Abstract
The Kaposi's sarcoma-associated herpesvirus (KSHV) contains several open reading frames (ORFs) that encode proteins capable of initiating and modulating cellular signaling pathways. Among them is ORF K15, encoding a 12-transmembrane-spanning protein with a cytoplasmic C-terminal domain. Through conserved binding motifs, such as Src homology 2 (SH2) and SH3 binding sites, K15 interacts with cellular proteins, activates the NF-κB, MEK/Erk, and Jun N-terminal protein kinase (JNK) pathways, and induces the expression of several inflammatory and angiogenic genes. In this study, we investigated the role of an SH3 domain binding site centered on a PPLP motif in K15. We screened libraries of cellular SH3 domains to identify signaling molecules interacting with the KSHV PPLP motif. We found its affinities for two Src kinase family members, Lyn and Hck, to exceed those of other viral proteins. While the SH2 binding motif YEEV is essential for the inflammatory response induced by KSHV K15, recruitment of Lyn and Hck to the K15 PPLP motif seems to be dispensable for this inflammatory response. However, the PPLP motif is essential for the decrease in B-cell receptor-mediated signaling induced by K15, as measured by calcium mobilization assays.
Kaposi's sarcoma (KS)-associated herpesvirus (KSHV), or human herpesvirus 8 (HHV-8), belongs to the gamma-2 herpesviruses (27, 32). It is the causative agent of all forms of KS (34) and the following two lymphoproliferative diseases: primary effusion lymphoma (PEL) (5) and the plasma cell variant of multicentric Castleman's disease (MCD) (5). KSHV infects the endothelial and spindle cells of KS lesions and the B cells of PEL and MCD (1, 8, 18, 29). Inflammatory cytokines were shown to play an important role in KSHV-associated pathogenesis (9). KSHV encodes several proteins, such as the viral G protein-coupled receptor (vGPCR) (28) and K1 (4), which are capable of initiating signal transduction cascades and cellular cytokine expression (19, 30, 36).
Another KSHV open reading frame (ORF), K15, consists of eight exons located at the “right” end of the KSHV genome, between ORF75 and the terminal repeat (TR) region. A group of integral membrane proteins with up to 12 transmembrane domains and a common hydrophilic C-terminal cytoplasmic region originates from this multiple, alternatively spliced gene during lytic replication (3, 7, 10, 15, 26, 31). The K15 full-length transcript (exon 1 to 8) is translated into a protein with a molecular mass of 45 kDa, which is localized in lipid rafts (2, 7, 10). Several highly diverged alleles of the K15 gene exist in different isolates; the two more frequent encountered alleles are termed predominant (P) and minor (M) and are almost identical concerning splicing pattern and predicted protein structure, but they share only 33% amino acid identity with one another. Sequence conservation between the P and M genotypes is concentrated in several motifs that are thought to be involved in the interaction with cellular proteins. Among these are putative binding motifs for tumor necrosis factor receptor-associated factors (TRAFs) (for K15-P, P449DQSGMS and A473TQPTDD; for K15-M, INQ464SGIS and PPPFQ484PADE), Src homology 2 binding (SH2-B) sites (for K15-P, Y431ASI and Y481EEV; for K15-M, Y444ASI and Y490EEV), and SH3-B sites (for K15-P, P387PLPP; for K15-M, P396PLP399) in the cytoplasmic C-terminal domain encoded by K15 exon 8 (2, 7, 10, 16, 31). Among the cellular proteins that have been reported to interact are TRAF1 to -3, several Src kinase family members, the antiapoptotic protein HAX-1, and others (2, 10, 35). Recently, several other cellular proteins, among them the endocytic adaptor protein intersectin 2 (ITSN2), were shown to interact with K15 through its proline-rich SH3 binding site, suggesting that K15 affects endocytic trafficking in KSHV-infected cells, presumably of cell surface receptors, such as the B-cell receptor (BCR) (21). This finding is in line with the study of Choi and coworkers, in which a chimeric protein, composed of the transmembrane domain of CD8α and the cytoplasmic tail of K15 (CD8-K15 chimera), was shown to downregulate BCR signaling (7). The putative SH2 binding site Y481EEV was found to be constitutively phosphorylated (7) and to be targeted for binding and phosphorylation by members of the Src family of protein tyrosine kinases (PTKs), such as Src, Hck, Lyn, Fyn, and Yes (2, 7). Activation of the transcription factors NF-κB and AP-1 and the MEK1/2-Erk2 and Jun N-terminal protein kinase 1 (JNK1) pathways was observed subsequent to phosphorylation of Y481 (2). Downstream cellular targets regulated by K15-induced signaling cascades include a broad range of inflammation-related genes, and mutation of the K15 YEEV SH2 binding site significantly reduced the activation of inflammatory genes (3, 38). These findings emphasize the importance of this YEEV SH2 binding motif. A second tyrosine-containing motif (YASI) was reported to be important for the interaction with HAX-1 (35), while the recent report from Lim and colleagues (21) identified several cellular SH3 domains that interacted with the K15 PPLP SH3 binding motif.
In this study, we attempted to investigate systematically the participation of putative binding motifs in signaling cascades induced by K15. We found that members of the Src family of PTKs, such as Hck and Lyn, specifically interact via their SH3 domain with the PPLP SH3 binding site of K15 and determined binding affinities for Hck and Lyn with K15 by isothermal titration calorimetry. Mutation of the putative K15 SH3 binding motif and the YASI motif did not lead to changes in the inflammatory transcriptome induced by K15, as measured by microarray analysis. In contrast, the PPLP SH3 binding site contributes to K15-mediated interference with B-cell receptor (BCR) signaling, as evidenced by calcium mobilization assays. Taken together, the conserved K15 SH3 binding site binds several cellular Src kinases with high affinity and has an important functional role in interference with BCR signaling rather than in induction of an inflammatory phenotype.
MATERIALS AND METHODS
Cell culture.
The epithelial cell lines HeLa and HEK (human embryonic kidney) 293-T were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum (FCS), 50 IU of penicillin, and 50 μg streptomycin per ml at 37°C in humidified air with 5% CO2. BJAB cells were cultured under the same conditions, except that cells were maintained in RPMI 1640 medium supplemented with 20% FCS.
Plasmids.
The plasmids pFJ-EA K15-P, pFJ-EA K15-P Y481F, pFJ-EA K15-M, and pFJ-EA K15-M Y490F have been described previously (2, 3, 38). The following plasmids (in parentheses) were generated, using a standard site-directed mutagenesis protocol, with pFJ-EA K15-P or pFJ-EA K15-M as the template and primers K15-P AALAA For, 5′-GGCAATAATGTAATTAGCCCTGCAGCATTAGCAGCTTTTTTTAGACAGCCTG-3′, and K15-P AALAA Rev, CAGGCTTAAAAAAAGCTGCTAATGCTGCAGGGCTAATTACATTATTGCC-3′ (pFJ-EA K15-P SH3); K15-M APLA For, 5′-GCTCGCCGTTTAGCACCTCTAGCATCTAGAAATGTTATAC-3′, and K15-M APLA Rev, 5′-GTATAACATTTCTAGATGCTAGAGGTGCTAAACGGCGAGC-3′ (pFJ-EA K15-M SH3); K15-P YASI For, 5′-CCCAATGTATTCGGGTTTGCCAGTATTTTAGTG-3′, and K15-P YASI Rev, CACTAAAATACTGGCAACCCGAATACATTGGG-3′ (pFJ-EA K15-P YASI); and K15-M YASI For, CGCACAGTCACCTTTGCTAGTATCTTGGGC-3′, and K15-M YASI Rev, 5′-GCCCAAGATACTAGCAAAGGTCACTGTGCG-3′ (pFJ-EA K15-M YASI). The double-mutant plasmid K15-P YF SH3 was generated with pFJ-EA K15-P Y481F as template and primers K15-P AALAA For and K15-P AALAA Rev. The double-mutant plasmid K15-M YF SH3 was generated with pFJ-EA K15-M Y490F as the template and primers K15-M APLA For and K15-M APLA Rev. The LMP2A expression plasmid pSVTP1 was a kind gift from A. Kieser (GSF Munich). The expression plasmids pcSrc and pEFHck were gifts from S. Lang (University of Erlangen-Nuremberg), and pcLyn was provided by J. C. Albrecht (University of Erlangen-Nuremberg). The monomeric red fluorescent protein (mRFP) expression plasmid was provided by B. Sodeik (Hannover Medical School). The glutathione S-transferase (GST)-K15-P fusion protein constructs were generated using pFJ-EA K15-P, pFJ-EA K15-P Y481F, pFJ-EA K15-P YASI, pFJ-EA K15-P SH3, and pFJ-EA K15-P YF SH3 as templates and primers K15-P ex8 BglII For, 5′-TATAGATCTGTAAATAGTTACCGACAG-3′, and K15-P ex8 EcoRI Rev, 5′-TATGAATTCTTAGTTCCTGGGAAATAA-3′, in a standard PCR and cloned into BamHI-EcoRI of pGEX-4T-1 (Amersham). The GST-K15-M fusion protein constructs were generated by cloning the entire cytoplasmic tails of the K15-M wild type (WT) and its mutants into BamHI-EcoRI sites of pGEX-4T-1.
Transfections.
One day prior to transfection, 8 × 104 HeLa cells or 5 × 105 HEK 293-T cells were plated per well in a six-well plate. At 24 h postseeding, cells were transfected using the FuGene 6 reagent (Roche), according to the manufacturer's instructions, with a FuGene 6/DNA ratio of 3 μl/1 μg per well. BJAB cells were split 1:3 at 1 day prior to transfection. Cells were transfected with a Gene Pulser Xcell system (Bio-Rad) at the following settings: 220 V and 950 μF. A total of 1.3 × 107 cells were resuspended in 400 μl Opti-MEM I (Gibco), mixed with 20 μg endotoxin-free K15 expression construct DNA and 3 μg mRFP plasmid DNA in a microcentrifuge tube, and incubated for 10 min at room temperature. The mix was transferred to 4-mm cuvettes, and cells were electroporated and cultured immediately afterward in 10 ml RPMI 1640 medium in T-25 flasks for 48 h.
Expression and purification of recombinant proteins and GST fusion protein binding assays.
For GST pulldown experiments, Escherichia coli Rosetta cultures transformed with the GST-K15 expression plasmids or GST alone were grown at 37°C in Luria broth medium plus ampicillin and chloramphenicol until an optical density at 600 nm of 0.4 to 0.6 was reached, induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and harvested by centrifugation at 6 h after induction. The pellet was resuspended in 500 μl of phosphate-buffered saline (PBS) plus protease inhibitors. After sonication for 1 min on ice, Triton X-100 was added to a 1% final concentration, and the cells were kept on ice for 30 min and then centrifuged for 10 min at 14,000 × g at 4°C. The supernatant was adsorbed onto 50 μl of glutathione-Sepharose beads overnight at 4°C, after which the beads were washed twice in PBS and then once in 1% Triton-X lysis buffer (20 mM Tris-HCl at pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100). Equal binding of GST proteins to glutathione beads was controlled by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining. HEK 293-T cells were transfected with the myc-tagged Src kinase Src, Hck, or Lyn cDNA expression constructs at 1 μg per well in a six-well plate. At 48 h posttransfection, cells were washed once with PBS on ice and lysed in 1% Triton X-100 lysis buffer containing protease inhibitors for 10 min on ice. Cell lysates were cleared by centrifugation at 14,000 × g for 10 min at 4°C, and supernatants were incubated overnight at 4°C with either GST control protein or GST-K15 fusion protein immobilized to glutathione beads. The beads were washed four times in 1% Triton X-100 lysis buffer, and proteins adsorbed to beads were incubated in 25 μl of SDS electrophoresis sample buffer for 10 min prior to analysis by SDS-PAGE and immunoblotting. Proteins were detected with a mouse anti-c-myc 9E10 antibody (Biomol) and a standard enhanced chemiluminescence (ECL) reaction.
Peptide array.
Peptides immobilized on cellulose were synthesized in the Cancer Research UK Protein and Peptide Chemistry Laboratory (N. O'Reilly; London, United Kingdom). Membranes were prewetted in ethanol and blocked overnight in Tris-buffered saline (TBS) containing 0.1% Tween 20, 5% skimmed milk, and 0.1% sodium azide. After being blocked, membranes were incubated for 3 h with recombinant GST protein at a concentration of 1.25 μg/ml in blocking buffer containing 5 mM dithiothreitol (DTT). Nonbound protein was removed by washing it three times in TBS containing 0.1% Tween 20. Bound protein was subsequently visualized with an anti-GST antibody (2C8-1-4; E. Kremmer; GSF Munich, Germany) and ECL detection. Membranes were then stripped of bound antibodies, blocked overnight, and probed for 3 h with 1.25 μg/ml of recombinant GST-Hck or GST-Lyn SH3 protein. GST fusion proteins were detected by ECL as previously described.
SH3 domain phage display experiments.
GST-K15 fusion proteins were expressed and purified as described above in “Expression and purification of recombinant proteins and GST fusion protein binding assays.” Phage display experiments, with the entire human SH3 proteome consisting of 296 SH3 domain sequences, were essentially performed as described previously (17). Three independent selection experiments were carried out for each target protein, and the identities of 34 or 44 individual K15-M- or K15-P-selected SH3 clones, respectively, were determined. The same methodology was also used to generate individual infectious SH3 phage preparations and to test their relative binding to different GST fusion proteins.
Isothermal titration calorimetry.
Measurements were performed on a VP-isothermal titration calorimetry (ITC) microcalorimeter (MicroCal, Northampton, MA), essentially as described previously (11). Briefly, 1.43 ml of affinity-purified GST SH3 domain in ITC buffer (25 mM HEPES-KOH at pH 7.5, 100 mM potassium acetate, 5 mM magnesium acetate) at a final concentration of 0.1 or 0.2 mM was clarified for 10 min at 20,800 × g and degassed for 10 min (ThermoVac; MicroCal) before being transferred into the sample chamber. Synthetic peptides were diluted to 1 or 2 mM (10× GST fusion protein concentration) in ITC buffer, clarified, and degassed, and 300 μl was loaded into the syringe. During the measurements, 5 and 10 μl aliquots of peptide solution were titrated once and 28 times, respectively, into the sample chamber at an equilibrium temperature of 25°C. The heat of dilution was negligible. Total heat generated and equilibrium dissociation constant (KD) values were calculated in ORIGIN (version 5.0). The binding affinity for each peptide was measured at least twice.
Low-density microarray experiments.
HeLa cells were transfected as described above in “Transfections.” At 32 h posttransfection, cells were lysed for RNA extraction, according to the manufacturer's instructions (RNeasy kit; Qiagen). For microarray experiments, the Human Inflammation Array (MWG Biotech) was used (now distributed by Ocimum Biosolutions as the human Inflammation OciChip). This microarray contains 155 validated oligonucleotide probes for 136 inflammatory and 19 housekeeping genes. Total RNA was purified with the RNeasy microkit, including on-column DNase I digestion (Qiagen). RNA was used to prepare Cy3-labeled cRNA (Amino Allyl MessageAmp II kit, catalog no. AM1795; Ambion), as directed by the company. cRNA yields and fluorescence incorporation efficiencies were determined photometrically.
A total of 8 μg of labeled cRNAs were fragmented, repurified, and hybridized to microarrays in preprepared hybridization solution (MWG Biotech) at 42°C overnight and then washed sequentially in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% SDS, 1× SSC, and 0.5× SSC. Hybridized arrays were scanned using the Agilent microarray scanner G2565CA at three different photomultiplier settings (90%, 30%, and 10%) to increase the dynamic range of the measurements. Fluorescence intensity values were processed by using ImaGene 5.0 software (BioDiscovery). In order to obtain maximal signal intensities without saturation effects, intensity values obtained from TIFF images were integrated into one value per probe by MAVI software (version Pro 2.5.1; MWG Biotech). After quality control and normalization (according to the expression of 19 housekeeping genes), data were used to calculate ratios of gene expression by using Microsoft Excel macros. Additional information on microarrays and methodology used can be obtained at the Cytokine Microarray Project website (http://microarray.med.uni-giessen.de/).
ELISA.
Conditioned medium of transfected HeLa cells was collected at 32 h posttransfection and cleared by centrifugation at 14,000 × g at 4°C for 10 min, and supernatants were analyzed by interleukin-8 (IL-8) enzyme-linked immunosorbent assay (ELISA), according to the manufacturer's instructions (Biosource).
Immunoblotting.
At 32 h posttransfection, HeLa cells were washed one time with cold PBS and lysed with 250 μl RIPA 100 buffer (20 mM Tris at pH 7.5, 1 mM EDTA, 100 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate) with protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 5 μM leupeptin, 100 U/ml aprotinin, 200 μM benzamidine, 1 μM pepstatin A) per well. Lysates were cleared by centrifugation (10,000 × g for 10 min at 4°C) and analyzed by immunoblotting. Proteins were detected with a rabbit anti-K15 (2), a mouse anti-Flag (Sigma), or a mouse anti-Actin antibody (Chemicon) and a standard ECL reaction.
Calcium mobilization analysis.
BJAB cells were cotransfected with expression plasmids for the K15 WT or mutants, as indicated in the figure legends, and a mRFP plasmid to identify successfully transfected cells. At 48 h posttransfection, cells were washed once in PBS and resuspended in 1 ml RPMI 1640 medium without FCS and antibiotics. Cells were loaded with 3 μM Indo-1 AM ester (Molecular Probes) for 30 min at 37°C in a water bath. Following a washing step in PBS, BJAB cells were resuspended in 1 ml RPMI 1640 medium without FCS and antibiotics. Baseline calcium levels were established 2 min prior to addition of the antibody. Cells were stimulated with 10 μg goat anti-human IgM antibody (Southern Biotech), and data were collected for 7 or 8 min. Experiments were performed on an LSR II flow cytometer (Becton Dickinson) and analyzed with FlowJo software.
RESULTS
Screening for interaction partners of the K15 SH3 binding site.
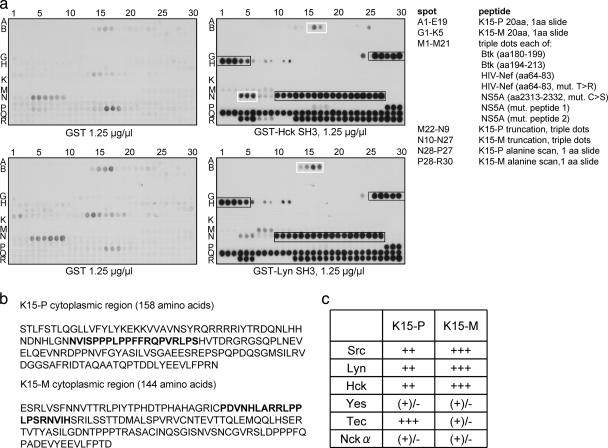
Initially, we screened a collection consisting of 35 SH3 domains of intracellular signaling proteins, such as adaptor proteins and kinases. These domains were expressed as GST fusion proteins, immobilized to glutathione-Sepharose beads, and tested in GST pulldown assays for their binding ability to both of the K15 genotypes. In this screen, we identified the K15-P and K15-M proteins as ligands for the SH3 domains of Hck and Lyn (data not shown). In addition, we found K15-M to interact with the SH3 domain of the kinase Src, although to a lesser extent. In order to map the epitopes within the K15 proteins responsible for the direct interactions with the SH3 domains of Hck and Lyn, we synthesized the cytoplasmic domains of K15-P (amino acids [aa] 331 to 489) and K15-M (354 to 498) as overlapping peptides of 20 amino acids, each shifted by one amino acid (Fig. 1 a, spots A1 to E19 for K15-P and spots G1 to K5 for K15-M). Peptides were synthesized on cellulose membranes and incubated with recombinantly expressed GST-Hck or GST-Lyn SH3 domain proteins (Fig. 1a). We found a strong interaction between K15-M and the Hck and Lyn SH3 domains (Fig. 1a, black boxes, spots G26 to H5). Interestingly, the PPLP sequence was present in all peptides represented by these spots. In contrast to this unambiguous interaction, we observed a weaker, but still detectable interaction between K15-P and Hck SH3 (Fig. 1a, top, white box, spots B16 and 17) and between K15-P and Lyn SH3 (Fig. 1a, bottom, white box, spots B14 to 17). Binding of Hck SH3 and Lyn SH3 to K15-M was much stronger than binding of them to the known Hck and Lyn interaction partners Btk (aa 180 to 199 and aa 194 to 213), HIV Nef (aa 64 to 83), and a hepatitis C virus (HCV) NS5A mutant (aa 2313 to 2332) (see longer exposure of Fig. 1 in Fig. S1 in the supplemental material).
FIG. 1.
The SH3 domains of Hck and Lyn bind to the putative SH3 binding sites of K15-P and K15-M. (a) Overlapping peptides corresponding to the cytoplasmic region of K15-P or K15-M and control peptides were immobilized via their C termini on cellulose membranes and were incubated with recombinant GST alone (left panels) or GST Hck SH3 or GST Lyn SH3 protein (right panels). Spots A1 to E19 (K15-P) and G1 to K5 (K15-M) represent peptides of 20 amino acids, with 1 amino acid slide per peptide to cover the entire cytoplasmic regions of the two K15 proteins. Spots M1 to 21 are peptides derived from proteins known to bind to Hck and Lyn as indicated in the list. Spots M22 to N9 (K15-P) and N10 to 27 (K15-M) represent truncation peptides of the K15 proteins to map the core binding regions within the K15 SH3-B sites (printed in boldface in panel b). Spots N28 to P27 (K15-P) and P28 to R30 represent an alanine scan through the identified K15 motifs (printed in boldface in panel b). (b) Amino acid sequences of the entire cytoplasmic regions of K15-P and K15-M. The identified SH3-B site within the cytoplasmic region of each K15 protein is printed in boldface. (c) Relative binding of individual SH3 domain-displaying phages to GST-K15 (P- and M-type)-coated wells. Wells were coated with recombinantly expressed fusion proteins consisting of GST fused to the cytoplasmic tail of K15-P or K15-M. Individual SH3 domain-displaying phages were incubated with GST fusion protein-coated wells, and the strength of binding was estimated based on the increase (1- to 10,000-fold) of phages bound compared with that in plates coated with plain GST. The table indicates semiquantitative binding strengths of the individual SH3-displaying phages (left vertical column) to GST-K15-P or GST-K15-M fusion proteins (top horizontal column). Binding strengths: +++, strong (>1,000-fold enrichment); ++, good (>100-fold enrichment); +, moderate (>10-fold enrichment); (+)/−, weak/none (<10-fold enrichment).
To further map the K15 core region responsible for binding to SH3 domains of Hck and Lyn, we spotted truncation peptides of K15 (see Fig. S2 in the supplemental material) on the cellulose membranes (Fig. 1a, spots M22 to N9 for K15-P and spots N10 to 27 for K15-M). We noticed an interaction among all K15-M truncation peptides, since all of them contained the PPLP motif (Fig. 1a, black box, spots N10 to 27). Furthermore, we detected binding of Hck SH3 to a K15-P truncation peptide (Fig. 1a, top, white box, spots N4 to 6), which seemed to be less strong. To determine which amino acids within the identified SH3 binding sites are important for K15 binding to Hck and Lyn SH3 domains, we performed a scan with spot-synthesized alanine-scanning mutant peptides on the membranes in triple dots; in each peptide, the substituted alanine was moved by one amino acid (Fig. 1a, spots N28 to P27 for K15-P and spots P28 to R30 for K15-M). We observed a nearly complete loss of the interaction between K15-M and SH3 domains of Hck and Lyn when the PPLP sequence was changed to APLP (Fig. 1a, spots R1 to 3), PPAP (spots R7 to 9), or PPLA (spots R10 to 12). This emphasizes the importance of the outer proline residues of this motif, since mutation of each of them inhibited complete binding of the Hck and Lyn SH3 domain proteins. In contrast to that, we could not detect any binding of the Hck and Lyn SH3 proteins to the K15-P alanine-scanning mutant peptides (Fig. 1a, spots N28 to P27).
To substantiate these data and further characterize putative K15-interacting SH3 domains, we performed an initial phage display-assisted global SH3 screen. This phage library comprised a virtually complete human SH3 proteome, consisting of 296 SH3 domain sequences (17). As target proteins in these experiments, we used fusion protein constructs, which comprised the entire cytoplasmic tail of wild-type K15-P or -M fused to GST (GST-K15-P/K15-M exon 8 [ex8] WT).
A single round of affinity selection of the SH3 phage library with the recombinant GST-K15 fusion proteins resulted in a significant enrichment of phages compared to that with mock selection with GST alone (data not shown). The GST-K15-M ex8 WT protein showed more than 100-fold SH3 phage binding compared to that of GST, while the enrichment with GST-K15-P ex8 WT was more modest but yet more than 10-fold higher than that with the GST control. When the identity of the selected SH3 clones was determined, we found that among the collection of 296 human SH3 domains, the GST-K15-M ex8 WT protein preferentially selected Src family SH3 domains of Lyn, Src, Hck, and Yes. K15-M most frequently selected Lyn (38% of all selected clones), Src (38%), and Hck (18%). The collection of K15-P-selected clones was to some extent more heterogeneous. Although Src family SH3 domains (37%), in particular Lyn (25%), were frequently selected, the single most commonly identified SH3 domain was the non-Src family tyrosine kinase Tec (36%). Unlike K15-M, K15-P also consistently selected the second SH3 domain (of three) of the adaptor protein Nckα (9%).
For a more quantitative idea of the relative affinities of the consistently selected SH3 domains, we generated individual phage preparations displaying these domains and compared their levels of binding to the GST-K15 fusion proteins (Fig. 1c). Based on the degree of the enrichment compared to that of the GST control, their binding was categorized as strong, good, moderate, or weak/none, corresponding on average to more than 1,000-fold, 100-fold, 10-fold, and less than 10-fold phage enrichment, respectively. The results are summarized in Fig. 1c and correlated well with the primary data obtained from the initial SH3 library screening, confirming relative SH3 binding selectivity of the different K15 proteins.
K15-M bound very well to Lyn, Src, and Hck but not significantly to the other SH3 domains. K15-P also bound to these three Src family SH3 domains, although less strongly. Unlike K15-M, K15-P also bound well to Tec. The binding of K15-M and K15-P to Nckα and of K15-P to Yes was found to be weak/borderline in this more quantitative assay (Fig. 1c).
High-affinity binding of K15 to Hck and Lyn SH3.
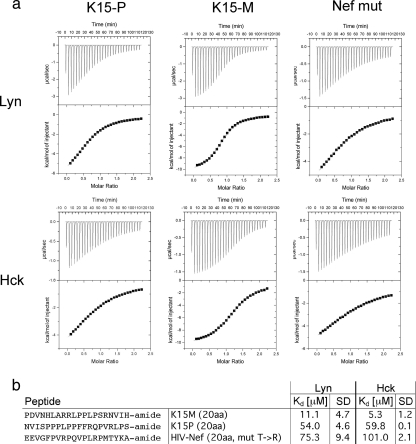
To measure the binding strength and selectivity of the interaction between the KSHV K15 protein and the SH3 domains of Hck and Lyn, we utilized isothermal titration calorimetry (ITC). As a positive control, we included an HIV Nef mutant corresponding to the HIV-1 NL4-3 Nef, with a change from a threonine to an arginine at position 71. This mimics the amino acid sequence that occurs in this region of most Nef alleles, which have been obtained directly from patients (14, 37). Arginine 71 is adjacent to the amino acid residues of Nef that are involved in SH3 binding and was shown to increase this interaction (20, 33). Amidated peptides of 20 amino acids in length corresponding to identified SH3 binding sites (Fig. 1b, boldface) of both of the K15 genotypes or the Nef mutant were used in these experiments (Fig. 2 b). The results obtained from the ITC measurements revealed that both of the K15 alleles bind with high affinities to the SH3 domains of Hck and Lyn (Fig. 2a). The KD values are summarized in Fig. 2b and show that K15-M binds with higher affinities to the Hck (KD, 5.3) and Lyn (KD, 11.1) SH3 domains than K15-P (Hck KD, 59.8; Lyn KD, 54) or the Nef mutant (Hck KD, 101; Lyn KD, 75.3), in keeping with the qualitative results obtained using the peptide array (Fig. 1a).
FIG. 2.
Measurement of binding strengths among K15 proteins and the Src kinases Hck and Lyn by isothermal titration calorimetry (ITC). (a) ITC experiments were performed as described in Materials and Methods. Amidated peptides consisting of 20 amino acids of K15-P and K15-M (printed in boldface in Fig. 1b) were used to determine binding affinities to Src kinases Hck and Lyn. The 20-amino-acid peptide of the HIV Nef mutant was included as a positive control. (b) Binding strengths (KD values), as calculated with the software ORIGIN (version 5.0), and standard deviations (SD) of the K15-P, -M, or HIV Nef peptides with the SH3 domains of the Src kinases Lyn and Hck are indicated.
K15 interacts with the kinases Hck, Lyn, and Src mainly via its SH3 binding site.
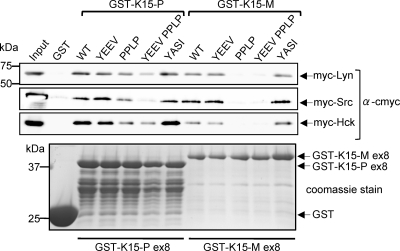
In order to confirm SH3 domain-mediated interactions between the kinases Hck, Lyn, or Src and K15 and to evaluate whether other conserved motifs may contribute to the interaction, we performed GST pulldown experiments. We used the GST-K15-P/K15-M fusion protein constructs mentioned above to generate GST fusion protein constructs containing mutations of the SH2 binding sites YASI and YEEV and the SH3 binding motif PPLP. The tyrosine residues in the SH2 binding motifs were changed to phenylalanine residues (K15-P, Y431F and Y481F; K15-M, Y444F and Y490F), and the proline residues in the SH3 binding sites were replaced by alanine residues (K15-P, PPLPP→AALAA; K15-M, PPLP→APLA). We also generated a K15-P and -M double mutant construct containing mutations of both the YEEV and PPLP motifs, since we had previously shown that the tyrosine residue in the K15-P YEEV motif is targeted by members of the Src kinase family for binding and phosphorylation (2). As shown in Fig. 3, we could precipitate Hck, Lyn, and Src full-length proteins with the wild-type (WT) K15-P and K15-M proteins immobilized on glutathione-Sepharose beads. Mutation of the K15-P or K15-M YEEV or YASI motif did not affect binding of Hck, Lyn, and Src (Fig. 3). When we compared the K15-P and K15-M PPLP mutant proteins for their ability to precipitate PTKs (Fig. 3), we observed a slight reduction of binding of all PTKs to the K15-P PPLP SH3 binding mutant and a complete loss of binding to the K15-M PPLP SH3 binding mutant. While the K15-M double mutant was not able to bind to PTKs at all, the K15-P double mutant showed only a reduction in binding to Lyn and Hck and a loss of binding to the PTK Src. This finding emphasizes the importance of an intact K15 PPLP SH3 binding motif for interaction with PTKs. Taken together, we found that the binding of these three PTKs, Hck, Lyn, and Src, seemed to be dependent on the PPLP SH3 binding motifs of both of the K15 genotypes but that the K15-M SH3 binding motif, possibly because of its high affinity for the PTK SH3 domain (Fig. 2), seems to play a stronger role in this interaction.
FIG. 3.
Binding of Src kinases to K15 mutants. HEK 293-T cells were transiently transfected with expression vectors for myc-tagged PTKs Lyn, Src, and Hck. Cell lysates were incubated overnight at 4°C with the GST-K15-P and GST-K15-M exon 8 (ex8) wild-type (WT) proteins, and their respective mutants (YEEV, PPLP, YEEV PPLP, and YASI), immobilized to glutathione-Sepharose beads. Precipitated Lyn, Src, and Hck proteins were analyzed by immunoblotting with an anti-c-myc antibody. Purified GST or GST-K15 fusion proteins separated on an SDS-PAGE gel were visualized by Coomassie blue staining to show that equal amounts of fusion protein were used for GST pulldown experiments (bottom). α, anti.
The SH3 binding motif is dispensable for the K15-induced inflammatory phenotype.
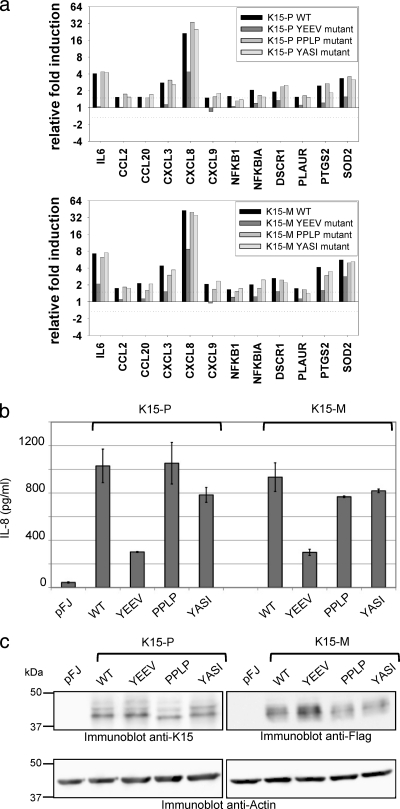
We have recently reported that K15, when expressed ectopically in epithelial cells, increases the expression of numerous inflammation- and angiogenesis-related genes and that these changes are dependent on an intact YEEV SH2 binding site (3, 38). In view of the role of the K15-M and K15-P SH3 binding sites in the recruitment of Src family PTKs, we were interested in their role in K15-induced signaling. We investigated the cellular transcriptome of HeLa cells transiently transfected with a full-length K15-P or K15-M expression construct or mutant constructs of the two SH2 binding sites (YASI and YEEV) and the SH3 binding motif (PPLP). Equivalent expression levels of wild-type and mutant K15 proteins were verified by immunoblotting (Fig. 4 c). RNA extracted from transfected HeLa cells was converted to cRNA, which was hybridized to an inflammatory low-density microarray (12, 13), and changes in mRNA levels were calculated for individual genes. As previously reported (3, 38), wild-type K15-P and K15-M induced the expression of inflammation- and angiogenesis-related genes, such as IL-6, IL-8, SOD2, PTGS2 (COX2), and DSCR1 (Fig. 4a) but not that of housekeeping genes (see Fig. S3 in the supplemental material). Most K15 WT-activated cellular genes were not induced by the YEEV motif mutant (YF), as previously reported (3, 38). In contrast, the K15 mutants lacking the intact YASI or PPLP motif were not or only moderately impaired in their ability to activate inflammatory genes (Fig. 4a), indicating that protein interactions with these motifs are not crucial for this K15-induced phenotype. To confirm the data obtained from microarray experiments at the protein level, we used an IL-8 ELISA, since the IL-8 gene was most strongly induced by the K15 WT. Supernatants from K15 WT-expressing HeLa cells contained amounts of secreted IL-8 similar to those of the K15 YASI mutant- or SH3 binding mutant-expressing cells (Fig. 4b). Consistent with our microarray data, the K15 YEEV mutant was significantly impaired in the induction of IL-8 secretion.
FIG. 4.
Contribution of conserved K15 SH2 and SH3 binding motifs to inflammatory signaling. (a) HeLa cells were transfected with expression vectors for the K15-P or K15-M wild-type (WT) protein or its mutant YEEV, PPLP, or YASI versions. Extracted RNA was subjected to low-density microarray analysis using a Human Inflammation Array (Ocimum Biosolutions). Ratios of relative gene expression were calculated by dividing signal intensity values originating from K15-P (top), K15-M (bottom), or mutant protein samples by values obtained for the empty vector control. The graphs show ratio values for genes that were induced at least 1.5-fold by both the K15-P WT and the K15-M WT proteins. (b) HeLa cells were transfected with expression vectors for the K15-P or K15-M WT or its mutant YEEV, PPLP, or YASI versions, as described for low-density microarrays, and IL-8 levels (pg/ml) in cleared supernatants were measured by ELISA. (c) Cleared lysates from transfected HeLa cells were analyzed with an anti-K15 antibody for equal K15-P expression, an anti-Flag antibody for equal K15-M expression, and an anti-actin antibody as a control.
Inhibition of BCR signaling by K15 depends on the SH2 and SH3 binding motifs.
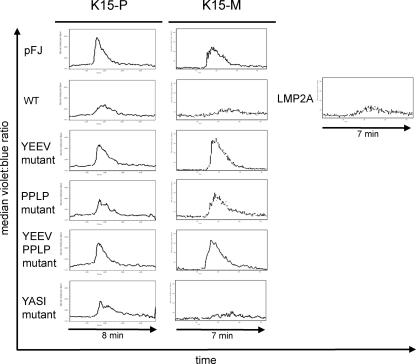
It had been previously demonstrated that the cytoplasmic tail of K15-P fused to the transmembrane and extracellular domain of CD8 is able to interfere with B-cell receptor signaling, as shown in calcium mobilization assays, but is not capable of initiating intracellular signaling upon anti-CD8 antibody stimulation (7). The downregulation of calcium mobilization seemed to be mediated by the K15-P YEEV SH2 binding site and the PPLP SH3 binding motif. Based on these observations, we investigated whether an intact full-length K15 protein could inhibit calcium mobilization in response to B-cell receptor ligation and whether this involved any of the three conserved protein interaction motifs (YEEV, YASI, and PPLP). To this end, we transiently cotransfected BJAB cells with full-length expression constructs for both of the wild-type K15 genotypes, mutants of the YEEV SH2 and PPLP SH3 binding sites, or the YEEV and PPLP double mutant, together with an mRFP expression plasmid (K15/mRFP, 7:1 ratio) to identify transfected cells. An expression plasmid for Epstein-Barr virus (EBV) LMP2A served as the positive control for inhibition of calcium mobilization (23, 25). After stimulation of BCR signaling with an anti-human IgM antibody, calcium mobilization was measured by Indo-1 staining in cells gated on mRFP-expressing cells. We detected a substantial inhibition of calcium mobilization in K15 WT (P- and M-type)-expressing B cells compared to that in empty vector (pFJ)-transfected cells (Fig. 5). The decrease of calcium mobilization was comparable to that of cells that were transfected with an EBV LMP2A expression plasmid (Fig. 5, right panel). When BJAB cells were transfected with K15-P expression constructs with mutations in individual motifs (SH2 or SH3 binding sites), we found that the YEEV mutant and the PPLP mutant, as well as the double mutant (YEEV and PPLP) constructs, were significantly impaired in their ability to inhibit calcium mobilization. The contribution of the PPLP motif to the inhibition of calcium mobilization by K15 was more clearly seen in the case of K15-M. In contrast, mutation of the YASI motif had no (in the case of K15-M) or a minimal (K15-P) impact on the inhibition of calcium mobilization by K15 (Fig. 5). We concluded from these results that interference with BCR signaling required both the YEEV and PPLP motifs.
FIG. 5.
Effect of K15 expression on intracellular free calcium mobilization induced by anti-human IgM antibody. BJAB cells were transiently cotransfected with K15 expression plasmids and an mRFP plasmid in a 7:1 ratio to monitor successful transfections. The EBV LMP2A expression plasmid was included as a positive control for inhibition of calcium mobilization. Cells were loaded with 3 μM Indo-1 dye and stimulated with 10 μg/ml of goat anti-human IgM antibody. K15- or LMP2A-expressing cells were gated for high levels of mRFP expression, and calcium mobilization was monitored by changes in the ratio of violet to blue (405 to 485 nm) fluorescence of cells (median value, y axis) over time (x axis). Baseline calcium levels were established for 2 min prior to addition of goat anti-human IgM antibody. All measurements were performed at room temperature.
DISCUSSION
Previous studies have shown that the KSHV K15 transmembrane protein has the ability to interact through its cytoplasmic domain with a range of cellular proteins and thereby triggers signaling cascades (2, 3, 6, 7, 10, 21, 35, 38). Several sequence motifs, such as the putative SH2 and SH3 binding motifs, are located in the C-terminal domain, and an involvement in K15-mediated signaling has been shown for some of these. These motifs are conserved between the two highly diverged P and M genotypes (10, 16, 31). The YEEV motif was shown to be targeted for binding and phosphorylation by PTKs, and phosphorylation of tyrosine 481 of K15-P is essential for activation of the NF-κB, MEK/Erk, and JNK pathways and for induction of inflammatory and angiogenic cytokines (2, 3, 7, 38). The PPLP SH3 binding site of K15-P has been shown to interact with a range of cellular SH3 domains (21), while the YASI motif was reported to interact with the cellular HAX-1 protein (35). Here we systematically investigated the participation of these three protein interaction motifs in signaling cascades induced by K15. We also determined the affinities of the interaction between cellular Src kinase family members and the PPLP SH3 binding sites in K15-P and K15-M. Initially, we took two approaches to identifying cellular SH3 domains that could be recruited by K15. We screened a collection of 35 SH3 domains from well-studied cellular proteins with GST-K15 proteins. In this screen, we identified members of the Src family of protein tyrosine kinases, Hck, Lyn, and Src in particular, as ligands of the cytoplasmic tails of both of the K15 genotype wild-type proteins. In addition, a screen of a phage display library for the entire set of human SH3 domains identified additional potential ligands for the K15 SH3 binding sites, including Tec, Nckα, and the Src family kinase Yes. Furthermore, we determined relative binding affinities of ligands with K15 SH3 binding sites using a semiquantitative phage display assay (Fig. 1c). These additional ligands were also identified by Lim and colleagues in a recently published similar screen of SH3 domains (21). To determine the epitope within the cytoplasmic tail of K15, which is responsible for binding to PTKs, we performed peptide arrays (Fig. 1a). We mapped the binding site within K15-M to the PPLP motif and showed, by alanine scanning, that the outer proline residues are necessary for binding to the SH3 domains of Hck and Lyn (Fig. 1a). To study the SH3 domain affinities of these Src kinases for the K15 SH3 binding sites, we used isothermal titration calorimetry (Fig. 2) and determined the equilibrium dissociation constants (KD) for both of the K15 genotypes and SH3 domains of Hck and Lyn (Fig. 2b). KD values for the interaction of the Lyn and Hck SH3 domains with K15-P and K15-M were lower than those for their interaction with an HIV-1 Nef mutant that is considered an example of a high-affinity interaction of a viral protein with cellular Src kinases, although in the case of HIV-1 Nef, the Hck interaction also involves residues outside the proline-rich motif, thus increasing the affinity (20). This result highlights the significance of the interaction between K15 and Lyn or between K15 and Hck. Interestingly, the affinity of the K15-M SH3 binding site for the Lyn and Hck binding sites was much higher than that of the K15-P SH3 binding site (Fig. 2b). This result agrees well with the stronger interaction of K15-M with Lyn and Hck seen on the peptide arrays (Fig. 1 a) and in the semiquantitative phage display assay (Fig. 1c). GST pulldown assays with the cytoplasmic domains of K15-P and K15-M, in which the three protein interaction motifs had been mutated (Fig. 3), indicated that in the case of K15-P, both the YEEV SH2 binding site and the PPLP SH3 binding site may contribute to the interaction with Lyn, Hck, and Src. However, the contribution of the PPLP SH3 binding site clearly exceeds that of the YEEV SH2 binding site (Fig. 3). In contrast, the interaction of the Src kinases Hck, Lyn, and Src with K15-M were dependent solely on the PPLP SH3 binding site. We have recently reported (38) that while K15-P and K15-M activate similar cellular signaling pathways, the activation of JNK by K15-M was less dependent on an intact YEEV motif than the activation of JNK by K15-P. We therefore surmise that the increased affinity of the K15-M SH3 binding site allows the recruitment of SH3 domain-containing cellular signaling components even when the YEEV motif is mutated, as seen in the GST pulldown assays (Fig. 3). In view of the higher affinity of the K15-M SH3 binding site for Lyn and Hck (Fig. 2) and its importance in the recruitment of these two Src kinases (Fig. 3), we were surprised to find that the induction of cellular inflammatory and angiogenic genes by K15 is not significantly affected by the mutation of this binding site (Fig. 4). In contrast, as reported previously (3, 38), mutation of the YEEV SH2 binding site abolished the induction of most K15-activated cellular genes, including the strongly activated angiogenic chemokine IL-8 (Fig. 4b). We therefore assume that phosphorylation of the YEEV motif, which is required for the induction of these cellular genes (Fig. 4), is not mediated by any of these Src kinase family members studied here, since they all required an intact K15-M SH3 binding site for efficient recruitment to K15-M (Fig. 3). The kinase responsible, yet to be identified, would not depend on the SH3 binding site to be recruited to a K15 signaling complex. Using small interfering RNA (siRNA) silencing of Syk and a chemical inhibitor of Syk (piceatannol) in luciferase reporter assays, we did not find any evidence for the involvement of Syk in K15-mediated activation of an AP-1 or NFκB reporter (data not shown).
In order to establish which of the K15-induced phenomena depended on the SH3 binding site and recruitment of Src kinase family members, we investigated the impact of K15 on calcium mobilization in B cells, first reported by Choi et al. (7). Those authors had reported that a CD8-K15 chimera interferes with B-cell receptor signaling (7). Since we had previously shown (2) that the 12 transmembrane domains of K15 are required for its ability to activate intracellular signaling cascades and cannot, for example, be replaced by the 6 transmembrane domains of EBV LMP1, we investigated the inhibition of calcium mobilization by K15 wild-type proteins. We found that both K15-P and K15-M inhibited calcium mobilization induced by triggering the B-cell receptor but that K15-M showed a stronger inhibition, which was comparable to that obtained with EBV LMP2A (Fig. 5). Mutation of either the YEEV SH2 binding site or the PPLP SH3 binding site reduced the ability of K15 to inhibit calcium mobilization (Fig. 5). These findings are in line with those reported by Choi et al. (7) and with the interpretation that recruitment of a B-cell Src kinase like Lyn via the K15 PPLP SH3 binding site could reduce the availability of this kinase for B-cell receptor-initiated signaling events. A possible effect of K15-mediated inhibition of intracellular calcium mobilization could be the maintenance of viral latency, as has been reported for the EBV LMP2A protein (22-24). Interestingly, it was reported that mobilization of free intracellular calcium reactivates KSHV from latency in BCBL-1 cells (40). Therefore, it is conceivable that K15 might inhibit KSHV reactivation by interference of BCR signaling. The very high affinity of the K15 SH3 binding site for the Lyn SH3 domain (Fig. 2), hence, might play a role in maintaining KSHV latency in infected B cells.
Supplementary Material
Acknowledgments
We thank Matthias Ballmaier, Lars Steinbrück, and Elena Grabski for their help with the acquisition and analysis of fluorescence-activated cell sorter (FACS) data.
This study was supported by SFB 566, TP11 from the Deutsche Forschungsgemeinschaft, and the European Union Integrated Project INCA (grant LSHC-CT-2005-018704).
Footnotes
Published ahead of print on 9 June 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Boshoff, C., T. F. Schulz, M. M. Kennedy, A. K. Graham, C. Fisher, A. Thomas, J. O. McGee, R. A. Weiss, and J. J. O'Leary. 1995. Kaposi's sarcoma-associated herpesvirus infects endothelial and spindle cells. Nat. Med. 1:1274-1278. [DOI] [PubMed] [Google Scholar]
- 2.Brinkmann, M. M., M. Glenn, L. Rainbow, A. Kieser, C. Henke-Gendo, and T. F. Schulz. 2003. Activation of mitogen-activated protein kinase and NF-kappaB pathways by a Kaposi's sarcoma-associated herpesvirus K15 membrane protein. J. Virol. 77:9346-9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brinkmann, M. M., M. Pietrek, O. Dittrich-Breiholz, M. Kracht, and T. F. Schulz. 2007. Modulation of host gene expression by the K15 protein of Kaposi's sarcoma-associated herpesvirus. J. Virol. 81:42-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brinkmann, M. M., and T. F. Schulz. 2006. Regulation of intracellular signalling by the terminal membrane proteins of members of the Gammaherpesvirinae. J. Gen. Virol. 87:1047-1074. [DOI] [PubMed] [Google Scholar]
- 5.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 6.Cho, N. H., Y. K. Choi, and J. K. Choi. 2008. Multi-transmembrane protein K15 of Kaposi's sarcoma-associated herpesvirus targets Lyn kinase in the membrane raft and induces NFAT/AP1 activities. Exp. Mol. Med. 40:565-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi, J. K., B. S. Lee, S. N. Shim, M. Li, and J. U. Jung. 2000. Identification of the novel K15 gene at the rightmost end of the Kaposi's sarcoma-associated herpesvirus genome. J. Virol. 74:436-446. [PMC free article] [PubMed] [Google Scholar]
- 8.Dupin, N., T. L. Diss, P. Kellam, M. Tulliez, M. Q. Du, D. Sicard, R. A. Weiss, P. G. Isaacson, and C. Boshoff. 2000. HHV-8 is associated with a plasmablastic variant of Castleman disease that is linked to HHV-8-positive plasmablastic lymphoma. Blood 95:1406-1412. [PubMed] [Google Scholar]
- 9.Ensoli, B., and M. Sturzl. 1999. HHV-8 and multistep tumorigenesis. Trends Microbiol. 7:310-312. [DOI] [PubMed] [Google Scholar]
- 10.Glenn, M., L. Rainbow, F. Aurade, A. Davison, and T. F. Schulz. 1999. Identification of a spliced gene from Kaposi's sarcoma-associated herpesvirus encoding a protein with similarities to latent membrane proteins 1 and 2A of Epstein-Barr virus. J. Virol. 73:6953-6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harkiolaki, M., M. Lewitzky, R. J. Gilbert, E. Y. Jones, R. P. Bourette, G. Mouchiroud, H. Sondermann, I. Moarefi, and S. M. Feller. 2003. Structural basis for SH3 domain-mediated high-affinity binding between Mona/Gads and SLP-76. EMBO J. 22:2571-2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoffmann, E., A. Thiefes, D. Buhrow, O. Dittrich-Breiholz, H. Schneider, K. Resch, and M. Kracht. 2005. MEK1-dependent delayed expression of Fos-related antigen-1 counteracts c-Fos and p65 NF-kappaB-mediated interleukin-8 transcription in response to cytokines or growth factors. J. Biol. Chem. 280:9706-9718. [DOI] [PubMed] [Google Scholar]
- 13.Holzberg, D., C. G. Knight, O. Dittrich-Breiholz, H. Schneider, A. Dorrie, E. Hoffmann, K. Resch, and M. Kracht. 2003. Disruption of the c-JUN-JNK complex by a cell-permeable peptide containing the c-JUN delta domain induces apoptosis and affects a distinct set of interleukin-1-induced inflammatory genes. J. Biol. Chem. 278:40213-40223. [DOI] [PubMed] [Google Scholar]
- 14.Huang, Y., L. Zhang, and D. D. Ho. 1995. Characterization of nef sequences in long-term survivors of human immunodeficiency virus type 1 infection. J. Virol. 69:93-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jenner, R. G., M. M. Alba, C. Boshoff, and P. Kellam. 2001. Kaposi's sarcoma-associated herpesvirus latent and lytic gene expression as revealed by DNA arrays. J. Virol. 75:891-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kakoola, D. N., J. Sheldon, N. Byabazaire, R. J. Bowden, E. Katongole-Mbidde, T. F. Schulz, and A. J. Davison. 2001. Recombination in human herpesvirus-8 strains from Uganda and evolution of the K15 gene. J. Gen. Virol. 82:2393-2404. [DOI] [PubMed] [Google Scholar]
- 17.Karkkainen, S., M. Hiipakka, J. H. Wang, I. Kleino, M. Vaha-Jaakkola, G. H. Renkema, M. Liss, R. Wagner, and K. Saksela. 2006. Identification of preferred protein interactions by phage-display of the human Src homology-3 proteome. EMBO Rep. 7:186-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katano, H., Y. Sato, T. Kurata, S. Mori, and T. Sata. 2000. Expression and localization of human herpesvirus 8-encoded proteins in primary effusion lymphoma, Kaposi's sarcoma, and multicentric Castleman's disease. Virology 269:335-344. [DOI] [PubMed] [Google Scholar]
- 19.Lee, B. S., S. H. Lee, P. Feng, H. Chang, N. H. Cho, and J. U. Jung. 2005. Characterization of the Kaposi's sarcoma-associated herpesvirus K1 signalosome. J. Virol. 79:12173-12184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, C. H., B. Leung, M. A. Lemmon, J. Zheng, D. Cowburn, J. Kuriyan, and K. Saksela. 1995. A single amino acid in the SH3 domain of Hck determines its high affinity and specificity in binding to HIV-1 Nef protein. EMBO J. 14:5006-5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim, C. S., B. T. Seet, R. J. Ingham, G. Gish, L. Matskova, G. Winberg, I. Ernberg, and T. Pawson. 2007. The K15 protein of Kaposi's sarcoma-associated herpesvirus recruits the endocytic regulator intersectin 2 through a selective SH3 domain interaction. Biochemistry 46:9874-9885. [DOI] [PubMed] [Google Scholar]
- 22.Miller, C. L., A. L. Burkhardt, J. H. Lee, B. Stealey, R. Longnecker, J. B. Bolen, and E. Kieff. 1995. Integral membrane protein 2 of Epstein-Barr virus regulates reactivation from latency through dominant negative effects on protein-tyrosine kinases. Immunity 2:155-166. [DOI] [PubMed] [Google Scholar]
- 23.Miller, C. L., J. H. Lee, E. Kieff, A. L. Burkhardt, J. B. Bolen, and R. Longnecker. 1994. Epstein-Barr virus protein LMP2A regulates reactivation from latency by negatively regulating tyrosine kinases involved in sIg-mediated signal transduction. Infect. Agents Dis. 3:128-136. [PubMed] [Google Scholar]
- 24.Miller, C. L., J. H. Lee, E. Kieff, and R. Longnecker. 1994. An integral membrane protein (LMP2) blocks reactivation of Epstein-Barr virus from latency following surface immunoglobulin crosslinking. Proc. Natl. Acad. Sci. U. S. A. 91:772-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller, C. L., R. Longnecker, and E. Kieff. 1993. Epstein-Barr virus latent membrane protein 2A blocks calcium mobilization in B lymphocytes. J. Virol. 67:3087-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakamura, H., M. Lu, Y. Gwack, J. Souvlis, S. L. Zeichner, and J. U. Jung. 2003. Global changes in Kaposi's sarcoma-associated virus gene expression patterns following expression of a tetracycline-inducible Rta transactivator. J. Virol. 77:4205-4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neipel, F., J. C. Albrecht, A. Ensser, Y. Q. Huang, J. J. Li, A. E. Friedman-Kien, and B. Fleckenstein. 1997. Human herpesvirus 8 encodes a homolog of interleukin-6. J. Virol. 71:839-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicholas, J. 2003. Human herpesvirus-8-encoded signalling ligands and receptors. J. Biomed. Sci. 10:475-489. [DOI] [PubMed] [Google Scholar]
- 29.Parravicini, C., B. Chandran, M. Corbellino, E. Berti, M. Paulli, P. S. Moore, and Y. Chang. 2000. Differential viral protein expression in Kaposi's sarcoma-associated herpesvirus-infected diseases: Kaposi's sarcoma, primary effusion lymphoma, and multicentric Castleman's disease. Am. J. Pathol. 156:743-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Polson, A. G., D. Wang, J. DeRisi, and D. Ganem. 2002. Modulation of host gene expression by the constitutively active G protein-coupled receptor of Kaposi's sarcoma-associated herpesvirus. Cancer Res. 62:4525-4530. [PubMed] [Google Scholar]
- 31.Poole, L. J., J. C. Zong, D. M. Ciufo, D. J. Alcendor, J. S. Cannon, R. Ambinder, J. M. Orenstein, M. S. Reitz, and G. S. Hayward. 1999. Comparison of genetic variability at multiple loci across the genomes of the major subtypes of Kaposi's sarcoma-associated herpesvirus reveals evidence for recombination and for two distinct types of open reading frame K15 alleles at the right-hand end. J. Virol. 73:6646-6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. U. S. A. 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saksela, K., G. Cheng, and D. Baltimore. 1995. Proline-rich (PxxP) motifs in HIV-1 Nef bind to SH3 domains of a subset of Src kinases and are required for the enhanced growth of Nef+ viruses but not for down-regulation of CD4. EMBO J. 14:484-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schulz, T. F. 2001. KSHV/HHV8-associated lymphoproliferations in the AIDS setting. Eur. J. Cancer 37:1217-1226. [DOI] [PubMed] [Google Scholar]
- 35.Sharp, T. V., H. W. Wang, A. Koumi, D. Hollyman, Y. Endo, H. Ye, M. Q. Du, and C. Boshoff. 2002. K15 protein of Kaposi's sarcoma-associated herpesvirus is latently expressed and binds to HAX-1, a protein with antiapoptotic function. J. Virol. 76:802-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shepard, L. W., M. Yang, P. Xie, D. D. Browning, T. Voyno-Yasenetskaya, T. Kozasa, and R. D. Ye. 2001. Constitutive activation of NF-kappa B and secretion of interleukin-8 induced by the G protein-coupled receptor of Kaposi's sarcoma-associated herpesvirus involve G alpha(13) and RhoA. J. Biol. Chem. 276:45979-45987. [DOI] [PubMed] [Google Scholar]
- 37.Shugars, D. C., M. S. Smith, D. H. Glueck, P. V. Nantermet, F. Seillier-Moiseiwitsch, and R. Swanstrom. 1993. Analysis of human immunodeficiency virus type 1 nef gene sequences present in vivo. J. Virol. 67:4639-4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang, L., M. M. Brinkmann, M. Pietrek, M. Ottinger, O. Dittrich-Breiholz, M. Kracht, and T. F. Schulz. 2007. Functional characterization of the M-type K15-encoded membrane protein of Kaposi's sarcoma-associated herpesvirus. J. Gen. Virol. 88:1698-1707. [DOI] [PubMed] [Google Scholar]
- 39.Reference deleted.
- 40.Zoeteweij, J. P., A. V. Moses, A. S. Rinderknecht, D. A. Davis, W. W. Overwijk, R. Yarchoan, J. M. Orenstein, and A. Blauvelt. 2001. Targeted inhibition of calcineurin signaling blocks calcium-dependent reactivation of Kaposi sarcoma-associated herpesvirus. Blood 97:2374-2380. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.