Abstract
Respiratory syncytial virus (RSV) is a major cause of virus-induced respiratory disease and hospitalization in infants. Palivizumab, an RSV-neutralizing monoclonal antibody, is used clinically to prevent serious RSV-related respiratory disease in high-risk infants. Motavizumab, an affinity-optimized version of palivizumab, was developed to improve protection against RSV. These antibodies bind RSV F protein, which plays a role in virus attachment and mediates fusion. Determining how these antibodies neutralize RSV is important to help guide development of new antibody drugs against RSV and, potentially, other viruses. This study aims to uncover the mechanism(s) by which palivizumab and motavizumab neutralize RSV. Assays were developed to test the effects of these antibodies at distinct steps during RSV replication. Pretreatment of virus with palivizumab or motavizumab did not inhibit virus attachment or the ability of F protein to interact with the target cell membrane. However, pretreatment of virus with either of these antibodies resulted in the absence of detectable viral transcription. These results show that palivizumab and motavizumab act at a point after F protein initiates interaction with the cell membrane and before virus transcription. Palivizumab and motavizumab also inhibited F protein-mediated cell-to-cell fusion. Therefore, these results strongly suggest that these antibodies block both cell-to-cell and virus-to-cell fusion, since these processes are likely similar. Finally, palivizumab and motavizumab did not reduce viral budding. Based on models developed from numerous studies of viral fusion proteins, our results indicate that these antibodies may prevent conformational changes in F protein required for the fusion process.
Respiratory syncytial virus (RSV) is classified into the subfamily Pneumovirinae within the Paramyxoviridae family of enveloped, single-stranded, and negative-sense RNA viruses (13). RSV infection can result in severe lower respiratory tract disease requiring hospitalization. Populations considered at high risk for developing severe RSV respiratory disease include premature infants and infants with chronic lung or congenital heart diseases (34). However, most patients hospitalized for RSV infection are healthy infants with no known risk factors (7). In addition to the potential development of RSV-induced respiratory disease upon acute infection, a history of RSV infection alone or in conjunction with other risk factors may predispose infants to chronic wheezing or asthma later in life, as reviewed by Hansbro et al. (21). RSV is highly prevalent, with yearly epidemics lasting five to 7 months and generally spanning the winter season of a particular region (13). Over half of all children are seropositive by 1 year of age, and nearly all children are seropositive by 2 years of age (41). Despite the presence of anti-RSV antibodies in the population, lifelong reinfection is a hallmark of RSV (13, 18).
RSV is considered an important target for antiviral development. Unfortunately, previous vaccine attempts have failed to elicit a long-lived protective immune response, and there is currently no approved vaccine against RSV (11). Treatment for RSV infection is limited to ribavirin, a nonspecific antiviral that interferes with virus transcription (30, 42). However, side effects associated with the use of ribavirin and historical debate surrounding its efficacy illustrate the need for more potent and safe therapeutics to treat RSV infection (30, 42). Palivizumab, a humanized monoclonal antibody, is approved for immunoprophylactic use to prevent RSV-induced respiratory tract disease in high-risk infants (27, 52). Motavizumab, an affinity-optimized monoclonal antibody developed from palivizumab, has been evaluated clinically (43, 51, 52). Preclinical in vitro studies show that palivizumab and motavizumab neutralize RSV replication in cell culture when virus is pretreated with either of these antibodies (27, 51). In addition, palivizumab reduces virus replication in cell culture when added after infection commences (27); such an effect for motavizumab has not been published yet. Preclinical in vivo studies show that prophylaxis with palivizumab or motavizumab reduces RSV replication in the lower respiratory tracts of cotton rats (27, 51). Furthermore, motavizumab reduces RSV replication in the upper respiratory tracts of cotton rats (51). Finally, clinical trials show prophylaxis with palivizumab or motavizumab helps reduce RSV-related hospitalizations of at risk infants (43, 50).
The precise molecular mechanisms of action of palivizumab or motavizumab are not known. Understanding the mechanism of action of these antibodies at a molecular level might guide development of better inhibitors of RSV F protein, as well as inhibitors of other similar viral fusion proteins (10, 28, 31, 35, 40, 44, 57). Palivizumab and motavizumab bind to the antigenic A site of the F protein (4), a glycoprotein found on the surface of RSV (13). The F protein participates in viral attachment (48) and mediates the process of fusion between the virus and cell membranes, as well as between infected cell membranes, resulting in syncytium formation (13). Therefore, it is most likely that one or more of these steps in RSV replication is inhibited by palivizumab and motavizumab. A study by Osiowy and Anderson shows that convalescent human serum contains antibodies that inhibit RSV attachment; however, the virus protein targeted by these antibodies was not determined and may have been the result of either anti-F protein, anti-G protein, or anti-SH protein antibodies (37). Small molecule inhibitors of RSV F protein have been shown to inhibit both attachment and fusion (23, 39) or fusion alone (12, 15). Therefore, assays were designed to individually test whether palivizumab or motavizumab inhibit virus attachment, virus-to-cell fusion, and cell-to-cell fusion. In addition to its role in virus attachment and fusion, F protein is one of the minimally required RSV proteins necessary to produce viral particles (49). Although there is no precedent for an antibody to inhibit virus budding, it is conceivable that these antibodies could inhibit budding, since both palivizumab (53) and motavizumab (data not published) bind F protein on the surfaces of infected cells. Therefore, an assay was designed to test this possibility as well.
We examined the step(s) in virus replication affected by palivizumab and motavizumab. The results show that palivizumab or motavizumab did not inhibit virus attachment or F protein interaction with the cell membrane; however, these antibodies did inhibit virus transcription. In addition, palivizumab and motavizumab inhibited cell-to-cell fusion but did not affect virus budding. These results suggest that palivizumab and motavizumab block fusion, most likely through inhibiting the conformational changes in F protein required for this process.
MATERIALS AND METHODS
Cells and viruses.
HEp-2 cells were maintained in minimal essential medium (MEM) supplemented with 5% heat-inactivated fetal bovine serum (FBS), 2 mM l-glutamine, 100 U of penicillin/ml, and 100 μg of streptomycin/ml (all from Invitrogen). Vero cells were maintained in MEM supplemented with 10% heat-inactivated FBS, 2 mM l-glutamine, 100 U of penicillin/ml, and 100 μg of streptomycin/ml. TREx-F is a 293-derived cell line that maintains stable, tetracycline-inducible expression of F protein created with the Flp-In TREx system available from Invitrogen. TREx-F cells were maintained on poly-lysine-coated flasks and plates (Becton Dickson) in Dulbecco modified Eagle medium containing high glucose and 2 mM glutamate (Invitrogen) supplemented with 10% tetracycline-approved FBS (Clontech), 100 U of penicillin/ml, 100 μg of streptomycin/ml, 15 μg of blasticidin (Invitrogen)/ml, and 150 μg of hygromycin (Invitrogen)/ml. For the induction of F protein expression, 15 μg of tetracycline (Teknova)/ml was added to the medium. All cultures were grown in 5% CO2 at 37°C.
TREx-F cells were modified to constitutively express either a green or red fluorescence protein. Cells, plated 1 day prior at 106 cells/well in six-well plates, were transfected with either ZsGreen-C1 or AsRed-C1 expression vectors (Clontech) by using 4 μg of DNA and 5 μl of Lipofectamine2000 (Invitrogen) according to Invitrogen's protocol. Transfected cells were placed under 500 μg of Geneticin (Invitrogen)/ml selection for the presence of the vector. Cells that survived selection were pooled and cloned by limiting dilution and fluorescence-activated cell sorting on an Aria II instrument (BD Biosciences). The resulting cell lines, TREx-F green and TREx-F red, were maintained in TRex-F growth medium containing 500 μg of Geneticin/ml.
The RSV A Long strain was propagated in HEp-2 cells by infecting a freshly prepared confluent monolayer of HEp-2 cells at a multiplicity of infection (MOI) of 0.2. Virus was harvested at 48 h postinfection by sonicating supernatants and scraped cells twice at 50% power, followed by centrifugation to pellet the cell debris. An equal volume of 50% sucrose in water solution (wt/vol) was added to the virus supernatant, divided into aliquots, flash frozen in a dry ice-ethanol bath, and stored at −80°C.
Recombinant RSV B 9320 and 9320ΔG (RSV B and RSV BΔG) was constructed and rescued in a manner similar to that previously reported for recombinant RSV A2 (26). Antigenomic cDNA spanning the entire RSV 9320 genome was assembled by sequential ligation of six cDNA fragments generated from reverse transcriptase (RT) and PCR of RNA isolated from supernatants of cells infected with RSV B 9320 using Pfu polymerase (Stratagene) and subcloned into the pET vector (Novagen), modified to contain unique restriction enzyme sites (XmaI, AvrII, SacI, BamHI, and NotI). To facilitate the cloning process, several modifications of restriction sites were performed without changing the corresponding amino acid sequences. SacI sites at nucleotides (nt) 2305, 10371, and 14946 and the BamHI site at nt 11684 were removed; a SacI site was introduced at nt 4498. In addition, a single C-to-G nucleotide change at the fourth position of the leader sequence was also obtained by mutagenesis. Hepatitis delta virus ribozyme (RBZ) and T7 RNA polymerase terminator sequences were amplified by PCR from RSV A2 antigenomic cDNA (26) and ligated to the trailer sequence of the RSV B 9320 antigenome. To construct 9320 antigenomic cDNA that lacked G protein, deletion mutagenesis was performed on the subcloned fragment that encoded the G protein by using an ExSite PCR-based site-directed mutagenesis kit (Stratagene) and cloned in place of the fragment that contained G protein in the antigenomic cDNA containing plasmid. To recover 9320 viruses by reverse genetics, the N, P, and L genes of 9320 were cloned into pCITE vector (Novagen) under the control of T7 RNA polymerase promoter. HEp-2 cells were infected with MVA-T7 (provided by Bernard Moss and amplified in CEK cells) at an MOI of 5.0 and transfected with 0.4 μg of pN, 0.4 μg of pP, 0.2 μg of pL, and 0.8 μg of plasmids encoding 9320 or 9320ΔG antigenomic cDNA by Lipofectamine 2000 (Invitrogen). Three days after transfection, culture supernatants were used to infect Vero cells to amplify recovered viruses. Six days after infection, culture supernatants were harvested, and virus-infected cells were identified by immunostaining with polyclonal anti-RSV A2 serum (Biogenesis). Recombinant viruses from culture supernatants were plaque purified and amplified in Vero cells. Stocks of both RSV B and RSV BΔG were propagated in Vero cells in a manner similar to that described for RSV A Long, except the virus was grown at 35°C for 3 to 4 days or until 70% or more of the cells exhibited a cytopathic effect.
Attachment assay.
The attachment assay used in the present study was adapted from the immunofluorescence assay of Budge et al. (9) and modified into a 96-well enzyme-linked immunosorbent assay (ELISA) format. The virus was mixed with HEp-2 medium containing palivizumab, motavizumab, or heparin (Sigma), a known inhibitor of virus attachment (20), in a 96-well plate, followed by incubation for 2 h at 37°C and then cooled for 1 h at 4°C. Working on ice, medium was removed from precooled HEp-2 cells, plated at 2 × 104 cells/well in a 96-well plate 48 h prior, and replaced by the virus mixtures. The cells and virus mixtures were incubated for 2 h at 4°C, which allowed for virus attachment but prevented fusion (45). Working on ice, the cells were washed three times with chilled phosphate-buffered saline (PBS). The cells were fixed with chilled 3.7% formalin in PBS for 15 min at 4°C, followed by one manual wash with chilled PBS prior to ELISA for detection of attached virus. Samples were blocked with 5% bovine serum albumin (BSA) in PBS for 1 h at 37°C. Mouse anti-RSV F protein (Chemicon catalogue no. 858-1) diluted 1:6,400 in a solution of 0.1% Tween 20 in PBS (PBST) containing 5% BSA and incubated at 37°C for 1 h following the block. The wells were manually washed two times with PBST containing 5% BSA and two times with PBST only, allowing 5 min for each wash with shaking to decrease background effects. A 100-μl portion of biotinylated goat anti-mouse IgG (Southern Biotech) diluted 1:10,000 in PBST was incubated at 37°C for 1 h. After a washing step with PBST using a plate washer, SA-HRP (Amersham-GE) diluted 1:80,000 in PBST was added to each well, followed by incubation at 37°C for 1 h. After a washing step with PBST using a plate washer, TMB microwell peroxidase substrate (KPL) was added to the wells, followed by incubation at room temperature for 20 min. The reaction was stopped with 2 N H2SO4, and the absorbance at 450 nm was read with a Spectromax 340PC plate reader (Molecular Devices). The assay was performed with RSV A Long, RSV B, and RSV BΔG at an MOI of 5, 0.02, and 0.16, respectively. The lower MOIs for the RSV B experiments were due to low titers of these virus stocks.
Lipid mixing assay.
A lipid mixing assay modified from several previous studies (36, 39, 45) was performed using RSV labeled with R18, a lipophilic dye. Fresh virus supernatant from infected HEp-2 cells was harvested as described earlier with the exception that sonicated supernatants were clarified by two consecutive centrifugation steps at 2,885 × g for 20 min. R18 (Invitrogen), a lipophilic dye that is self-quenching at saturating concentrations, was solubilized in ethanol and added to the virus supernatant at a final concentration of 10 μM. This concentration was determined to be the maximum amount of R18 that could be used to label virus without significant loss of infectivity. The virus and R18 mixture was rocked gently for 1 h at room temperature. To remove excess R18 and labeled debris, the labeled virus was purified once over a sucrose gradient. The labeled virus was pelleted by centrifugation at 8,000 × g at 4°C overnight. The virus pellet was resuspended in 21 ml of Opti-MEM containing 50 mM HEPES and 100 mM MgSO4 and then layered onto 10 ml of a 30% sucrose solution (wt/vol) containing 50 mM HEPES, 100 mM MgSO4, and 150 mM NaCl, followed by 4 ml of 60% sucrose solution containing 50 mM HEPES, 100 mM MgSO4, and 150 mM NaCl. The sucrose gradient was spun in an ultracentrifuge (Beckman Coulter) using an SW28 rotor for 100 min at 25,000 rpm (average, 82,705 × g) at 4°C. The virus band at the interphase between the 30 and 60% sucrose solutions was collected, and an equal volume of Opti-MEM containing 50 mM HEPES and 100 mM MgSO4 was added so that the virus stocks contained ≤25% sucrose, which is standard. The labeled virus was divided into aliquots, frozen in a dry ice-ethanol bath, and stored at −80°C. Labeled mock stocks were generated from uninfected HEp-2 cells in 10 T150 flasks in the same manner. A band at the interphase between the 30 and 60% sucrose solutions was evident in the sucrose gradient preparation of the labeled mock stock, indicating some excess R18 and/or R18 associated with other cell contents were copurified with virus. Therefore, the labeled mock preparation accounts for the background amount of fluorescence due to excess dye or copurified cellular content associated with R18.
To detect virus-to-cell fusion, purified labeled virus was mixed with medium containing palivizumab, motavizumab, or heparin and incubated at 37°C for 2 h. The virus mixtures were added to HEp-2 cells plated the day before at 2.5 × 105 cell/well in a 24-well plate at an MOI of 0.02. Infections were carried out at room temperature or 4°C with gentle rocking for 1 h. The infections performed at 4°C were washed at this point. For all other infections, the inoculum was replaced by growth medium containing palivizumab, motavizumab, or heparin, and the cells were incubated for 3 h at 37°C to allow for complete virus-to-cell fusion (48). The cells were treated with trypsin, washed, and prepared for flow cytometric analysis in PBS containing 0.5% BSA and 5 mM EDTA. Flow cytometry was performed on an LSR II instrument (BD Biosciences) using FACSDiva software (BD Biosciences). Fluorescence detected in the phycoerythrin channel was collected, and the data analyzed by using FlowJo software (Tree Star, Inc.).
Assay for detection of virus transcription.
One million PFU of RSV and inhibitors were incubated at 37°C for 2 h. HEp-2 cells, seeded the day before at 106 cells/well in a six-well plate, were infected with the virus mixtures at room temperature for 1 h with gentle rocking. Inoculum was removed, and growth medium containing inhibitors was added to the cells. Infections carried out at 4°C, as described earlier, were also included in the experiment. At 6 and 24 h postinfection (hpi), the cells were washed three times with of PBS, RLT from the Qiagen RNeasy Plus kit was added, and the samples were frozen at −80°C until total RNA was purified from the samples by using the Qiagen RNeasy Plus kit. These time points were chosen to observe an increase in newly synthesized viral transcripts over the course of infection. To specifically target transcripts in a two-step quantitative PCR, the first-strand synthesis was carried out using the adapter primer, AP, from the 3′RACE (3′ rapid amplification of cDNA ends) kit available from Invitrogen. This reaction was carried out by using the AccuScript high fidelity kit from Stratagene according to the manufacturer's protocol with ∼1 μg of RNA. A relative quantitative PCR for RSV NS1 gene transcript was achieved using a forward primer specific for NS1 (5′-CACAACAATGCCAGTGCTACAA-3′) and the abridged universal adapter primer, AUAP, from the 3′RACE kit as the reverse primer. Reactions were set up by using the SYBR green core reagents kit (Applied Biosystems) with 2.5 μl of cDNA, 2 mM MgCl2, and 10 pmol of each primer in a 25-μl reaction. Only a single band was detected by agarose gel analysis of reactions performed as described previously (data not shown). For normalization, a quantitative PCR was carried out simultaneously for GAPDH (glyceraldehyde-3-phosphate dehydrogenase) using an “assay-on-demand” primer-probe set available from Applied Biosystems, a TaqMan universal PCR master mix (Applied Biosystems), and 2.5 μl of cDNA in a 25-μl reaction. Reactions were run on an Applied Biosystems 7900HT instrument on fast mode using a thermal cycling profile that included a 10-min initial denaturation step at 95°C, followed by 40 cycles of a 10-s denaturation step at 95°C and a 30-s annealing and polymerization step at 67°C. All reactions were run in duplicate, and data were collected analyzed using SDS software version 2.2.2 (Applied Biosystems).
Cell-to-cell fusion assay.
A cell-to-cell fusion assay was developed much like other assays that have been reported for RSV (8, 16, 19), as well as for HIV (24). TREx-F green and red cells were plated together at 5 × 105 each/well in a six-well plate. After 24 h, the medium was replaced by medium containing palivizumab or motavizumab and tetracycline, to induce F protein expression and syncytium formation. Cultures were analyzed 55 h postinduction by fluorescence microscopy for visualization of syncytium formation. This time point was chosen because the maximum amount of double positive cells was observed at 55 h postinduction in preliminary time course experiments (data not shown). All images were acquired by using a Nikon TE2000-E microscope (Nikon Instruments, Inc.) with a 10× NA 0.25 objective. The system was equipped with a Cool Snap ES2 camera (Photometrics) driven by NIS-Elements AR 3.0 (Nikon). A Nikon B-2 E/C fluorescein isothiocyanate (FITC) filter with an excitation of 465 to 495 nm was used for visualizing green fluorescence and a Nikon G-2E/C filter with an excitation of 528 to 553 nm was used for visualizing the red fluorescence. These images were enhanced by altering the contrast, brightness, and CMY color curves using Adobe Photoshop, and these alterations were applied equally to all images. Cells were released from the plate using nonenzymatic cell dissociation buffer (Invitrogen) and then washed and prepared for flow cytometric analysis in PBS containing 5 mM EDTA and 0.5% BSA. Flow cytometry was performed on an LSR II instrument (BD) using FACSDiva software (BD). The samples were gated on forward scatter and side scatter to include only intact cells and eliminate debris. The remaining cells were analyzed for fluorescence detection in the FITC and phycoerythrin (PE) channels, followed by data analysis with FlowJo software (Tree Star, Inc.).
Assay to detect virus budding.
A PCR approach to detect genomes in the supernatants of virus-infected cultures was developed. HEp-2 cells were infected at an MOI of ∼0.5 with RSV Long at 37°C for 1 h with gentle rocking. The inoculum was removed, and growth medium was added to the cells. The medium was replaced at 6 hpi by medium containing palivizumab, motavizumab, or ribavirin. Infections were further incubated for 21 to 23 more hours, until syncytia were present, but minimal cell lysis had occurred. Budded virus was collected as supernatants, which were centrifuged to remove debris, divided into aliquots, and frozen at −80°C. Supernatants were thawed, and RNA was purified from 140 μl by using a QiaAMP viral RNA kit (Qiagen) with an elution volume of 50 μl. A cDNA synthesis was carried using the AccuScript high-fidelity kit from Stratagene according to the manufacturer's protocol for random primers amplification with 5 μl of purified RNA. A relative quantitative PCR for RSV genome and antigenome was achieved with a primer and probe set designed and manufactured by Applied Biosystems that targets the intergenic region of SH and G (forward primer 5′-GCAAACCACCATCCATACTATAAAGTAGT-3′; reverse primer, 5′-TTTGCATTTGCCCCAACGTTATT-3′; FAM-labeled probe, 5′-ATGAACTAGGATATCAAGACTAAC-3′). The intergenic region between the SH and G genes was chosen because it experiences a relatively low level of readthrough phenomena (22). In addition, a quantitative PCR for GAPDH was included by using an “assay-on-demand” primer and probe set from Applied Biosystems. The GAPDH quantitative PCR serves as the endogenous control for cell lysis, resulting in the release of viral transcripts that were not actually budded. The reactions for both genome and GAPDH included the primer and probe sets with TaqMan universal PCR master mix (Applied Biosystems) and 4 μl of cDNA in 20-μl reactions and were carried out simultaneously. The quantitative PCR was run on an Applied Biosystems 7900HT instrument with a thermal cycling profile consisting of an initial 10-min denaturation step at 95°C, followed by 40 cycles of a 10-s denaturation step at 95°C and a 30-s annealing and polymerization step at 60°C. All reactions were run in duplicate, and the data collected were analyzed for relative quantitation using SDS software version 2.2.2 (Applied Biosystems).
RESULTS
Attachment of RSV to target cells is not inhibited by pretreatment with palivizumab or motavizumab.
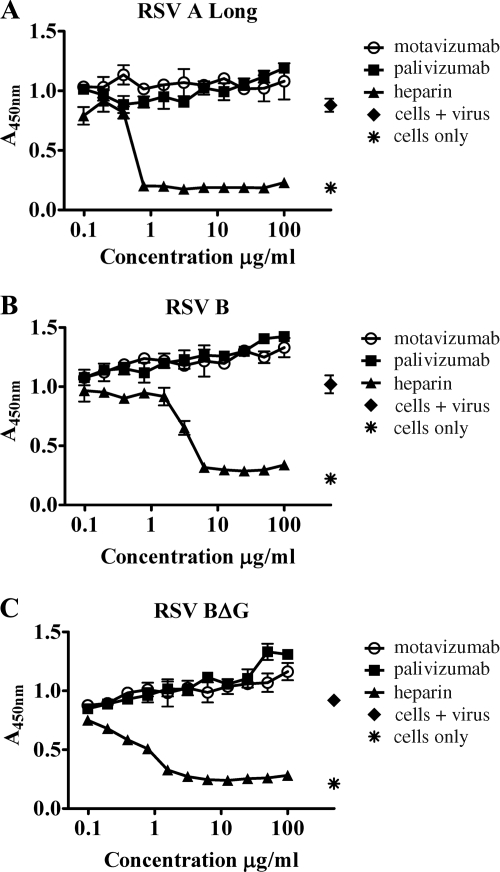
The stages of RSV replication potentially affected by neutralizing antibodies directed against F protein are most likely those that occur on the outside of the cell, such as virus attachment, virus-to-cell fusion, cell-to-cell fusion, and virus budding. Therefore, we first tested whether palivizumab or motavizumab can inhibit virus attachment. To this end, the immunofluorescence assay of Budge et al. (9) was adapted into an ELISA format. This assay was performed at 4°C, which allows virus attachment but prohibits virus-to-cell fusion (45). HEp-2 cells were used as the target for attachment since this cell line is permissive for RSV replication. Heparin, a known inhibitor of virus attachment (20), was included as a control. Antibodies and heparin were titrated from 100 μg/ml down to 0.1 μg/ml (Fig. 1). This assay was first performed with RSV A Long (Fig. 1A). As expected, heparin inhibited virus attachment in a dose-dependent manner (Fig. 1A). However, neither motavizumab nor palivizumab inhibited virus attachment, even at concentrations 2 to 3 logs above their reported respective 50% inhibitory concentrations (IC50s), 0.02 and 0.363 μg/ml (51) (Fig. 1A).
FIG. 1.
Attachment of RSV to HEp-2 cells is not inhibited by palivizumab or motavizumab. Attachment assays were performed with RSV A Long (A), RSV B (B), and RSV BΔG (C) pretreated with palivizumab, motavizumab, or heparin. The dose-response curves are plotted to show the absorbance values obtained, indicating the presence of virus, at each concentration of antibody tested. The plots shown are from representative experiments, and the standard deviations (SD) are indicated.
Three glycoproteins are found on the surface of RSV and include the F protein, the G protein, and the SH protein (13). RSV attachment is primarily mediated by the G protein; however, recombinant viruses lacking G protein or both SH and G proteins are capable of binding and infecting target cells, indicating that F protein alone is capable of specific attachment (48). Alternatively, it is possible that a host transmembrane protein the virus acquires during the budding process mediates attachment in the absence of G protein; however, there is no precedent for this in the literature. It should be noted that the SH protein does not appear to contribute to binding to target cells in vitro (48). Therefore, it was possible that these antibodies could inhibit F protein-mediated attachment, and yet specific attachment could still occur through the G protein. To address this possibility, this assay was performed with recombinant RSV B viruses with (Fig. 1B) and without G protein (Fig. 1C). Similar results were observed with these viruses as were observed with RSV A Long, indicating that neither motavizumab nor palivizumab inhibit virus attachment.
Transfer of lipids from RSV to target cells is not inhibited by pretreatment with palivizumab or motavizumab.
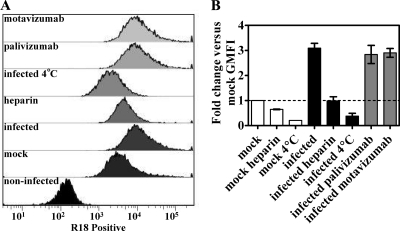
A lipid-mixing assay to assess virus-to-cell fusion was modified from several reports (36, 39, 45) and included a labeled mock control to account for non-virus-associated events. In this assay, RSV A Long was labeled with R18, a self-quenching lipophilic dye. When the labeled virus fuses with cells, R18 diffuses within the cellular plasma membrane, relieving the self-quenching and resulting in a fluorescent signal that can be detected by flow cytometry. In contrast to previous reports (36, 39, 45), we found that removal of non-virus-associated R18 within the labeled virus stock to be a problem. This issue was revealed by comparison of infections with virus and mock supernatants labeled with R18 (data not shown). Purification of labeled virus and mock stocks on a sucrose gradient was a purification method we found that resulted in a greater signal with virus infection versus mock infection (Fig. 2). Mock infection resulted in a significant shift in the fluorescent signal detected by flow cytometry compared to uninfected cells, as shown in representative histograms (Fig. 2A). However, the geometric mean fluorescence intensity (GMFI) for virus infection was 3-fold greater than the GMFI for mock infection (Fig. 2B). The GMFI for virus infection was reduced by heparin, an attachment inhibitor (20), and by infection carried out at 4°C, which allows attachment but not fusion (45) (Fig. 2), as expected. The GMFI for infections treated with heparin or carried out at 4°C was similar to mock infections treated with heparin or carried out at 4°C (Fig. 2), indicating that these inhibitors reduced the transfer of R18 from virus to cells to background levels. In addition, these controls show that the same amount of background fluorescence was detected in both mock and virus stocks, which indicates that a similar amount of lipids were added with the mock and virus infections. Infections with virus treated with palivizumab or motavizumab at 100 μg/ml (2 to 3 logs above their respective IC50s [51]) resulted in a GMFI equal to infections with untreated virus (Fig. 2), indicating the transfer of R18 from virus to HEp-2 cells was not inhibited by these antibodies.
FIG. 2.
Transfer of R18 dye from RSV to HEp-2 cells is not inhibited by palivizumab or motavizumab. Lipid-mixing assays were performed with purified R18-labeled RSV A Long pretreated with neutralizing concentrations of palivizumab, motavizumab, or heparin. (A) Infected cells were prepared for flow cytometry, as described earlier, and analyzed by flow cytometry. The analysis included a gating step for size to exclude debris. Representative histograms are shown, and the x axis denotes the mean fluorescence intensity. (B) The fold change in geometric mean fluorescence intensity (GMFI) is plotted relative to mock-infected samples. The graph represents duplicate (4°C infections only) or triplicate experiments, along with the standard errors of the mean.
Virus transcription is inhibited by pretreatment with palivizumab or motavizumab.
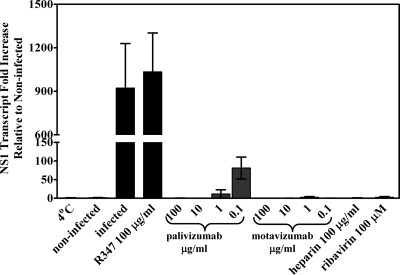
To further assess virus entry, a PCR approach designed to exclusively detect transcript and not to detect genome and antigenome was developed. For this assay, total RNA was purified 6 and 24 hpi from HEp-2 cells infected with RSV A Long (MOI of 0.5) pretreated with either palivizumab or motavizumab. In addition, total RNA was purified from infections performed at 4°C and 6 and 24 hpi from control infections with virus pretreated with heparin, ribavirin, or R347, a negative control antibody that does not bind RSV (1). Purified RNAs were converted into cDNA and analyzed by quantitative PCR for the abundance of NS1 gene transcripts using a 3′RACE approach. This approach relies on the use of a first-strand synthesis primer consisting of oligo(dT), which recognizes polyadenylated transcripts, flanked on the 5′ end by an adaptor primer sequence. This adaptor primer sequence is recognized in the second strand synthesis by a complementary primer, and with the addition of a gene-specific primer, the amplification of specific transcripts is accomplished. The NS1 transcript is the most abundant transcript found in infected cells (14), which lends greater sensitivity to PCR detection of newly synthesized transcripts. As expected, 350-fold more NS1 transcripts were detected at 24 hpi compared to 6 hpi (data not shown). The results of the virus transcription assay are plotted as the fold increase relative to uninfected HEp-2 cells at 24 hpi (Fig. 3). Viral transcripts in RNA isolated from HEp-2 cells infected with RSV A Long at 4°C, where virus was attached but not fused, were not detected in 40 cycles of PCR. As expected, and in agreement with published data, little to no NS1 transcripts were detected in RNA isolated from cells infected with virus pretreated with heparin and from cells infected with virus pretreated with ribavirin (Fig. 3). Infections with virus alone or virus pretreated with control antibody R347 (1) resulted in an ∼1,000-fold increase in NS1 transcripts over uninfected cultures (Fig. 3). However, little to no NS1 transcripts were detected in RNA isolated from cells infected with virus pretreated with palivizumab at 10 and 100 μg/ml and with motavizumab at all concentrations ranging from 0.1 to 100 μg/ml (Fig. 3). These results, taken together with the results of the attachment assay and lipid-mixing assay, indicate that palivizumab and motavizumab inhibit a step in virus replication that occurs after initial F protein interaction with the target cell membrane and before virus transcription. In addition, these data correlate to previous reports that show motavizumab is more potent than palivizumab (51).
FIG. 3.
Virus transcription is absent in the presence of neutralizing amounts of palivizumab and motavizumab. A quantitative PCR for RSV NS1 transcripts normalized to GAPDH transcripts was performed on cDNA generated from total cellular RNA of cells infected with RSV A Long pretreated with palivizumab, motavizumab, heparin, or ribavirin. The fold change in NS1 transcripts is plotted relative to uninfected controls. The graph represents triplicate data points, along with the SD.
RSV F protein mediated cell-to-cell fusion is inhibited by palivizumab and motavizumab.
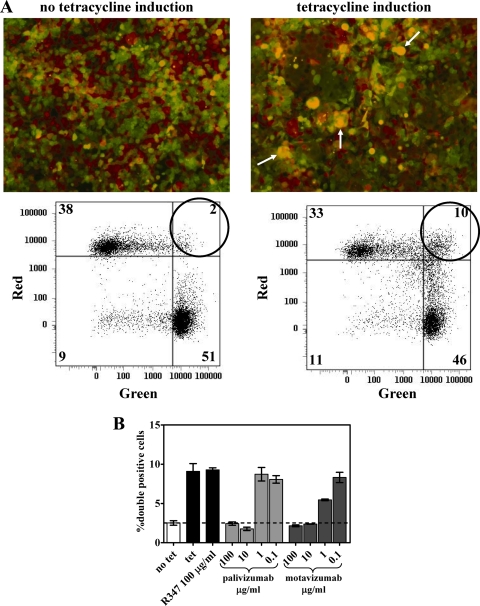
In order to better assess the process of fusion by F protein, a quantitative assay to measure F protein mediated cell-to-cell fusion was developed in a manner similar to that described in previous reports for RSV and other viruses (8, 16, 19, 24). The TREx-F cell line was created, which maintains tetracycline-inducible expression of RSV F protein. This cell line was modified by stable transfection with vectors that encode either a green fluorescent protein or a red fluorescent protein to produce the cell lines TREx-F green and TREx-F red. Tetracycline induction of F protein expression led to cell-to-cell fusion in TRex-F red and green cell coculture within 50 h. Fluorescence microscopic examination showed syncytium formation containing both green and red fluorescent proteins, appearing as yellow in merged images, in the presence of tetracycline, but not in TRex-F red and green cell coculture that did not receive tetracycline treatment (Fig. 4 A). Cell-to-cell fusion was quantitated by flow cytometry, measured as cells double positive for both green and red fluorescent proteins. A 5-fold increase in the amount of fused, double-positive cells was detected between cultures with or without tetracycline treatment (Fig. 4B). This assay was also performed using a TREx-CAT (cells transfected with a control vector that encodes the CAT gene) green cell line at 1:1, 2:1, 5:1, and 10:1 ratios with the TREx-F red cell line. This configuration of the assay resulted in 50% fewer cell-to-cell fusion events detected at the 1:1 ratio and, as the ratio increased, the amount of fusion events decreased to undetectable (data not shown). These results indicate that a 1:1 coculture of the TREx-F green and TREx-F red cell lines is the optimal configuration for this assay. Titrated amounts of palivizumab or motavizumab were added to the cultures at the same time tetracycline was added to determine whether these antibodies inhibit cell-to-cell fusion. Flow cytometric analysis revealed both palivizumab and motavizumab reduced the amount of fused, double-positive cells to background levels in a dose-dependent manner (Fig. 4B). These results indicate palivizumab and motavizumab inhibit F protein-mediated fusion. Again, motavizumab had greater potency than palivizumab in this assay, as previously reported (51).
FIG. 4.
RSV-mediated cell-to-cell fusion is inhibited by palivizumab and motavizumab. TREx-F green and TREx-F red were cocultured in equal proportion, followed by induction of F protein expression with tetracycline (tet). The cultures were treated with palivizumab or motavizumab at the time of tetracycline addition. The cultures were analyzed 55 h posttreatment by fluorescence microscopy and flow cytometry. (A) Representative images and corresponding dot plots obtained through flow cytometric analysis are shown. (B) The plots show the average percentage of fused, double-positive cells detected by flow cytometry for each treatment condition. The dashed line indicates the level of background double-positive cells observed in these cultures. The plot represents duplicate data points, along with the SD.
Neither palivizumab nor motavizumab inhibits virus budding.
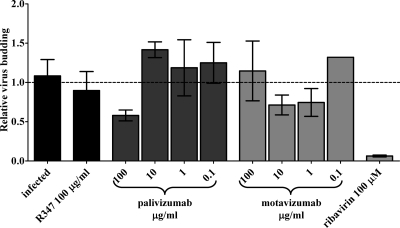
Although not a classically defined mechanism of viral neutralization by antibodies (28), virus budding is another possible step in RSV replication that these antibodies may impact. Palivizumab (53) and motavizumab (data not published) bind to F protein on the surface of RSV-infected cells. In addition, F protein is one of the minimally required RSV-encoded proteins necessary to produce infectious virus particles (49). Therefore, binding of antibodies to F protein may interfere with the budding process. An assay was developed to test this possibility. In this assay, HEp-2 cells were infected with RSV A Long at an MOI of 0.5 for 1 h, at which point the inoculum was replaced with normal growth medium. Treatments were added to the infections 6 hpi, a time point at which virus fusion is complete and viral transcription is initiating as previously shown (6, 48). Supernatants were collected at 27 to 29 hpi, before extensive cell lysis was apparent based on microscopic examination of the infected, untreated cultures. As determined earlier in the present study, the presence of palivizumab or motavizumab prevents virus fusion, and therefore any assay that relies on infection, such as a plaque assay, was prohibited by the presence of these antibodies. Therefore, a quantitative PCR approach was developed to detect genome and antigenome, both of which are encapsidated and presumably found in budded virus particles (38), but not viral gene transcripts. There was a significant difference in the extent of syncytia formation between antibody treated and untreated infections in these cultures (data not shown), which was most likely due to the inhibition of fusion by palivizumab and motavizumab. This was a concern in the development of this assay, since this observed difference in syncytium formation, and the extent of cell lysis, which seems to occur simultaneously, could result in a difference in the release of nucleic acids, including viral nucleic acids, into the supernatant that are not associated with virus budding and potentially skew the results. To account for this, the assay was performed as a relative quantitation for RSV genome and antigenome normalized to the amount of GAPDH in the reaction. It should be noted that GAPDH transcripts were more abundant in the supernatants of infected cells compared to the supernatants of infected cells treated with motavizumab or palivizumab (data not shown). This observation suggests that even at these early stages of syncytium formation, a certain level of cell lysis does occur. As expected, a 17-fold reduction in cell-free virus was observed with this assay in the supernatants of infected cultures treated with 100 μM ribavirin (twice its IC50) (56) (Fig. 5). However, there was only a 1.7-fold reduction in cell-free virus, and no dose-dependent effect was observed in infected cultures treated at 6 hpi with palivizumab or motavizumab (Fig. 5). Therefore, these antibodies do not appear to inhibit virus budding; however, it is possible that a specific type of viral particle morphology that is strictly cell associated may be blocked by these antibodies and are of interest for future studies.
FIG. 5.
Palivizumab and motavizumab do not inhibit virus budding. A quantitative PCR for RSV genome and antigenome normalized to GAPDH was performed on cDNA generated from viral RNA in the supernatants of cells infected with RSV A Long and treated with palivizumab, motavizumab, R347, or ribavirin at 6 hpi. The fold change in the RSV genome and antigenome is plotted relative to infected controls. The graph represents triplicate data points, along with the SD.
DISCUSSION
The objective of this study was to determine the mechanism of action of palivizumab and motavizumab on specific step(s) in RSV replication. The classically defined mechanisms by which antibodies neutralize virus include blocking virus attachment or inhibiting virus entry (29). In the present study, the attachment assay performed with RSV and RSVΔG showed palivizumab and motavizumab did not inhibit RSV attachment to target cells in the presence or absence of G protein (Fig. 1). A lipid-mixing assay was chosen to measure RSV fusion with target cells, since this assay has been used to measure fusion for a variety of enveloped viruses including vesicular stomatitis virus, influenza, Sendai virus, human immunodeficiency virus, mumps virus, vaccinia virus, Epstein-Barr virus, and respiratory syncytial virus as reviewed by Blumenthal et al. (5). The results of the lipid-mixing assay showed that neither palivizumab nor motavizumab prevented the transfer of R18 from virus to target cells during infection, indicating that lipid mixing occurred (Fig. 2). These results were surprising, since virus attachment was not blocked by these antibodies and it was anticipated that a block in virus fusion would be detected. However, detection of lipid mixing may only indicate that the process of fusion initiated and may not account for complete virus fusion based on current modeling of the fusion process as reviewed previously (29). Although these results indicate that palivizumab and motavizumab permit the transfer of R18 from virus to target cell, it was still possible that these antibodies may block completion of virus fusion. Therefore, it was important to test whether these antibodies block virus entry.
An assay to detect newly synthesized viral nucleic acids was developed to further assess whether palivizumab or motavizumab block virus entry. The results of the virus transcription assay showed virus pretreated with neutralizing concentrations of palivizumab or motavizumab (51) reduced NS1 transcripts in a dose-dependent manner (down to undetectable levels) (Fig. 3). These results, in addition to the results of the virus attachment assay and the lipid-mixing assay, indicate palivizumab and motavizumab block a step in virus replication after virus attachment and before virus transcription. This is in contrast to a previous report showing that viral transcripts were detected in cells when virus is pretreated with up to 600 μg of palivizumab/ml (6). However, differences between how the assays were performed most likely explain the contrasting results. In the study by Boukhvalova et al. (6), pretreated virus was allowed to adsorb to the cells for 1 h, the inoculum was removed, the cells were washed twice with PBS, and medium that did not contain palivizumab was added to the cells for the remainder of the infection (M. Boukhvalova, unpublished data). In the present study, pretreated virus was allowed to adsorb to the cells for 1 h, the inoculum was removed, and medium containing antibody was added to the cells for the remainder of the infection. The washes and addition of medium that did not contain palivizumab in the Boukhvalova et al. study (6) most likely reduced the concentration of the antibody below its effective concentration. Similar to the work by Boukhvalova et al. (6), we observed loss of neutralization when virus pretreated with neutralizing amounts of palivizumab or motavizumab was subsequently diluted in medium that does not contain antibody and used to infect cells (data not shown).
A cell-to-cell fusion assay was developed as another approach to examine fusion. The results of the cell-to-cell fusion assay showed that palivizumab and motavizumab inhibit cell-to-cell fusion in a dose-dependent manner (Fig. 4B). In addition, motavizumab maintained a greater potency than palivizumab in this assay (Fig. 4B), which is consistent with previous reports (51). The only difference between virus-to-cell fusion and cell-to-cell fusion reported to date is that RhoA signaling is required for F protein-mediated cell-to-cell fusion but dispensable for virus-to-cell fusion (19). It appears that cell-to-cell fusion most likely proceeds in a manner similar to that of virus-to-cell fusion, and therefore we conclude that these results also suggest that the block to virus entry is virus-to-cell fusion.
The results of the present study illustrate that, aside from a difference in potency, both palivizumab and motavizumab neutralize RSV by blocking virus-to-cell and cell-to-cell fusion but do not affect virus attachment or virus budding. Inhibition of virus entry, either through blocking attachment or blocking fusion, is a common mechanism of action reported for virus-neutralizing antibodies, as previously reviewed (29). In addition, neutralizing antibodies to similar viral fusion proteins such as influenza virus and HIV also block fusion (2, 3, 17, 25, 32, 47). Another common mechanism of action reported for virus-neutralizing antibodies is through Fc-related functions of the antibody (29). However, it remains to be determined whether the Fc portion of the antibody also contributes to virus neutralization. Finally, the stoichiometry of antibody and virus required for neutralization seems to play a role in the mechanism of action for virus-neutralizing antibodies (29). Future studies are needed to address the stoichiometric requirements for neutralization of RSV by motavizumab and palivizumab, which are anticipated to be complicated by the pleiomorphic nature of RSV virions (13).
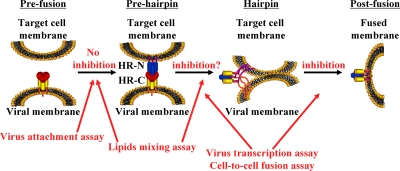
When considered in the context of an adapted model for F protein-mediated fusion (28), which is based on the proposal that conformational changes in the F protein fuel this process, as reviewed by Lamb and Jardetzky (28) and as suggested by structural data for the RSV F protein (10, 31, 35, 40, 44, 57), the results of the attachment assay and lipid-mixing assay indicate that the transition from prefusion to prehairpin conformations of F protein is not inhibited by palivizumab or motavizumab and that these antibodies block a conformational change in F protein beyond this stage of fusion (Fig. 6). The lipid-mixing assay may indicate that the hairpin conformation, which is the point at which lipid mixing occurs, is achieved in the presence of these antibodies and that palivizumab and motavizumab block the transition from a hairpin conformation to a postfusion conformation. However, several reports suggest that R18 can passively transfer from virus to target cell by shear proximity of the membranes (5, 36, 46, 54, 55). Therefore, it is possible that R18 may transfer from virus to target cell upon the initial interaction of the F protein with the target cell membrane or the prehairpin conformation. Further studies are needed to examine whether these antibodies block the transition from prehairpin to hairpin conformations or between hairpin and postfusion conformations, which could also add to the understanding of what drives F protein-mediated fusion. In this context, these results are consistent with the conclusion, drawn from the recently reported crystal structure of motavizumab bound to an epitope-derived peptide, that motavizumab prevents the conformational changes of F protein required for fusion (33). Overall, the results of our study help to shed light on the mechanism by which palivizumab and motavizumab neutralize RSV.
FIG. 6.
Model for the proposed mechanism of action of palivizumab and motavizumab. The diagram shows proposed progression of the conformational changes in F protein that are proposed to fuel the process of fusion (28). The attachment assay results showed that palivizumab and motavizumab did not inhibit the transition from the prefusion to prehairpin conformation. The lipid-mixing assay results suggest that palivizumab and motavizumab do not inhibit the transition to either the prehairpin or hairpin conformations, as described in the text. The virus transcription assay and the cell-to-cell fusion assay showed that palivizumab and motavizumab inhibited fusion at either the transition from the prehairpin-to-hairpin conformation or the hairpin-to-postfusion conformation. HR-N and HR-C denote the F protein N-terminal and C-terminal heptad repeats, respectively.
Acknowledgments
We thank Bernard Moss for providing MVA-T7 and Randall MacGill, JoAnn Suzich, Mike McCarthy, Andriani Patera, and Subramaniam Krishnan for critical reviews of the manuscript.
This study was supported by MedImmune.
Footnotes
Published ahead of print on 2 June 2010.
REFERENCES
- 1.Bachelder, R. E., J. Bilancieri, W. Lin, and N. L. Letvin. 1995. A human recombinant Fab identifies a human immunodeficiency virus type 1-induced conformational change in cell surface-expressed CD4. J. Virol. 69:5734-5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbas, C. F., III, E. Bjorling, F. Chiodi, N. Dunlop, D. Cababa, T. M. Jones, S. L. Zebedee, M. A. Persson, P. L. Nara, and E. Norrby. 1992. Recombinant human Fab fragments neutralize human type 1 immunodeficiency virus in vitro. Proc. Natl. Acad. Sci. U. S. A. 89:9339-9343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbato, G., E. Bianchi, P. Ingallinella, W. H. Hurni, M. D. Miller, G. Ciliberto, R. Cortese, R. Bazzo, J. W. Shiver, and A. Pessi. 2003. Structural analysis of the epitope of the anti-HIV antibody 2F5 sheds light into its mechanism of neutralization and HIV fusion. J. Mol. Biol. 330:1101-1115. [DOI] [PubMed] [Google Scholar]
- 4.Beeler, J. A., and K. van Wyke Coelingh. 1989. Neutralization epitopes of the F glycoprotein of respiratory syncytial virus: effect of mutation upon fusion function. J. Virol. 63:2941-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blumenthal, R., S. A. Gallo, M. Viard, Y. Raviv, and A. Puri. 2002. Fluorescent lipid probes in the study of viral membrane fusion. Chem. Phys. Lipids 116:39-55. [DOI] [PubMed] [Google Scholar]
- 6.Boukhvalova, M. S., G. A. Prince, and J. C. Blanco. 2007. Respiratory syncytial virus infects and abortively replicates in the lungs in spite of preexisting immunity. J. Virol. 81:9443-9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyce, T. G., B. G. Mellen, E. F. Mitchel, Jr., P. F. Wright, and M. R. Griffin. 2000. Rates of hospitalization for respiratory syncytial virus infection among children in Medicaid. J. Pediatr. 137:865-870. [DOI] [PubMed] [Google Scholar]
- 8.Branigan, P. J., C. Liu, N. D. Day, L. L. Gutshall, R. T. Sarisky, and A. M. Del Vecchio. 2005. Use of a novel cell-based fusion reporter assay to explore the host range of human respiratory syncytial virus F protein. Virol. J. 2:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Budge, P. J., Y. Li, J. A. Beeler, and B. S. Graham. 2004. RhoA-derived peptide dimers share mechanistic properties with other polyanionic inhibitors of respiratory syncytial virus (RSV), including disruption of viral attachment and dependence on RSV G. J. Virol. 78:5015-5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calder, L. J., L. Gonzalez-Reyes, B. Garcia-Barreno, S. A. Wharton, J. J. Skehel, D. C. Wiley, and J. A. Melero. 2000. Electron microscopy of the human respiratory syncytial virus fusion protein and complexes that it forms with monoclonal antibodies. Virology 271:122-131. [DOI] [PubMed] [Google Scholar]
- 11.Castilow, E. M., and S. M. Varga. 2008. Overcoming T cell-mediated immunopathology to achieve safe RSV vaccination. Future Virol. 3:445-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cianci, C., N. Meanwell, and M. Krystal. 2005. Antiviral activity and molecular mechanism of an orally active respiratory syncytial virus fusion inhibitor. J. Antimicrob. Chemother. 55:289-292. [DOI] [PubMed] [Google Scholar]
- 13.Collins, P. L., and J. E. Crowe, Jr. 2007. Respiratory syncytial virus and metapneumovirus, p. 1601-1645. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed., vol. II. Lippincott/The Williams & Wilkins Co., Philadelphia, PA. [Google Scholar]
- 14.Collins, P. L., and G. W. Wertz. 1983. cDNA cloning and transcriptional mapping of nine polyadenylylated RNAs encoded by the genome of human respiratory syncytial virus. Proc. Natl. Acad. Sci. U. S. A. 80:3208-3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Douglas, J. L., M. L. Panis, E. Ho, K. Y. Lin, S. H. Krawczyk, D. M. Grant, R. Cai, S. Swaminathan, X. Chen, and T. Cihlar. 2005. Small molecules VP-14637 and JNJ-2408068 inhibit respiratory syncytial virus fusion by similar mechanisms. Antimicrob. Agents Chemother. 49:2460-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Douglas, J. L., M. L. Panis, E. Ho, K. Y. Lin, S. H. Krawczyk, D. M. Grant, R. Cai, S. Swaminathan, and T. Cihlar. 2003. Inhibition of respiratory syncytial virus fusion by the small molecule VP-14637 via specific interactions with F protein. J. Virol. 77:5054-5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edwards, M. J., and N. J. Dimmock. 2000. Two influenza A virus-specific Fabs neutralize by inhibiting virus attachment to target cells, while neutralization by their IgGs is complex and occurs simultaneously through fusion inhibition and attachment inhibition. Virology 278:423-435. [DOI] [PubMed] [Google Scholar]
- 18.Glezen, W. P., L. H. Taber, A. L. Frank, and J. A. Kasel. 1986. Risk of primary infection and reinfection with respiratory syncytial virus. Am. J. Dis. Child. 140:543-546. [DOI] [PubMed] [Google Scholar]
- 19.Gower, T. L., M. K. Pastey, M. E. Peeples, P. L. Collins, L. H. McCurdy, T. K. Hart, A. Guth, T. R. Johnson, and B. S. Graham. 2005. RhoA signaling is required for respiratory syncytial virus-induced syncytium formation and filamentous virion morphology. J. Virol. 79:5326-5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hallak, L. K., P. L. Collins, W. Knudson, and M. E. Peeples. 2000. Iduronic acid-containing glycosaminoglycans on target cells are required for efficient respiratory syncytial virus infection. Virology 271:264-275. [DOI] [PubMed] [Google Scholar]
- 21.Hansbro, N. G., J. C. Horvat, P. A. Wark, and P. M. Hansbro. 2008. Understanding the mechanisms of viral induced asthma: new therapeutic directions. Pharmacol. Ther. 117:313-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hardy, R. W., S. B. Harmon, and G. W. Wertz. 1999. Diverse gene junctions of respiratory syncytial virus modulate the efficiency of transcription termination and respond differently to M2-mediated antitermination. J. Virol. 73:170-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hosoya, M., J. Balzarini, S. Shigeta, and E. De Clercq. 1991. Differential inhibitory effects of sulfated polysaccharides and polymers on the replication of various myxoviruses and retroviruses, depending on the composition of the target amino acid sequences of the viral envelope glycoproteins. Antimicrob. Agents Chemother. 35:2515-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huerta, L., E. Lamoyi, A. Baez-Saldana, and C. Larralde. 2002. Human immunodeficiency virus envelope-dependent cell-cell fusion: a quantitative fluorescence cytometric assay. Cytometry 47:100-106. [DOI] [PubMed] [Google Scholar]
- 25.Imai, M., K. Sugimoto, K. Okazaki, and H. Kida. 1998. Fusion of influenza virus with the endosomal membrane is inhibited by monoclonal antibodies to defined epitopes on the hemagglutinin. Virus Res. 53:129-139. [DOI] [PubMed] [Google Scholar]
- 26.Jin, H., D. Clarke, H. Z. Zhou, X. Cheng, K. Coelingh, M. Bryant, and S. Li. 1998. Recombinant human respiratory syncytial virus (RSV) from cDNA and construction of subgroup A and B chimeric RSV. Virology 251:206-214. [DOI] [PubMed] [Google Scholar]
- 27.Johnson, S., C. Oliver, G. A. Prince, V. G. Hemming, D. S. Pfarr, S. C. Wang, M. Dormitzer, J. O'Grady, S. Koenig, J. K. Tamura, R. Woods, G. Bansal, D. Couchenour, E. Tsao, W. C. Hall, and J. F. Young. 1997. Development of a humanized monoclonal antibody (MEDI-493) with potent in vitro and in vivo activity against respiratory syncytial virus. J. Infect. Dis. 176:1215-1224. [DOI] [PubMed] [Google Scholar]
- 28.Lamb, R. A., and T. S. Jardetzky. 2007. Structural basis of viral invasion: lessons from paramyxovirus F. Curr. Opin. Struct. Biol. 17:427-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Law, M., and L. Hangartner. 2008. Antibodies against viruses: passive and active immunization. Curr. Opin. Immunol. 20:486-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leyssen, P., E. De Clercq, and J. Neyts. 2008. Molecular strategies to inhibit the replication of RNA viruses. Antivir. Res. 78:9-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lopez, J. A., R. Bustos, C. Orvell, M. Berois, J. Arbiza, B. Garcia-Barreno, and J. A. Melero. 1998. Antigenic structure of human respiratory syncytial virus fusion glycoprotein. J. Virol. 72:6922-6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McInerney, T. L., L. McLain, S. J. Armstrong, and N. J. Dimmock. 1997. A human IgG1 (b12) specific for the CD4 binding site of HIV-1 neutralizes by inhibiting the virus fusion entry process, but b12 Fab neutralizes by inhibiting a postfusion event. Virology 233:313-326. [DOI] [PubMed] [Google Scholar]
- 33.McLellan, J. S., M. Chen, A. Kim, Y. Yang, B. S. Graham, and P. D. Kwong. 2010. Structural basis of respiratory syncytial virus neutralization by motavizumab. Nat. Struct. Mol. Biol. 17:248-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meissner, H. C., M. B. Rennels, L. K. Pickering, and C. B. Hall. 2004. Risk of severe respiratory syncytial virus disease, identification of high risk infants and recommendations for prophylaxis with palivizumab. Pediatr. Infect. Dis. J. 23:284-285. [DOI] [PubMed] [Google Scholar]
- 35.Morton, C. J., R. Cameron, L. J. Lawrence, B. Lin, M. Lowe, A. Luttick, A. Mason, J. McKimm-Breschkin, M. W. Parker, J. Ryan, M. Smout, J. Sullivan, S. P. Tucker, and P. R. Young. 2003. Structural characterization of respiratory syncytial virus fusion inhibitor escape mutants: homology model of the F protein and a syncytium formation assay. Virology 311:275-288. [DOI] [PubMed] [Google Scholar]
- 36.Ohki, S., J. Z. Liu, J. Schaller, and R. C. Welliver. 2003. The compound DATEM inhibits respiratory syncytial virus fusion activity with epithelial cells. Antivir. Res. 58:115-124. [DOI] [PubMed] [Google Scholar]
- 37.Osiowy, C., and R. Anderson. 1995. Neutralization of respiratory syncytial virus after cell attachment. J. Virol. 69:1271-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peeples, M. E., and P. L. Collins. 2000. Mutations in the 5′ trailer region of a respiratory syncytial virus minigenome which limit RNA replication to one step. J. Virol. 74:146-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Razinkov, V., A. Gazumyan, A. Nikitenko, G. Ellestad, and G. Krishnamurthy. 2001. RFI-641 inhibits entry of respiratory syncytial virus via interactions with fusion protein. Chem. Biol. 8:645-659. [DOI] [PubMed] [Google Scholar]
- 40.Ruiz-Arguello, M. B., D. Martin, S. A. Wharton, L. J. Calder, S. R. Martin, O. Cano, M. Calero, B. Garcia-Barreno, J. J. Skehel, and J. A. Melero. 2004. Thermostability of the human respiratory syncytial virus fusion protein before and after activation: implications for the membrane-fusion mechanism. J. Gen. Virol. 85:3677-3687. [DOI] [PubMed] [Google Scholar]
- 41.Shay, D. K., R. C. Holman, R. D. Newman, L. L. Liu, J. W. Stout, and L. J. Anderson. 1999. Bronchiolitis-associated hospitalizations among US children, 1980-1996. JAMA 282:1440-1446. [DOI] [PubMed] [Google Scholar]
- 42.Sidwell, R. W., and D. L. Barnard. 2006. Respiratory syncytial virus infections: recent prospects for control. Antivir. Res. 71:379-390. [DOI] [PubMed] [Google Scholar]
- 43.Simoes, E., X. Carbonell, G. Losonsky, M. Hultquist, B. Harris, and E. Connor. 2007. Phase III trial of motavizumab, an enhanced potency respiratory syncytial virus (RSV)-specific monoclonal antibody for the prevention of serious RSV disease in high risk infants. Abstr. 48th Annu. Meet. Eur. Soc. Paediatr. Res.
- 44.Smith, B. J., M. C. Lawrence, and P. M. Colman. 2002. Modelling the structure of the fusion protein from human respiratory syncytial virus. Protein Eng. 15:365-371. [DOI] [PubMed] [Google Scholar]
- 45.Srinivasakumar, N., P. L. Ogra, and T. D. Flanagan. 1991. Characteristics of fusion of respiratory syncytial virus with HEp-2 cells as measured by R18 fluorescence dequenching assay. J. Virol. 65:4063-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stegmann, T., P. Schoen, R. Bron, J. Wey, I. Bartoldus, A. Ortiz, J. L. Nieva, and J. Wilschut. 1993. Evaluation of viral membrane fusion assays. Comparison of the octadecylrhodamine dequenching assay with the pyrene excimer assay. Biochemistry 32:11330-11337. [DOI] [PubMed] [Google Scholar]
- 47.Sui, J., W. C. Hwang, S. Perez, G. Wei, D. Aird, L. M. Chen, E. Santelli, B. Stec, G. Cadwell, M. Ali, H. Wan, A. Murakami, A. Yammanuru, T. Han, N. J. Cox, L. A. Bankston, R. O. Donis, R. C. Liddington, and W. A. Marasco. 2009. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct. Mol. Biol. 16:265-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Techaarpornkul, S., N. Barretto, and M. E. Peeples. 2001. Functional analysis of recombinant respiratory syncytial virus deletion mutants lacking the small hydrophobic and/or attachment glycoprotein gene. J. Virol. 75:6825-6834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Teng, M. N., and P. L. Collins. 1998. Identification of the respiratory syncytial virus proteins required for formation and passage of helper-dependent infectious particles. J. Virol. 72:5707-5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.The IMpact-RSV Study Group. 1998. Palivizumab, a humanized respiratory syncytial virus monoclonal antibody, reduces hospitalization from respiratory syncytial virus infection in high-risk infants. Pediatrics 102:531-537. [PubMed] [Google Scholar]
- 51.Wu, H., D. S. Pfarr, S. Johnson, Y. A. Brewah, R. M. Woods, N. K. Patel, W. I. White, J. F. Young, and P. A. Kiener. 2007. Development of motavizumab, an ultra-potent antibody for the prevention of respiratory syncytial virus infection in the upper and lower respiratory tract. J. Mol. Biol. 368:652-665. [DOI] [PubMed] [Google Scholar]
- 52.Wu, H., D. S. Pfarr, G. A. Losonsky, and P. A. Kiener. 2008. Immunoprophylaxis of RSV infection: advancing from RSV-IGIV to palivizumab and motavizumab. Curr. Top. Microbiol. Immunol. 317:103-123. [DOI] [PubMed] [Google Scholar]
- 53.Wu, H., D. S. Pfarr, Y. Tang, L. L. An, N. K. Patel, J. D. Watkins, W. D. Huse, P. A. Kiener, and J. F. Young. 2005. Ultra-potent antibodies against respiratory syncytial virus: effects of binding kinetics and binding valence on viral neutralization. J. Mol. Biol. 350:126-144. [DOI] [PubMed] [Google Scholar]
- 54.Wunderli-Allenspach, H., M. Gunthert, and S. Ott. 1993. Inactivation of PR8 influenza virus through the octadecylrhodamine B chloride membrane marker. Biochemistry 32:900-907. [DOI] [PubMed] [Google Scholar]
- 55.Wunderli-Allenspach, H., and S. Ott. 1990. Kinetics of fusion and lipid transfer between virus receptor containing liposomes and influenza viruses as measured with the octadecylrhodamine B chloride assay. Biochemistry 29:1990-1997. [DOI] [PubMed] [Google Scholar]
- 56.Xie, Y. Y., X. D. Zhao, L. P. Jiang, H. L. Liu, L. J. Wang, P. Fang, K. L. Shen, Z. D. Xie, Y. P. Wu, and X. Q. Yang. 2006. Inhibition of respiratory syncytial virus in cultured cells by nucleocapsid gene targeted deoxyribozyme (DNAzyme). Antivir. Res. 71:31-41. [DOI] [PubMed] [Google Scholar]
- 57.Zhao, X., M. Singh, V. N. Malashkevich, and P. S. Kim. 2000. Structural characterization of the human respiratory syncytial virus fusion protein core. Proc. Natl. Acad. Sci. U. S. A. 97:14172-14177. [DOI] [PMC free article] [PubMed] [Google Scholar]