of F triggering and/or fusion kinetics induced by B2 or B3 receptors is unknown. To address these questions, we first performed a cell-cell fusion assay by transfecting NiV-F and -G expression plasmids into CHOB2 or CHOB3 cells. NiV-F/G-transfected CHOB2 cells produced about 4-fold more syncytia than did CHOB3 cells (Fig. 3 A), despite having similar levels of cell surface F, G (Fig. 3A), and receptor (Fig. 3B) proteins.
FIG. 3.
EphrinB2 receptor engagement triggers NiV-F to a greater extent than that with ephrinB3. (A) Levels of syncytia produced in CHOB2 or CHOB3 cells transfected with equal amounts of F and G glycoproteins. F and G cell surface expression (CSE) levels were measured with anti-F and anti-G polyclonal antisera. Values were normalized to the surface expression and syncytium levels from the CHOB2 cell experiment. Data are averages ± SD for 3 experiments. (B) Relative amounts of F triggering induced by B2 versus B3 receptors. Similar levels of B2 and B3 receptors were expressed in CHOB2 and CHOB3 cells, as measured by soluble EphB3-Fc binding. Values were normalized to the surface expression and triggering levels from the CHOB2 cell experiment. Data are averages ± SD for 3 experiments. (C) Relative real-time fusion kinetics profiles obtained with CHOB2 versus CHOB3 cells (see Materials and Methods). Blue/green fluorescence ratios were measured every 3 min and were plotted after subtracting the blue/green ratio obtained using effector cells lacking the F protein. Data shown are averages for 3 independent experiments. Regression curves were obtained using GraphPad Prism.
Next, we performed the F-triggering assay as described above (Fig. 1D) and found that ephrinB2 triggered NiV-F more efficiently than ephrinB3 did, as the levels of HR2 peptide binding induced by CHOB3 cells were only about 58% of those induced by CHOB2 cells (Fig. 3B). Since these F-triggering differences were not due to differences in F or G or in B2 or B3 cell surface expression levels (Fig. 3A and B), our results suggest that B3 binding to NiV-G triggers a smaller proportion of NiV-F proteins to undergo the fusion peptide exposure and PHI transitions. This would likely result in a lower rate and extent of fusion. To determine if this was true, we performed a real-time fusion kinetics assay using β-lactamase-mediated cleavage of CCF2-AM as a measure of cytosolic mixing (see Materials and Methods). Figure 3C shows that fusion occurred at a higher rate (6 × 10−3 versus 2 × 10−3 relative fusion units/min) and progressed to a greater extent (higher plateau) with CHOB2 than with CHOB3 cells, consistent with B2 binding to G being a more efficient trigger of F than B3 binding to G (Fig. 3B).
NiV-F hyperfusogenic mutants affect F triggering differently and affect distinct steps in the fusion cascade.
Next, we sought to determine if the F-triggering assay could discern how particular fusogenic mutants in NiV-F itself affect different steps of the fusion cascade. Thus, we selected two previously published NiV-F hyperfusogenic mutants that produce similarly increased levels of syncytia but different fusion kinetics, namely, the F3F5 ectodomain N-glycan mutant and the K1A cytoplasmic tail mutant (2, 3). For construction of the F3F5 mutant, two N-glycans (F3 and F5) were removed via conservative mutagenesis of their glycosylation sequences from NX(S/T) to QX(S/T) (3). For construction of the NiV-F K1A mutant, the first lysine in a membrane-proximal polybasic KKR motif was mutated to alanine (2). Importantly, both mutants yielded syncytium levels that were 6- to 7-fold higher than those produced by the wild-type F protein (2, 3). However, the F3F5 mutant had a different fusion kinetics profile from that of the K1A mutant, as shown schematically in Fig. 4 A (based on data from references 2 and 3). Compared to the wild-type protein, the F3F5 mutant showed an increased rate of fusion from the very beginning, and it reached much higher levels of fusion by the end of the time course. In contrast, the K1A mutant started with the same fusion kinetics rate as the wild-type F protein and displayed only moderately increased fusion kinetics later in the time course (Fig. 4A) (2, 3). Thus, we hypothesized that the F3F5 and K1A mutants affect F triggering differently and modulate fusogenicity at distinct steps during the fusion cascade.
FIG. 4.
NiV-F hyperfusogenic mutants affect distinct steps in the fusion cascade. (A) Schematic representation of the fusion kinetics profiles of the wild-type and indicated mutant NiV-F proteins obtained in real-time fusion kinetics assays (based on data from references 2 and 3). (B) Flow cytometry histograms showing the amounts of binding of biotinylated HR2 peptide (F triggering) to the wild-type and hyperfusogenic F3F5 and K1A mutant proteins. Data were obtained exactly as described in the legend to Fig. 2A and B. (C) The cell surface expression (CSE) levels of F, F3F5, and K1A were measured using polyclonal anti-F (834) antiserum. The levels of F triggering obtained with these proteins were measured exactly as described in the legend to Fig. 2C and were normalized to those of the wild-type F protein. Data shown are averages ± SD for 3 experiments. (D) Time course model of the membrane fusion cascade (not to scale). The time between the prefusion and PHI conformations (tA) and the time between the PHI and 6HB conformations (tB) are depicted. The total time between the prefusion and 6HB conformations (tT) is the sum of tA and tB. (E) Time-of-addition experiment. In this experiment, the HR2 peptide was added 0, 5, 10, 15, 20, 30, 60, or 90 min after the switch of the temperature from 4°C to 37°C (t0) (step 2 in Fig. 1D) to allow the fusion cascade to proceed. Measurements of HR2 binding are shown, normalized to HR2 binding at t0. Data shown are averages ± SD for 3 experiments. Data were graphed and t1/2 values calculated using GraphPad Prism. (F) Time-of-stopping experiment. In this experiment, the HR2 peptide was added at the time of temperature switch from 4°C to 37°C (t0) (step 2 in Fig. 1D). However, 0, 2.5, 5, 7.5, 10, 15, 20, 30, or 60 min after t0, the cells were taken from their 37°C conditions, placed in ice, washed with cold wash buffer, and fixed with 2% paraformaldehyde. Polypropylene tubes were used for fast energy transfer. The amounts of HR2 binding are shown as normalized averages for 3 separate experiments ± SD, normalized to the maximum HR2 binding values.
Figure 4B and C show that the overall extent of F triggering was 3-fold greater for the F3F5 mutant protein than for the wild-type F or K1A mutant protein (representative histograms are shown in Fig. 4B, and data for multiple experiments are quantified and summarized in Fig. 4C). The increased level of HR2 peptide binding to the F3F5 protein was not due to increased levels of cell surface expression, as both mutant proteins had similar cell surface expression levels to that of the wild-type F protein, as shown previously by flow cytometry and biotinylated Western blot analysis (2, 3) and corroborated here by flow cytometry (Fig. 4C). Additionally, processing levels of the F3F5 and K1A mutant proteins were similar to those of the wild-type NiV-F protein (2, 3) and thus did not account for differences in their F-triggering levels.
In order to further address our hypothesis that our hyperfusogenic mutant F proteins affect F triggering differently and modulate fusogenicity at distinct steps during the fusion cascade, we devised two different time course experiments based on our F-triggering assay. In the first one, after synchronization of the fusion event by binding of effector and target cells for 2 h at 4°C, we subjected the cells to a 90-min incubation period at 37°C, adding the HR2 peptide at different time points after the switch to 37°C (step 2 in Fig. 1D; times of HR2 addition, 0, 5, 10, 15, 20, 30, 60, and 90 min). We called this the “time-of-addition” experiment. We had previously observed that the HR1 domain is not available for binding of the HR2 peptide at 4°C, but only after F is triggered at 37°C (1) to undergo fusion peptide exposure and PHI formation. Furthermore, once F completes the transition from the PHI to the 6HB form, the HR2 peptide is also no longer capable of binding to F. Thus, in the time-of-addition assay, the longer we wait before adding the HR2 peptide, the smaller is the fraction of F available for HR2 peptide binding, as more of the F in the PHI conformation transitions to the 6HB state.
In essence, the relative time of availability of HR1 in F for binding to the HR2 peptide added is a measure of the amount of F protein triggered but not yet folded into the 6HB. We calculated the half-life of this time by using GraphPad Prism and called this time tT, as it represents the total time that the F protein is in the PHI conformation(s) (Fig. 4D). The results of this time-of-addition experiment for wild-type F and the F3F5 and K1A mutants are shown in Fig. 4E. The calculated half-lives of the total time that fusion intermediates were available for HR2 peptide binding for the wild-type F protein and the F3F5 and K1A mutant proteins were 7.6, 8.3, and 9.0 min, respectively, which were not significantly different from each other. However, since tT could be defined as the total transition time from the PF stage to the PHI stage (tA) and from the PHI stage to the 6HB stage (tB), we designed a second modification to our experiment to measure the transition time for the first part of the fusion cascade (tA) (Fig. 4D).
In the second experiment, also after synchronization of the fusion event by binding of effector and target cells for 2 h at 4°C, we subjected the cell mixture to a 60-min incubation period at 37°C. However, in this case, the HR2 peptide was added at the time of the temperature switch (time zero) in all cases (step 2 in Fig. 1D), and then the fusion cascade was stopped by shifting the cells from 37°C to 4°C and fixing them at different time points during the 60-min incubation period (0, 2.5, 5, 7.5, 10, 15, 20, 30, and 60 min). We called this the “time-of-stopping” experiment. Essentially, at various points in this time-of-stopping experiment, we simply “froze” the fusion cascade and determined the amount of HR2 peptide already bound. If an F protein was already at the PHI stage, stopping the fusion cascade would not affect its binding to HR2. However, if the F protein was not yet triggered to form the PHI conformation, where HR1 is available for HR2 peptide binding, stopping the fusion cascade would prevent any additional HR2 peptide binding. Therefore, what we measured in this experiment was the half-life of the transition time between the PF and PHI conformations (tA) (Fig. 4D). We termed this phase I of the fusion cascade.
The results from the time-of-stopping experiment are shown in Fig. 4F, and the calculated tA half-lives for the wild-type, F3F5, and K1A proteins were 3.5, 1.9, and 7.5 min, respectively. Thus, for the F3F5 and K1A mutants, our data suggest that the prefusion-to-PHI conformational changes occurred significantly faster (tA = 1.9 min) and slower (tA = 7.5 min), respectively, than those of the wild-type protein (tA = 3.5 min) (P = 0.0005 for WT F versus F3F5 and P = 0.03 for WT F versus K1A; paired Student's t test). In summary, while both the F3F5 and K1A proteins resulted in hyperfusogenic phenotypes, our F-triggering assays revealed that these hyperfusogenic mutants were triggered to different extents and also manifested their hyperfusogenicity by affecting distinct steps of the fusion cascade.
NiV-F hypofusogenic mutants affect distinct steps of the fusion cascade.
To determine the various modes by which hypofusogenicity could be effected, we similarly selected two hypofusogenic NiV-F mutants based on their previously published syncytium formation levels and fusion kinetics, namely, the K2A and R3A cytoplasmic tail mutants (2). For these mutants, the second and third residues of the same membrane-proximal polybasic KKR motif that was mutated for the K1A mutant were both mutated to Ala. However, our previous results indicated that the K2A and R3A mutants yielded syncytium levels that were 20% and 30%, respectively, of those produced by the wild-type F protein. Interestingly, despite their similar levels of hypofusogenicity, they produced distinct fusion kinetics profiles, schematically illustrated in Fig. 4A (based on data from reference 2). The K2A mutant had a markedly reduced rate of fusion from the beginning and reached lower levels of cell-cell fusion by the end of the time course (Fig. 4A) (2). In contrast, the R3A mutant started with the same fusion kinetics as the wild-type F protein but significantly diverged later in the time course, displaying much slower fusion kinetics and reaching a plateau at lower levels of fusion than those for wild-type F (Fig. 4A) (2). Thus, we hypothesized that the K2A and R3A mutants manifest their hypofusogenicity by affecting the fusion cascade at distinct steps.
Therefore, we subjected these hypofusogenic mutant proteins to the same two F-triggering experiments described above for the hyperfusogenic mutants and calculated their tT and tA half-lives. First, Fig. 5 A and B show that the proportions of K2A and R3A mutant proteins that were triggered in 90 min were only approximately 60% of the level for the wild-type F protein. These data are consistent with the lower levels of cell-cell syncytium formation produced by these mutants than by the wild-type protein. Next, Fig. 5C shows that the tT half-lives obtained for the K2A and R3A mutants in the time-of-addition experiment were 18.1 min and 6.7 min, respectively. These data indicate that the half-life of the transition time between the prefusion and 6HB conformations for the K2A mutant (tT = 18.1 min) was significantly longer (∼2.5-fold; P = 0.02) than the corresponding half-life for either the wild-type (tT = 7.6 min) or R3A (tT = 6.7 min) protein. Thus, these data suggest an overall slower fusion cascade for the K2A mutant, consistent with its reduced rate and extent of fusion in the fusion kinetics assay (Fig. 4A) (2).
FIG. 5.
NiV-F hypofusogenic mutants distinctly affect the fusion cascade. (A) Flow cytometry histograms showing the amounts of HR2 peptide binding (F triggering) to the wild-type and hypofusogenic K2A and R3A mutant proteins, obtained as described in the legend to Fig. 2A and B. (B) Quantification of cell surface expression (CSE) and F-triggering levels for the wild type and the K2A and R3A mutants, obtained as described in the legend to Fig. 4C. (C) Time-of-addition experiment for the wild-type and K2A and R3A hypofusogenic mutant F proteins, performed as described in the legend to Fig. 4E. (D) Time-of-stopping experiment for the hypofusogenic mutants K2A and R3A, performed as described in the legend to Fig. 4F.
Next, we subjected these hypofusogenic mutants to a time-of-stopping experiment to obtain their tA half-lives. Figure 5D shows that the K2A and R3A mutants had tA half-lives of 2.5 and 1.9 min, respectively, compared to 3.5 min for the wild-type F protein. Since both hypofusogenic mutants transitioned from the prefusion state to the PHI conformation (tA) at an equivalent or even higher rate than that for wild-type F, our data suggest that their extrapolated half-lives of the transition time from the PHI to the 6HB conformation (tB = tT − tA) must be longer than that for the wild-type protein (summarized in Fig. 6). Overall, our results suggest that the hypofusogenic K2A and R3A cytoplasmic tail mutants slow down the fusion cascade predominantly after the PHI conformation, with K2A showing the longest transition times between the PHI and 6HB conformations. These data are consistent with the fusion kinetics profiles of these hypofusogenic mutants (Fig. 4A) (2). In addition, comparison of the R3A and wild-type protein data suggests that in order to account for the hypofusogenic phenotype of the R3A mutant, the R3A mutation likely affects a step subsequent to 6HB formation, such as fusion pore enlargement. This notion is also supported by the fusion kinetics profiles of the wild type and the R3A mutant (Fig. 4A) (2), in which the R3A mutant exhibited a lower kinetic rate than the wild-type protein only during the latter part of the time curve. The half-lives of all F proteins analyzed are summarized in Fig. 6, where tB was calculated as the difference between tT and tA.
FIG. 6.
Fusion cascade time course schematic for the wild-type F protein and hyperfusogenic and hypofusogenic F mutant proteins. The t1/2 for the time between the PF and PHI conformations (tA) and the t1/2 for the time between the PHI and 6HB conformations (tB) are depicted (top panel) and shown as values (bottom panel). tT values were obtained from Fig. 4E and 5C, while tA values were obtained from Fig. 4F and 5D. tB values were estimated by subtracting tA from tT for each wild-type or mutant F protein.
DISCUSSION
We optimized a FACS-based F-triggering assay that utilizes HR2 peptide binding as a measure of the extent of PHI formation during the fusion cascade. Using this assay in combination with real-time fusion kinetics to study Nipah virus membrane fusion, we first found that B2 binding to NiV-G induced greater levels of F triggering (Fig. 3B) and faster fusion kinetics (Fig. 3C) than did B3 binding. These results are consistent with B2 having a greater binding avidity for G (34) and producing higher levels of syncytia (Fig. 3A) than those with B3. Our data implicate the avidity of receptor/attachment protein binding as a strong determining factor for the extent of F triggering induced in NiV. This notion is consistent with our previously published data showing that receptor-induced conformational changes in NiV-G, detected with monoclonal antibody Mab45, were induced to a lesser extent by soluble B3 than B2 at an equivalent concentration (1). Somewhat parallel results have been observed for the HIV-1 class I fusion protein gp41, although for HIV-1, attachment and fusion functions exist in two subunits of a single protein (gp120 and gp41, respectively). For HIV-1, distinct coreceptors induce distinct levels of gp41 triggering to undergo the fusion cascade, and those levels correspond to distinct sensitivities to HR2 peptide inhibition (32). Therefore, we speculate that our results for NiV may have implications for the use of HR2 reagents for inhibition of infection of the henipaviruses and/or other paramyxoviruses.
The rate of fusion kinetics for HIV-1 seems to inversely correlate with relative sensitivities to HR2 peptide inhibition (32). In addition, for the paramyxoviruses, a peptide containing the HR2 sequence of human parainfluenza virus 3 (HPIV3) inhibited NiV and HeV viral infection more efficiently than homologous HPIV3 infection, despite a stronger homotypic HR1-HR2 interaction for HPIV3 (36, 37). This rather unexpected finding was attributed to relatively slower fusion kinetics for NiV and HeV than for HPIV3 (38). Therefore, it will be of interest to determine whether the lower levels of F triggering and slower fusion kinetics induced by ephrinB3 correlate with higher sensitivities to HR2 peptide inhibition and whether differential usage of B2 and B3 receptors by NiV and HeV may result in differential sensitivities to HR2 peptide inhibition for these two viruses in vitro and in vivo.
Our F-triggering assay was developed not only to determine the extent of F triggering but also to discern at least two phases of the fusion cascade affected by hyper- and hypofusogenic F mutants. We defined the half-life of the total time the PHI is available for HR2 peptide binding (tT) as the sum of the half-life of the transition time from the prefusion state to the PHI conformation (tA) and the half-life of the transition time from the PHI state to the 6HB conformation (tB) (Fig. 4D). Our F-triggering assays allowed us to directly measure tT and tA, and since tT = tA + tB, tB was then calculated with the equation tB = tT − tA. The transition time half-lives of the various F proteins analyzed are summarized in Fig. 6. Notably, tA and tB for wild-type F were almost equivalent (3.5 versus 4.1 min), although there is no a priori reason to expect them to be so. Also, for all the mutants analyzed, tA was always smaller than tT, consistent with our notion of what the time-of-addition and time-of-stopping experiments measure. It is also reassuring that our tT half-life (7.6 min) was comparable to the half-life of triggered HIV-1 gp41 (roughly 10 min), obtained from binding of monoclonal antibody NC-1 to the N-terminal trimer in the PHI of triggered gp41 (15).
Our results indicate that the hyperfusogenic F3F5 and K1A F mutants are triggered to different extents and affect distinct steps of the fusion cascade, even though both mutants exhibit similar levels of increased syncytium formation (2, 3). Significant differences were observed between the wild-type and hyperfusogenic mutant proteins, with shorter and longer tA half-lives for the F3F5 (tA = 1.9 min) and K1A (tA = 7.5 min) mutants, respectively, than for the wild-type protein (tA = 3.5 min). These data suggest that for the F3F5 protein, the PHI is formed more efficiently than for the wild-type and K1A proteins. We hypothesize that this may be attributable to the lack of bulky hydrophilic N-glycans at the F3 and F5 positions in the F3F5 mutant protein, thereby facilitating the conformational transitions from the prefusion to the PHI stage.
The transition time between the PHI and 6HB conformations (tB) for the F3F5 mutant (6.4 min) was longer than that for the wild-type protein (4.1 min). These data are consistent with the total amount of HR2 peptide binding (F triggering) for the F3F5 mutant being greater than that for the wild-type protein or the K1A protein (Fig. 4B and C). Proteins that reach the PHI state faster and stay in the PHI conformation longer will have a higher chance of HR2 binding. The higher absolute levels of HR2 peptide binding for the F3F5 mutant may also indicate that a larger proportion of F proteins are triggered than that for the wild-type protein, although we cannot rule out that the lack of F3F5 N-glycans allows for greater accessibility to HR2 peptide binding. Additionally, the higher rate and extent of fusion observed for the F3F5 mutant in the fusion kinetics assay are also consistent with a shorter tA time (Fig. 4F) and with more F molecules undergoing the triggering event (Fig. 4B and C). In sum, our data suggest that the F3F5 mutation manifests its hyperfusogenicity by speeding up the transition time from the prefusion to the PHI conformation (tA), allowing for a greater proportion of F proteins to be triggered.
In contrast, the transition time between the prefusion and PHI conformations for the hyperfusogenic K1A cytoplasmic tail mutant (tA) was longer than that for the wild-type protein (7.5 min versus 3.5 min; P = 0.03), which implied that the corresponding transition time between the PHI and 6HB conformations (tB) was shorter than that for wild-type F (Fig. 6). Interestingly, while this mutant resulted in increased syncytium formation similar to that of the F3F5 mutant (6- to 7-fold more than that of wild-type F) (2, 3), its fusion kinetics profile did not escalate nearly as much as the profile for the F3F5 mutant (Fig. 4A). Indeed, the K1A mutant fused at a similar rate to that of the wild-type protein during the first 30 min of the fusion kinetics assay (Fig. 4A) (2, 3) and fused to an increasingly greater extent only thereafter, in contrast to the F3F5 mutant, which fused at a higher rate and to a greater extent throughout the course of the fusion kinetics assay. These data suggest that the K1A mutant increased fusion by a different mechanism from that for the F3F5 mutant. Unlike the F3F5 mutant, the K1A mutant had no overall increased level of F triggering (Fig. 4C), and while it had a longer tA and shorter tB than the wild-type protein, the tT was not significantly different from that for wild-type F. Since our F-triggering assay cannot measure parameters past the transition from the PHI to 6HB conformation, our data suggest that the K1A mutant is hyperfusogenic due to steps beyond the PHI conformation (and possibly even later stages, such as fusion pore enlargement).
Our results also indicate that the K2A and R3A hypofusogenic mutants affect different steps of the fusion cascade. Our published results indicate that K2A, but not R3A, is significantly more sensitive than wild-type NiV-F to inhibition by HR2-Fc (2). This differential sensitivity to HR2-Fc inhibition is consistent with real-time fusion kinetics data, which showed that for the first 40 min, R3A fused at the same rate and extent as wild-type NiV-F, while K2A fused much more slowly and to a lesser extent from the very beginning (Fig. 4A) (2). However, after 40 min, R3A exhibited slower fusion kinetics, and it fused to a lesser extent than wild-type F by 90 min (Fig. 4A). As mentioned previously, it is likely that the defect in fusion in the R3A mutant is due to steps post-6HB formation, such as fusion pore enlargement (2), which cannot be measured by our F-triggering assay. However, our F-triggering data are consistent with both the real-time fusion kinetics and HR2-Fc inhibition data for the K2A mutant. We speculate that K2A is hypofusogenic due to a much longer PHI-to-6HB transition time (tB) than that for wild-type F (15.6 min versus 4.1 min; P = 0.02) and that above a certain threshold time limit, extended tB times negatively affect the proportion of F-triggered events that proceed to productive fusion pore formation.
Our results suggest that one parameter, e.g., tA, tB, or tT, is not enough to account for the final fusion phenotype but can complement data from other assays to elucidate the complex fusion cascade. Fusion intermediates have been characterized biochemically and captured by HR1 or HR2 peptides for different families of class I fusion proteins (32, 39, 43-45, 48). Thus, our assays should facilitate studies of how distinct steps in the fusion cascade modulate fusogenicity for diverse class I fusion proteins.
The knowledge of how receptor usage affects the kinetics of HR2 peptide inhibition for NiV and other paramyxoviruses should be valuable for the design of HR2 peptide use as a therapeutic tool, as suggested by studies on HIV receptor usage and HR2 peptide inhibition (18, 32, 40, 41). In addition, further studies of other fusion mutants in NiV-F and NiV-G that utilize the assays described here will likely enhance our understanding of the fusion process and pathobiology not only of NiV but also of other paramyxoviruses, to which these methods can likely be adapted.
Acknowledgments
This work was supported by NIH grants AI060694, AI069317, AI065359, and AI07495 to B.L. K.A. was supported by the Beckman Scholars Program. We acknowledge support from the UCLA AIDS Institute and the CFAR flow cytometry core, supported by NIH grants CA016042 and AI028697.
We thank the anonymous reviewer who made stylistic suggestions that increased the clarity of this report.
Footnotes
Published ahead of print on 2 June 2010.
REFERENCES
- 1.Aguilar, H. C., Z. A. Ataman, V. Aspericueta, A. Q. Fang, M. Stroud, O. A. Negrete, R. A. Kammerer, and B. Lee. 2009. A novel receptor-induced activation site in the Nipah virus attachment glycoprotein (G) involved in triggering the fusion glycoprotein (F). J. Biol. Chem. 284:1628-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguilar, H. C., K. A. Matreyek, D. Y. Choi, C. M. Filone, S. Young, and B. Lee. 2007. Polybasic KKR motif in the cytoplasmic tail of Nipah virus fusion protein modulates membrane fusion by inside-out signaling. J. Virol. 81:4520-4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aguilar, H. C., K. A. Matreyek, C. M. Filone, S. T. Hashimi, E. L. Levroney, O. A. Negrete, A. Bertolotti-Ciarlet, D. Y. Choi, I. McHardy, J. A. Fulcher, S. V. Su, M. C. Wolf, L. Kohatsu, L. G. Baum, and B. Lee. 2006. N-glycans on Nipah virus fusion protein protect against neutralization but reduce membrane fusion and viral entry. J. Virol. 80:4878-4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anonymous. 28 April 2004, posting date. Nipah virus breaks out in Bangladesh: mortality rates of 60% to 74%. Human-to-human transmission may be implicated. E-wire, New York, NY. www.ewire.com/display.cfm/Wire_ID/2117.
- 5.Anonymous. 2004. Nipah virus outbreak(s) in Bangladesh, January-April 2004. Wkly. Epidemiol. Rec. 79:168-171. [PubMed] [Google Scholar]
- 6.Baker, K. A., R. E. Dutch, R. A. Lamb, and T. S. Jardetzky. 1999. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell 3:309-319. [DOI] [PubMed] [Google Scholar]
- 7.Bishop, K. A., A. C. Hickey, D. Khetawat, J. R. Patch, K. N. Bossart, Z. Zhu, L. F. Wang, D. S. Dimitrov, and C. C. Broder. 2008. Residues in the stalk domain of the Hendra virus G glycoprotein modulate conformational changes associated with receptor binding. J. Virol. 82:11398-11409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonaparte, M. I., A. S. Dimitrov, K. N. Bossart, G. Crameri, B. A. Mungall, K. A. Bishop, V. Choudhry, D. S. Dimitrov, L. F. Wang, B. T. Eaton, and C. C. Broder. 2005. Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus. Proc. Natl. Acad. Sci. U. S. A. 102:10652-10657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bossart, K. N., B. A. Mungall, G. Crameri, L. F. Wang, B. T. Eaton, and C. C. Broder. 2005. Inhibition of henipavirus fusion and infection by heptad-derived peptides of the Nipah virus fusion glycoprotein. Virol. J. 2:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bowden, T. A., A. R. Aricescu, R. J. Gilbert, J. M. Grimes, E. Y. Jones, and D. I. Stuart. 2008. Structural basis of Nipah and Hendra virus attachment to their cell-surface receptor ephrin-B2. Nat. Struct. Mol. Biol. 15:567-572. [DOI] [PubMed] [Google Scholar]
- 11.Carr, C. M., and P. S. Kim. 1993. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell 73:823-832. [DOI] [PubMed] [Google Scholar]
- 12.Chua, K. B., W. J. Bellini, P. A. Rota, B. H. Harcourt, A. Tamin, S. K. Lam, T. G. Ksiazek, P. E. Rollin, S. R. Zaki, W. Shieh, C. S. Goldsmith, D. J. Gubler, J. T. Roehrig, B. Eaton, A. R. Gould, J. Olson, H. Field, P. Daniels, A. E. Ling, C. J. Peters, L. J. Anderson, and B. W. Mahy. 2000. Nipah virus: a recently emergent deadly paramyxovirus. Science 288:1432-1435. [DOI] [PubMed] [Google Scholar]
- 13.Corey, E. A., and R. M. Iorio. 2007. Mutations in the stalk of the measles virus hemagglutinin protein decrease fusion but do not interfere with virus-specific interaction with the homologous fusion protein. J. Virol. 81:9900-9910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diederich, S., M. Moll, H. D. Klenk, and A. Maisner. 2005. The Nipah virus fusion protein is cleaved within the endosomal compartment. J. Biol. Chem. 280:29899-29903. [DOI] [PubMed] [Google Scholar]
- 15.Dimitrov, A. S., J. M. Louis, C. A. Bewley, G. M. Clore, and R. Blumenthal. 2005. Conformational changes in HIV-1 gp41 in the course of HIV-1 envelope glycoprotein-mediated fusion and inactivation. Biochemistry 44:12471-12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doucet, N., P. Y. De Wals, and J. N. Pelletier. 2004. Site-saturation mutagenesis of Tyr-105 reveals its importance in substrate stabilization and discrimination in TEM-1 beta-lactamase. J. Biol. Chem. 279:46295-46303. [DOI] [PubMed] [Google Scholar]
- 17.Fass, D., S. C. Harrison, and P. S. Kim. 1996. Retrovirus envelope domain at 1.7 angstrom resolution. Nat. Struct. Biol. 3:465-469. [DOI] [PubMed] [Google Scholar]
- 18.Gallo, S. A., J. D. Reeves, H. Garg, B. Foley, R. W. Doms, and R. Blumenthal. 2006. Kinetic studies of HIV-1 and HIV-2 envelope glycoprotein-mediated fusion. Retrovirology 3:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halpin, K., P. L. Young, H. E. Field, and J. S. Mackenzie. 2000. Isolation of Hendra virus from pteropid bats: a natural reservoir of Hendra virus. J. Gen. Virol. 81:1927-1932. [DOI] [PubMed] [Google Scholar]
- 20.Iorio, R. M., and P. J. Mahon. 2008. Paramyxoviruses: different receptors-different mechanisms of fusion. Trends Microbiol. 16:135-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lam, S. K. 2003. Nipah virus—a potential agent of bioterrorism? Antiviral Res. 57:113-119. [DOI] [PubMed] [Google Scholar]
- 22.Lamb, R. A., and T. S. Jardetzky. 2007. Structural basis of viral invasion: lessons from paramyxovirus F. Curr. Opin. Struct. Biol. 17:427-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamb, R. A., R. G. Paterson, and T. S. Jardetzky. 2006. Paramyxovirus membrane fusion: lessons from the F and HN atomic structures. Virology 344:30-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, J. K., A. Prussia, T. Paal, L. K. White, J. P. Snyder, and R. K. Plemper. 2008. Functional interaction between paramyxovirus fusion and attachment proteins. J. Biol. Chem. 283:16561-16572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levroney, E. L., H. C. Aguilar, J. A. Fulcher, L. Kohatsu, K. E. Pace, M. Pang, K. B. Gurney, L. G. Baum, and B. Lee. 2005. Novel innate immune functions for galectin-1: galectin-1 inhibits cell fusion by Nipah virus envelope glycoproteins and augments dendritic cell secretion of proinflammatory cytokines. J. Immunol. 175:413-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lineberger, J. E., R. Danzeisen, D. J. Hazuda, A. J. Simon, and M. D. Miller. 2002. Altering expression levels of human immunodeficiency virus type 1 gp120-gp41 affects efficiency but not kinetics of cell-cell fusion. J. Virol. 76:3522-3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lou, Z., Y. Xu, K. Xiang, N. Su, L. Qin, X. Li, G. F. Gao, M. Bartlam, and Z. Rao. 2006. Crystal structures of Nipah and Hendra virus fusion core proteins. FEBS J. 273:4538-4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melanson, V. R., and R. M. Iorio. 2006. Addition of N-glycans in the stalk of the Newcastle disease virus HN protein blocks its interaction with the F protein and prevents fusion. J. Virol. 80:623-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melanson, V. R., and R. M. Iorio. 2004. Amino acid substitutions in the F-specific domain in the stalk of the Newcastle disease virus HN protein modulate fusion and interfere with its interaction with the F protein. J. Virol. 78:13053-13061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melikyan, G. B., R. M. Markosyan, H. Hemmati, M. K. Delmedico, D. M. Lambert, and F. S. Cohen. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 151:413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meulendyke, K. A., M. A. Wurth, R. O. McCann, and R. E. Dutch. 2005. Endocytosis plays a critical role in proteolytic processing of the Hendra virus fusion protein. J. Virol. 79:12643-12649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyauchi, K., M. M. Kozlov, and G. B. Melikyan. 2009. Early steps of HIV-1 fusion define the sensitivity to inhibitory peptides that block 6-helix bundle formation. PLoS Pathog. 5:e1000585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Negrete, O. A., E. L. Levroney, H. C. Aguilar, A. Bertolotti-Ciarlet, R. Nazarian, S. Tajyar, and B. Lee. 2005. EphrinB2 is the entry receptor for Nipah virus, an emergent deadly paramyxovirus. Nature 436:401-405. [DOI] [PubMed] [Google Scholar]
- 34.Negrete, O. A., M. C. Wolf, H. C. Aguilar, S. Enterlein, W. Wang, E. Muhlberger, S. V. Su, A. Bertolotti-Ciarlet, R. Flick, and B. Lee. 2006. Two key residues in ephrinB3 are critical for its use as an alternative receptor for Nipah virus. PLoS Pathog. 2:e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paal, T., M. A. Brindley, C. St. Clair, A. Prussia, D. Gaus, S. A. Krumm, J. P. Snyder, and R. K. Plemper. 2009. Probing the spatial organization of measles virus fusion complexes. J. Virol. 83:10480-10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Porotto, M., P. Carta, Y. Deng, G. E. Kellogg, M. Whitt, M. Lu, B. A. Mungall, and A. Moscona. 2007. Molecular determinants of antiviral potency of paramyxovirus entry inhibitors. J. Virol. 81:10567-10574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Porotto, M., L. Doctor, P. Carta, M. Fornabaio, O. Greengard, G. E. Kellogg, and A. Moscona. 2006. Inhibition of Hendra virus fusion. J. Virol. 80:9837-9849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Porotto, M., C. C. Yokoyama, G. Orefice, H. S. Kim, M. Aljofan, B. A. Mungall, and A. Moscona. 2009. Kinetic dependence of paramyxovirus entry inhibition. J. Virol. 83:6947-6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poveda, E., V. Briz, and V. Soriano. 2005. Enfuvirtide, the first fusion inhibitor to treat HIV infection. AIDS Rev. 7:139-147. [PubMed] [Google Scholar]
- 40.Reeves, J. D., S. A. Gallo, N. Ahmad, J. L. Miamidian, P. E. Harvey, M. Sharron, S. Pohlmann, J. N. Sfakianos, C. A. Derdeyn, R. Blumenthal, E. Hunter, and R. W. Doms. 2002. Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics. Proc. Natl. Acad. Sci. U. S. A. 99:16249-16254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reeves, J. D., F. H. Lee, J. L. Miamidian, C. B. Jabara, M. M. Juntilla, and R. W. Doms. 2005. Enfuvirtide resistance mutations: impact on human immunodeficiency virus envelope function, entry inhibitor sensitivity, and virus neutralization. J. Virol. 79:4991-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reeves, J. D., J. L. Miamidian, M. J. Biscone, F. H. Lee, N. Ahmad, T. C. Pierson, and R. W. Doms. 2004. Impact of mutations in the coreceptor binding site on human immunodeficiency virus type 1 fusion, infection, and entry inhibitor sensitivity. J. Virol. 78:5476-5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Russell, C. J., T. S. Jardetzky, and R. A. Lamb. 2001. Membrane fusion machines of paramyxoviruses: capture of intermediates of fusion. EMBO J. 20:4024-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shigeta, S., and T. Yamase. 2005. Current status of anti-SARS agents. Antivir. Chem. Chemother. 16:23-31. [DOI] [PubMed] [Google Scholar]
- 45.Skehel, J. J., and D. C. Wiley. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531-569. [DOI] [PubMed] [Google Scholar]
- 46.Smith, E. C., A. Popa, A. Chang, C. Masante, and R. E. Dutch. 2009. Viral entry mechanisms: the increasing diversity of paramyxovirus entry. FEBS J. 276:7217-7227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan, C. T., and K. T. Wong. 2003. Nipah encephalitis outbreak in Malaysia. Ann. Acad. Med. Singapore 32:112-117. [PubMed] [Google Scholar]
- 48.White, J. M., S. E. Delos, M. Brecher, and K. Schornberg. 2008. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 43:189-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolf, M. C., Y. Wang, A. N. Freiberg, H. C. Aguilar, M. R. Holbrook, and B. Lee. 2009. A catalytically and genetically optimized beta-lactamase-matrix based assay for sensitive, specific, and higher throughput analysis of native henipavirus entry characteristics. Virol. J. 6:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wong, K. T., W. J. Shieh, S. Kumar, K. Norain, W. Abdullah, J. Guarner, C. S. Goldsmith, K. B. Chua, S. K. Lam, C. T. Tan, K. J. Goh, H. T. Chong, R. Jusoh, P. E. Rollin, T. G. Ksiazek, and S. R. Zaki. 2002. Nipah virus infection: pathology and pathogenesis of an emerging paramyxoviral zoonosis. Am. J. Pathol. 161:2153-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu, K., K. R. Rajashankar, Y. P. Chan, J. P. Himanen, C. C. Broder, and D. B. Nikolov. 2008. Host cell recognition by the henipaviruses: crystal structures of the Nipah G attachment glycoprotein and its complex with ephrin-B3. Proc. Natl. Acad. Sci. U. S. A. 105:9953-9958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu, Y., Z. Lou, Y. Liu, D. K. Cole, N. Su, L. Qin, X. Li, Z. Bai, Z. Rao, and G. F. Gao. 2004. Crystallization and preliminary crystallographic analysis of the fusion core from two new zoonotic paramyxoviruses, Nipah virus and Hendra virus. Acta Crystallogr. D Biol. Crystallogr. 60:1161-1164. [DOI] [PubMed] [Google Scholar]
- 53.Yin, H. S., R. G. Paterson, X. Wen, R. A. Lamb, and T. S. Jardetzky. 2005. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc. Natl. Acad. Sci. U. S. A. 102:9288-9293. [DOI] [PMC free article] [PubMed] [Google Scholar]