Abstract
Overall, the time to AIDS after HIV-2 infection is longer than with HIV-1, and many individuals infected with HIV-2 virus remain healthy throughout their lives. Multiple HLA and KIR gene products have been implicated in the control of HIV-1, but the effect of variation at these loci on HIV-2 disease is unknown. We show here for the first time that HLA-B*1503 is associated significantly with poor prognosis after HIV-2 infection and that HLA-B*0801 is associated with susceptibility to infection. Interestingly, previous data indicate that HLA-B*1503 is associated with low viral loads in HIV-1 clade B infection but has no significant effect on viral load in clade C infection. In general, alleles strongly associated with HIV-1 disease showed no effect in HIV-2 disease. These data emphasize the unique nature of the effects of HLA and HLA/KIR combinations on HIV-2 immune responses relative to HIV-1, which could be related to their distinct clinical course.
Since the first report of this virus in 1986, HIV-2 remains largely confined to West Africa (11). It shares between 30 and 60% nucleotide and amino acid homology with HIV-1 but differs greatly in pathogenicity and transmissibility (20). Studies on HIV-2 patients across West Africa have shown that some people remain uninfected despite repeated exposure (36), and a substantial proportion of infected people remain relatively healthy for a very long time with low plasma viral load and normal CD4+ T-cell counts, a characteristic of long-term nonprogressors (LTNPs) infected with HIV-1 (37). This is perhaps a reflection of an effective immune response mounted against the virus, including a vigorous CD8+ T-cell response (28), maintenance of HIV-specific CD4+ T-cell function (15), and the presence of a strong neutralizing antibody response in many subjects (4), features that are highly desirable for a successful HIV-1 vaccine. Thus, HIV-2 disease course provides a natural model for investigating mechanisms that control HIV infection, and a better understanding of these mechanisms might inform new strategies for HIV prevention and treatment.
HLA class I molecules present antigenic epitopes to cytotoxic T cells and are central to the acquired immune response. A number of associations between HLA class I alleles and HIV disease outcomes have been reported (10), the most consistent being B*57 and B*27, which show strong protection across studies, and certain subtypes of B*35, which associate with more rapid progression (19). While several mother-infant studies have implicated sharing of certain HLA alleles in transmission of the virus from mother to infant (29, 30), there is no convincing data that particular HLA class I alleles protect against HIV infection in general.
HLA class I allotypes also serve as ligands for killer cell immunoglobulin-like receptors (KIRs), which modulate natural killer (NK) cell function. KIRs are structurally similar to one another and can be divided into activating and inhibitory receptors. NK cells are key components of the innate immune system and constantly survey host cell surfaces for appropriate levels of HLA class I molecules through a network of NK cell receptors, including KIRs (26). Upon engagement with their ligand, inhibitory KIR suppress NK cell activity, but if the ligand is missing or has been downregulated on target cells, the threshold for NK cell activation is lowered, thus allowing for activation signals to dominate (23).
HLA and KIR genes are found on chromosomes 6 and 19, respectively, so they segregate independently. As such, the genes/alleles for the corresponding receptor-ligand pair must be present to confer functionality, whereas presence of one without the other results in a null phenotype. A number of HLA and KIR gene products either individually or collectively has been implicated in the control of HIV-1 (9), but nothing is known of their role in HIV-2.
Epidemiological data from Caio and other cohorts in West Africa (3, 39) indicate that HIV-2 infection in a substantial proportion of infected individuals is compatible with normal survival and without signs of immunodeficiency, suggesting distinct viral pathogenic mechanisms and protective host factors against HIV-2 relative to HIV-1. Here, we determined the HLA class I and KIR gene profiles of the Caio population (>95% Manjako) from Guinea-Bissau and investigated their effects on susceptibility to HIV-2 infection and disease progression.
MATERIALS AND METHODS
Study populations.
The Medical Research Council (UK) unit in The Gambia has monitored for 2 decades an open cohort of HIV-2-infected, HIV-1-infected, and dually infected adults, and a similar number of age- and sex-matched uninfected controls in the rural village of Caio, Guinea-Bissau, a relatively isolated community of the Manjako tribe, an animist tribe in which marriages usually occur within the community. The Caio cohort was established in 1989 and a demographic census and sero-survey of the area was conducted in 1989 and 1990, in which 7.9% of the adult population was shown to be infected with HIV-2 (45). This was followed by a case-control study in 1991 (35), a resurvey in 1996 to 1998, and reexamination of the cases and controls in 1996 and 2003. Individuals who participated in the 1991 case-control study were recruited by matching cases and controls as stringently as possible for demographic factors such as age, sex, and location in the village. They are still being monitored to date, and the loss to follow-up is small in this cohort (<7% in both the infected and the uninfected groups). CD4+ T-cell counts and HIV-2 viral load measurements were obtained at four time points: in 1991, 1996, 2003, and 2006.
A total of 513 samples from HIV-2-infected and uninfected adults, predominantly of the Manjako tribe, were typed for HLA class I (HLA-A, -B, and -C) and KIR genes. Table 1 shows the characteristics of all of the study participants. There were more females than males in each category and females were also slightly older: this is consistent with previous epidemiological data from Africa, in which older women appear to be more susceptible to HIV infection in general and particularly to HIV-2 (1, 21). The median age difference between males and females singly infected with HIV-2 and their uninfected counterparts was statistically significant (P < 0.001), perhaps reflecting the fact that HIV-2 infection is decreasing across West Africa. Dually infected (HIV-1 and HIV-2) individuals were excluded from all HIV-2-related analyses because of the potential for HIV-1 to confound our results. Precise dates of seroconversion are unknown for most of the infected participants in our study. Although it is difficult to estimate HIV exposure in the control group accurately, previous anthropological studies in Caio strongly suggest that the unique traditional way of life of the Manjakos in Caio lead to most adults having been exposed to sexually transmitted diseases, including HIV, from multiple partners during their 4-year initiation period into adulthood (6, 40).
TABLE 1.
Characteristics of the study populationa
| Infection group | No. of subjects (%) | Median age in yr (IQR) |
|---|---|---|
| HIV negative | ||
| Male | 114 (35) | 36.2 (25.8-52.5) |
| Female | 213 (65) | 46.2 (30.8-62.8) |
| HIV-2 infected | ||
| Male | 49 (32) | 53.7 (43.0-62.9) |
| Female | 102 (68) | 59.5 (49.5-69.4) |
| HIV-1/HIV-2 dually infected | ||
| Male | 6 (17) | 37.0 (34.7-49.3) |
| Female | 29 (83) | 52.8 (42.8-61.7) |
n = 513, Caio, 2003 to 2007. IQR, interquartile range.
The study was approved by the Gambian Government/MRC joint Ethical Committee, the Open University's Life and Biomolecular Sciences Management Group, United Kingdom, and the Office of Human Subjects Research, National Institutes of Health.
KIR and HLA class I genotyping.
Genomic DNA was genotyped for presence or absence of the following KIR genes: 2DL1, 2DL2, 2DL3, 2DL4, 2DL5, 2DS1, 2DS2, 2DS3, 2DS4, 2DS5, 3DL1, 3DL2, 3DL3, 3DS1, and 2DP1. Genotyping was performed by using PCR amplification with two pairs of primers specific for each locus (PCR-SSP) as previously described (31). HLA class I genotyping was done by sequencing. Briefly, locus-specific primers flanking exons 2 and 3 were used to amplify HLA-A, -B, and -C loci. Purified PCR products were sequenced in both directions using exon-specific primers and a BigDye Terminator version 3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA) in an ABI-3130XL DNA analyzer (Applied Biosystems). Sequence traces were analyzed by using Assign 400 software (Conexio Genomics, Western Australia).
Statistical analysis.
Allele and genotype frequencies were calculated by using SPSS v16.0.1 (SPSS, Inc., Chicago, IL) and STATA v9.2 (Stata Corp., College Station, TX). Haplotype reconstruction, testing for Hardy-Weinberg equilibrium, and haplotype frequency estimations were performed by using Arlequin version 3.11. A χ2 test was used to examine differences in allele and genotype frequencies between Caio and other West African populations. All other statistical analyses were performed using SAS 9.1 (SAS Institute). PROC FREQ was used to compute frequencies of individual variables. PROC NPAR1WAY was used to test the median age difference between HIV-2-infected and uninfected subjects. PROC LOGISTIC was used for categorical analyses to obtain odds ratios and 95% confidence intervals. PROC GLM was used for analyzing continuous variables. Viral loads were transformed into log scale, and CD4 counts are shown as square root values in order to provide an approximate normal distribution. Both logistic and linear regression models were adjusted for age and gender.
RESULTS
HLA class I allele frequencies in Caio show significant differences compared to other West African populations.
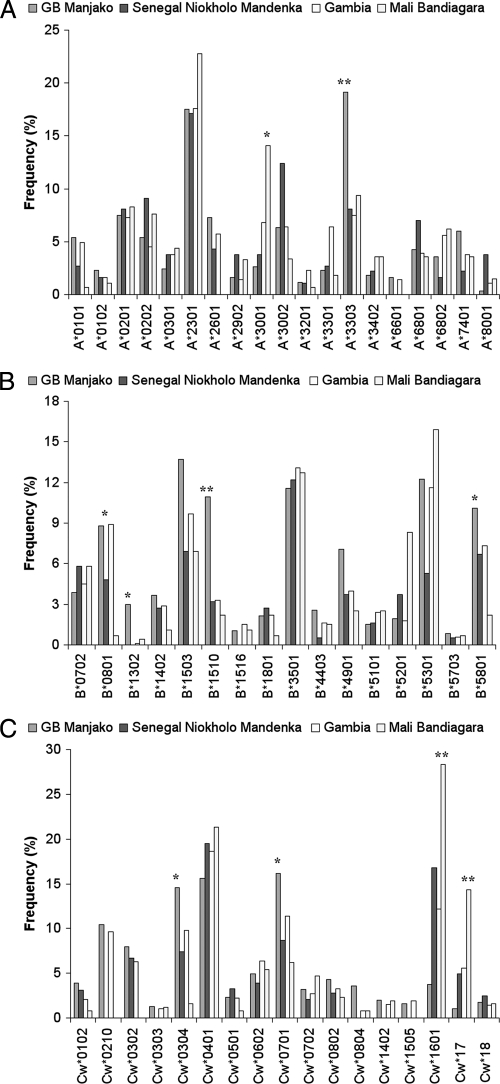
We first compared the frequencies of each HLA class I allele obtained from 327 HIV negative individuals from Caio with data from neighboring West African populations in Senegal (38), Mali (7), and The Gambia (L.-M. Yindom, unpublished data) (Fig. 1). Frequencies of HLA-A, -B, and -C alleles in Caio (n = 327) differed significantly for some alleles from those in neighboring countries, where HLA-A*3303 was more common and HLA-A*3001 was less common (Fig. 1A). HLA-B*0801, -B*1302, -B*1503, and -B*1510 were also relatively common in Caio as was HLA-B*5801, which has previously been shown to be protective against HIV-1 (41) (Fig. 1B). HLA-Cw*1601 and -Cw*17 allele frequencies were lower, whereas HLA-Cw*0304, -Cw*0701, and -Cw*0804 were higher in Caio than elsewhere (Fig. 1C).
FIG. 1.
HLA allele frequencies in Caio, Guinea Bissau, compared to frequencies in neighboring West African Countries. HLA-A (A), HLA-B (B), and HLA-C (C) allele frequencies were determined in Guinea-Bissau (327 samples) and the neighboring countries Senegal (165 samples), The Gambia (592 samples), and Mali (138 samples). GB Manjako, Guinea-Bissau Manjako HIV-uninfected individuals. Alleles with frequencies of <1% in all populations are not shown. *, P < 0.05; **, P < 0.001.
KIR genotypes in Caio contain more activating KIRs than reported in other African populations.
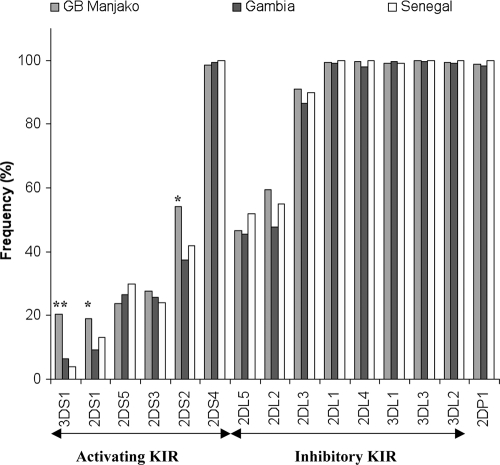
We also determined KIR gene frequencies in our samples and compared those from the HIV negative Caio control group to those from neighboring populations (Fig. 2). There were no significant differences in frequencies between the populations for any of the inhibitory KIR genes. On the other hand, activating receptors were present at higher frequencies in Caio compared to neighboring populations (Fig. 2). Interestingly, KIR3DS1, which is relatively rare in people of African ancestry, was not uncommon in Caio. KIR3DS1 has previously been shown to confer protection against rapid progression to AIDS after HIV-1 infection in combination with its putative ligand HLA-B Bw4-80I (32).
FIG. 2.
Frequencies of activating and inhibitory KIR genes in Caio, Guinea-Bissau, compared to those in The Gambia and Senegal. GB Manjako, Guinea-Bissau Manjako HIV-uninfected individuals. *, P < 0.05; **, P < 0.001.
HLA predicts HIV-2 disease progression and is associated with resistance to infection.
In order to determine whether certain HLA class I alleles could predict HIV-2 disease progression, we analyzed the CD4+ T-cell counts and viral load data from 136 HIV-2-infected individuals, who provided a blood sample in 2003, adjusting for age and gender. The overall mean square root (sqrt) CD4+ T-cell count was 24.65 (standard error [SE] = 0.57), and the mean log HIV-2 viral load was 3.06 (SE = 0.10). Analysis of HLA-B genotypes, CD4+ T-cell counts (n = 132) and viral load (n = 136) data from HIV-2-infected subjects revealed that HLA-B*15+ individuals had significantly lower mean sqrt CD4+ T-cell counts (22.72, SE = 0.89, P = 0.001) and higher mean log HIV-2 viral loads (3.57, SE = 0.15, P = 0.001) compared to those without this genotype (26.36 [SE = 0.72] and 2.94 [SE = 0.12], respectively) (Table 2). This association appeared to be due completely to the frequent B*1503 allele, which showed lower mean sqrt CD4+ T-cell counts (21.32, SE = 1.22, P = 0.001) and higher mean log HIV-2 viral loads (3.64, SE = 0.20, P = 0.010) compared to that of B*1503-negative individuals (25.88 [SE = 0.63] and 3.06 [SE = 0.11], respectively). No susceptibility effect was seen with the other common B*1510 allele. Not surprisingly, HLA-Cw*02, which is in linkage disequilibrium with B*15, also associated with lower mean sqrt CD4+ T-cell counts (20.49, SE = 1.47, P = 0.002) and trended toward high mean log HIV-2 viral loads. Other alleles weakly associated with markers of disease progression are shown in Table 2. Interestingly, most of the protective effects observed in the present study are mediated by relatively rare alleles (B*57 and B*14), as has also been observed for HIV-1 infection (44). However, given the limited numbers and weak effects in most of these analyses, further studies will be required to verify these associations.
TABLE 2.
Effects of HLA-B and HLA-C on CD4+ T-lymphocyte counts and viral load
| HLA allelea | CD4 |
HIV-2 VL |
||||
|---|---|---|---|---|---|---|
| Mean sqrt CD4 (n) | SE | P | Mean log HIV-2 VL (n) | SE | P | |
| B*14 (P) | 26.93 (14) | 1.70 | 0.220 | 2.56 (14) | 0.28 | 0.020 |
| B*15 (S) | 22.72 (51) | 0.89 | 0.001 | 3.57 (52) | 0.15 | 0.001 |
| B*1503 (S) | 21.32 (26) | 1.22 | 0.001 | 3.64 (28) | 0.20 | 0.010 |
| B*1510 | 24.91 (24) | 1.33 | 0.970 | 3.37 (25) | 0.22 | 0.330 |
| B*1516 | 19.94 (3) | 3.68 | 0.170 | 3.72 (2) | 0.77 | 0.480 |
| B*49 (S) | 21.45 (17) | 1.54 | 0.020 | 3.48 (20) | 0.24 | 0.180 |
| B*57 (P) | 30.00 (7) | 2.39 | 0.030 | 3.47 (7) | 0.41 | 0.470 |
| Cw*02 (S) | 20.49 (19) | 1.47 | 0.002 | 3.53 (20) | 0.24 | 0.170 |
(P), protective allele with either a mean sqrt CD4+ T-cell count of >24.7 (average mean sqrt CD4+ T-cell counts) or mean logVL of <3.1 (average mean HIV-2 VL) and a P value of <0.05; (S), susceptible allele with either a mean sqrt CD4+ T-cell count of <24.7 (average mean CD4+ T-cell counts) and/or mean logVL of >3.1 (average mean HIV-2 VL) and a P value of <0.05.
Next, we compared two-digit HLA class I genotype frequencies for alleles present in at least 5% of the study population, between cases and controls to examine the effect of HLA on HIV-2 acquisition (Table 3). All three loci were in Hardy-Weinberg equilibrium (P = 0.18517, 0.25241, and 0.10134 for HLA-A, -B, and -C loci, respectively). HLA-B*08 was more common in HIV-2-infected individuals than in controls (26.8% versus 14.8%), suggesting that it is associated with susceptibility to HIV-2 infection in this population (odds ratio [OR] = 2.27, P = 0.003). HLA-A*74, on the other hand, was associated with protection from infection (OR = 0.39, P = 0.02). However, due to the limited numbers of individuals being compared, these results will need to be substantiated in other cohorts. None of the HLA-C alleles were associated with susceptibility or resistance to HIV-2 infection.
TABLE 3.
HLA-A, -B, and -C genotypes and HIV-2 antibody status in the Caio communitya
| HLA allele | HIV-2+ (n) | HIV negative (n) | All (n) | P | OR | 95% CI |
|---|---|---|---|---|---|---|
| A*01 | 12.3 (18) | 14.0 (42) | 13.5 (60) | 0.58 | 0.84 | 0.45-1.56 |
| A*02 | 18.5 (27) | 24.3 (73) | 22.4 (100) | 0.37 | 0.79 | 0.47-1.32 |
| A*23 | 33.6 (49) | 31.3 (94) | 32.1 (143) | 0.69 | 1.09 | 0.70-1.70 |
| A*26 | 16.4 (24) | 13.7 (41) | 14.6 (65) | 0.28 | 1.37 | 0.77-2.44 |
| A*30 | 24.6 (36) | 17.3 (52) | 19.7 (88) | 0.25 | 1.34 | 0.20-2.22 |
| A*33 | 38.4 (56) | 39.0 (117) | 38.8 (173) | 0.98 | 1.01 | 0.66-1.54 |
| A*68 | 17.8 (26) | 14.3 (43) | 15.5 (69) | 0.32 | 1.33 | 0.76-2.32 |
| A*74 | 6.9 (10) | 12.3 (37) | 10.5 (47) | 0.02 | 0.39 | 0.18-0.83 |
| B*07 | 4.9 (7) | 7.0 (16) | 6.2 (23) | 0.30 | 0.61 | 0.24-1.55 |
| B*08 | 26.8 (38) | 14.8 (34) | 19.4 (72) | 0.003 | 2.27 | 1.32-3.90 |
| B*13 | 5.6 (8) | 5.7 (13) | 5.7 (21) | 0.80 | 1.13 | 0.44-2.89 |
| B*14 | 9.9 (14) | 8.3 (19) | 8.9 (33) | 0.75 | 1.13 | 0.53-2.39 |
| B*15 | 38.7 (55) | 45.2 (104) | 42.7 (159) | 0.11 | 0.69 | 0.44-1.08 |
| B*18 | 6.3 (9) | 4.4 (10) | 5.1 (19) | 0.64 | 1.26 | 0.48-3.33 |
| B*35 | 14.1 (20) | 21.3 (49) | 18.6 (69) | 0.10 | 0.61 | 0.34-1.11 |
| B*44 | 5.6 (8) | 5.2 (12) | 5.4 (20) | 0.79 | 1.14 | 0.43-2.98 |
| B*49 | 14.1 (20) | 13.5 (31) | 13.7 (51) | 0.88 | 0.95 | 0.51-1.79 |
| B*53 | 16.2 (23) | 23.9 (55) | 21.0 (78) | 0.09 | 0.62 | 0.35-1.08 |
| B*58 | 22.5 (32) | 18.7 (43) | 20.2 (75) | 0.19 | 1.43 | 0.84-2.46 |
| Cw*01 | 9.0 (13) | 6.9 (19) | 7.6 (32) | 0.34 | 1.45 | 0.67-3.11 |
| Cw*02 | 15.2 (22) | 21.1 (58) | 19.1 (80) | 0.17 | 0.68 | 0.39-1.19 |
| Cw*03 | 40.0 (58) | 44.7 (123) | 43.1 (181) | 0.46 | 0.85 | 0.56-1.30 |
| Cw*04 | 29.7 (43) | 30.2 (83) | 30.0 (126) | 0.51 | 0.86 | 0.54-1.36 |
| Cw*05 | 8.3 (12) | 4.4 (12) | 5.7 (24) | 0.37 | 1.49 | 0.62-3.58 |
| Cw*06 | 7.6 (11) | 9.8 (27) | 9.1 (38) | 0.64 | 0.83 | 0.39-1.78 |
| Cw*07 | 38.6 (56) | 33.1 (91) | 35.0 (147) | 0.10 | 1.46 | 0.94-2.27 |
| Cw*08 | 13.8 (20) | 15.3 (42) | 14.8 (62) | 0.61 | 0.86 | 0.47-1.56 |
| Cw*16 | 4.8 (7) | 7.3 (20) | 6.4 (27) | 0.23 | 0.56 | 0.22-1.43 |
“All” represents the percentage of individuals positive for the indicated allele in the community; n, the number of individuals carrying the allele of interest; P, P values uncorrected for multiple comparisons; OR, the odds ratio derived by comparing the number of individuals positive for the indicated allele versus those without the allele between case (HIV-2+) and control (HIV-negative) groups. P values and ORs were calculated by logistic regression, controlling for gender and age. Only alleles with an overall frequency of at least 5% are shown. P values of <0.05 are shown in bold.
We also estimated haplotype frequencies using Arlequin version 3.11 (16), and the most common two- and three-locus haplotypes were HLA-B*1503-Cw*0210 (9.8%), HLA-A*330301-B*1510-Cw*0304 (6.2%), and HLA-A*2301-B*1503-Cw*0210 (5.3%) (data not shown). None of the individual haplotypes had any significant influence on HIV-2 acquisition or disease progression.
HLA and KIR compound genotypes and resistance to HIV-2 infection.
KIR molecules are known to interact with their HLA class I ligands to modulate NK cell activity. The ligands for KIR2DL are HLA-C alleles, which are classified as C group 1 (C1) if the amino acid at position 80 is asparagine or C group 2 (C2) if lysine occupies that position. The inhibitory KIR2DL2 and 2DL3 (which are alleles of the same locus) recognize C1, whereas KIR2DL1 recognizes C2 allotypes. KIR3DL1 recognizes HLA-B Bw4 allotypes, particularly those with an isoleucine at position 80 (8). The activating receptors KIR2DS2, -2DS1, and -3DS1 share high sequence similarity in their extracellular domains with the corresponding inhibitory receptors KIR2DL2/3, -2DL1, and -3DL1, respectively. KIR2DS1 and -2DS2 appear to bind the same set of HLA class I ligands as their inhibitory counterparts, although with much lower affinity (43). Epidemiological data are consistent with a receptor-ligand relationship between KIR3DS1 and HLA-B Bw4-80I, but this has never been shown formally.
To determine the effect of HLA/KIR genotypes on susceptibility or resistance to HIV-2 infection, we grouped individuals based on whether they had appropriate ligands for their KIR genetic profile and compared the various HLA/KIR combinations between HIV-2-infected and uninfected groups. We also looked at HIV-2 viral load and CD4+ T-cell counts in the infected group. There was no significant effect of individual KIR genes on infection or markers of disease progression (data not shown). Individuals carrying either the activating KIR2DS2 or the inhibitory KIR2DL2 (these two KIR genes are in very strong linkage disequilibrium with one another), with at least one copy of their corresponding HLA ligands (C1), were more likely to be HIV-2 negative compared to those without these compound genotypes, suggesting that the compound genotypes 2DS2:HLA-C1/x and/or 2DL2:HLA-C1/x (where x could be C1 or C2) might protect against HIV-2 acquisition, although the differences were not statistically significant after adjusting for age and gender (OR = 0.72 [P = 0.15] and OR = 0.74 [P = 0.17], respectively; data not shown). We emphasize that these findings should be interpreted cautiously pending validation in other HIV-2 cohorts. Analysis of the various HLA/KIR gene profiles and HIV-2 viral load and CD4+ T-cell counts revealed no trend indicative of an influence of these markers on disease progression.
DISCUSSION
The HIV epidemic in West Africa is caused by two closely related retroviruses HIV-1 and HIV-2. The latter, which is the focus of the present study, was first reported to have infected healthy Senegalese prostitutes in 1985 (2) and was isolated the following year from two West-African patients with AIDS: one from Cape Verde Islands and the other from Guinea-Bissau (11). HIV-2 is structurally very similar to HIV-1 and shares between 30 to 60% homology at the nucleotide level.
Although both HIV-1 and HIV-2 use the same repertoire of coreceptors and target similar cell populations, their rate of infection and the resultant pathogenesis are quite different. The majority of HIV-2-infected people in Caio show high rates of long-term nonprogression (LTNP). Indeed, recent studies in Caio have shown that the majority of HIV-2-infected subjects who presented in 1991 (when viral load measurements were first made) with viral loads below detection have maintained undetectable viral loads for at least 15 years: these subjects have the same mortality risk as HIV uninfected subjects (L. F. Schim van der Loeff et al., unpublished data). However, in contrast to the ancestral virus (SIVsm) infecting its natural host, the sooty mangabey, a significant proportion of HIV-2-infected subjects later progress to AIDS, with a low CD4 count, high HIV-2 viral load, and clinical features that are indistinguishable from that observed in HIV-1 infection when CD4 count falls below 200 cells/μl (12). The observation that the majority of HIV-2-infected individuals are LTNPs suggests that HIV-2 infection presents a unique human model of naturally attenuated HIV infection that could provide considerable insights into the mechanism(s) of protective immunity needed for HIV vaccine development. The mechanisms underlying the attenuated course of HIV-2 infection and lack of disease progression in a substantial proportion of HIV-2-infected individuals are yet to be uncovered, but previous studies in Caio have shown a strong correlation between T-cell responses to a conserved region of HIV-2 gag and low plasma viral load (28).
Accumulating epidemiological evidence indicates that a range of distinct viral and host genetics and possibly some environmental factors contribute to the observed differences in clinical outcome after HIV infection. Among the host genetic factors are the HLA and KIR genes that are highly polymorphic and are very important antiviral innate and adaptive immune response regulators in humans. Epidemiological studies have implicated certain HLA alleles in rate of progression to AIDS in HIV-1 disease (10) and provided evidence that HLA genes are involved in the control of viral replication (17, 24), but their role in HIV-2 infection and disease progression is not clear. A small study of 62 female commercial sex workers infected with HIV-2, using p26 antigen exposure as a surrogate marker for disease progression, showed that HLA-B*35 was associated with the lack of p26 antibodies and a higher risk of disease progression (14). However, a detailed analysis of the effects of HLA and KIR genes on susceptibility to HIV-2 and disease progression is largely missing in the literature. In the present study, we determine for the first time the frequencies of HLA class I and KIR genes simultaneously from a predominantly Manjako community (>95%) in Caio, Guinea-Bissau, and demonstrated that specific HLA alleles can influence disease progression and risk of HIV-2 acquisition.
Comparison of HLA allele frequencies between Caio and neighboring populations showed distinct differences in allele frequencies that are consistent with previous anthropological findings, suggesting that Manjakos from the Caio sector are a unique set of people forming a close-knit community that tends to be isolated sexually from other ethnic groups in the subregion. Thus, it is unlikely that there is any significant population genetic stratification. The most common mode of HIV transmission in sub-Saharan Africa is through heterosexual contact. In HIV-1 infection, it has been reported that male-to-female transmission is more efficient than female-to-male transmission (5). A review of data from population-based surveys on discordant couples in East, Central, and West Africa strongly suggests that males generally introduce infection to the couples and, in 30 to 40% of cases, the female partner will acquire the infection from the infected male partner (25). In Caio, however, anthropological features (6, 40) suggest that most adults in Caio are exposed to multiple partners by virtue of their unique traditional way of life, mostly during the 4-year period of initiation into adulthood (40). Additional data from other HIV-2 cohorts suggest that older women are more vulnerable to HIV-2 infection than younger women (1, 22, 27). However, it would not be expected that the higher levels of infection of women would confound the interpretation of our data, since neither of these genetic factors are sex linked.
Recent studies on sequence analysis of the gag and nef genes in members of the Caio cohort showed uniform infection with closely related strains of clade A HIV-2 (34; J. Feldmann et al., unpublished data). Preliminary results from a similar study looking at the effect of HLA and KIR genes in a multiethnic cohort in Fajara, The Gambia, revealed that the Manjakos living in The Gambia exhibit HLA class I allele frequencies similar to those found in the present study, suggesting that there is little admixture between Manjakos and other tribes in the subregion irrespective of their geographical location (L. M. Yindom, unpublished).
Analysis of our KIR gene pool comparing frequencies of individual KIR genes with the scant available data from other West African populations showed that most of the activating KIR genes were present at higher frequencies in Caio (Fig. 2). In particular, KIR3DS1, which is rarely found in people of African ancestry, was significantly more common in the Caio population than in other West African populations in Senegal (13) and Nigeria (42). KIR3DS1 has previously been shown to confer protection against rapid progression to AIDS after HIV-1 infection in combination with its putative ligand HLA-B Bw4-80I (32).
Data on specific dates of seroconversion were not available for this cohort, as is usually the case in most African cohorts. We relied on information collected on known markers of HIV disease progression, such as CD4 counts and viral load measurements, to predict the effect of HLA class I and KIR gene variants on progression to AIDS following HIV-2 infection. We analyzed the HLA-A, -B, and -C allele and genotype data from individuals singly infected with HIV-2 and found that individuals carrying the common B*1503 allele were more likely to progress to AIDS than those without this allele. No susceptibility effect was seen with the other common B*15 allotype, B*1510. B*1503 and B*1510 differ by only three amino acids, all at critical sites in the peptide binding groove: positions 63 and 67, which reside in the alpha helix and contribute to the P2 pocket, and position 116, which resides on the floor of the peptide binding groove and is part of the P9 pocket. A single amino acid change at position 116 in B*3501 versus B*3503 has previously been implicated in differential susceptibility to HIV-1 disease progression (19), emphasizing the importance of this position in distinguishing the functional activity of HLA class I molecules. HLA-B*1503 has been associated with low viral loads in individuals infected with HIV-1 clade B, but no protective effect of this allele was observed in a C clade cohort from Durban, South Africa (18). HLA-B*1503 and the haplotype B*1503-Cw*0210 are unique markers confined to populations of sub-Sahara African origins (33). Therefore, the effect of HLA-B*1503 is more likely to be observed in these populations than in populations with a low frequency of this allele. Some minor or rare alleles were found to weakly associate with high CD4 count and a trend toward low viral load, indicating that they may offer a protective effect against rapid progression to AIDS after HIV-2 infection. This supports the notion proposed by Trachtenberg et al. that rare HLA alleles may have a selective advantage in protecting against rapid progression to AIDS in populations where HIV-1 has adapted to the most frequent HLA alleles (44).
Concluding remarks.
We determined here the HLA class I and KIR gene profiles of a relatively isolated population in Caio, Guinea-Bissau, which has one of the highest prevalence rates of HIV-2 infection in the world. The HLA-B*15 alleles B*1503 and B*1510 were observed at a relatively high frequency in this community compared to neighboring populations. Furthermore, HIV-2-infected individuals with B*1503 (but not B*1510) had significantly higher HIV-2 viral loads and lower CD4 counts compared to those without this allele, suggesting that this allele might be linked to poor control of viral replication and more rapid disease progression. Notably, none of the strongest HLA associations with HIV-1 were observed in our HIV-2 cohort. The frequencies of activating KIR genes were higher than those reported for other populations in West Africa. The frequency of KIR3DS1, which was previously shown to be protective in HIV-1 disease progression, was significantly higher in the Manjako group than that reported in any other African population. In general, the strongest associations in our study conferred susceptibility to HIV-2 outcomes, while protective factors were quite weak. This observation is contrary to that observed in HIV-1 disease, where the strongest HLA associations confer protection. Perhaps this reflects the less pathogenic nature of HIV-2 and the ability of most HLA class I allotypes to effectively control the virus, compared to HIV-1, where most allotypes are unable to maintain viral restriction. Our study is the first to provide a detailed analysis of the effects of HLA and KIR genetic variation on resistance and susceptibility to HIV-2 infection and disease progression. Since the present study was largely exploratory, corrected P values were not reported. Nevertheless, we were encouraged by the fact that the effect of B*15 remained significant after Bonferroni correction (corrected P values: 0.04 and 0.02 for the effect on CD4 levels and viral load, respectively). Thus, it will be of great interest to determine whether these effects can be validated in the other rare HIV-2 cohorts collected to date.
Acknowledgments
This study was funded by the European and Developing Countries Clinical Trials Partnership (EDCTP) and by the Medical Research Council (UK) The Gambia. This project was funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261200800001E. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
We are very grateful to all participants in this study. We also thank all Caio past investigators, the Caio field staff, and all MRC Fajara staff (past or present) who contributed to the success of this work. We especially thank David Jeffries and Matthew Cotten for useful comments and advice and Darlene Marti and Fuh-Mei Duh for technical assistance.
Footnotes
Published ahead of print on 2 June 2010.
REFERENCES
- 1.Aaby, P., K. Ariyoshi, M. Buckner, H. Jensen, N. Berry, A. Wilkins, D. Richard, O. Larsen, F. Dias, M. Melbye, and H. Whittle. 1996. Age of wife as a major determinant of male-to-female transmission of HIV-2 infection: a community study from rural West Africa. AIDS 10:1585-1590. [DOI] [PubMed] [Google Scholar]
- 2.Barin, F., S. M'Boup, F. Denis, P. Kanki, J. S. Allan, T. H. Lee, and M. Essex. 1985. Serological evidence for virus related to simian T-lymphotropic retrovirus III in residents of west Africa. Lancet ii:1387-1389. [DOI] [PubMed] [Google Scholar]
- 3.Berry, N., S. Jaffar, M. Schim van der Loeff, K. Ariyoshi, E. Harding, P. T. N′Gom, F. Dias, A. Wilkins, D. Ricard, P. Aaby, R. Tedder, and H. Whittle. 2002. Low level viremia and high CD4% predict normal survival in a cohort of HIV type-2-infected villagers. AIDS Res. Hum. Retrovir. 18:1167-1173. [DOI] [PubMed] [Google Scholar]
- 4.Bjorling, E., G. Scarlatti, A. von Gegerfelt, J. Albert, G. Biberfeld, F. Chiodi, E. Norrby, and E. M. Fenyo. 1993. Autologous neutralizing antibodies prevail in HIV-2 but not in HIV-1 infection. Virology 193:528-530. [DOI] [PubMed] [Google Scholar]
- 5.Boily, M. C., R. F. Baggaley, L. Wang, B. Masse, R. G. White, R. J. Hayes, and M. Alary. 2009. Heterosexual risk of HIV-1 infection per sexual act: systematic review and meta-analysis of observational studies. Lancet Infect. Dis. 9:118-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buckner, M. 2000. Manjako sex and gender. Ph.D. thesis. University of Paris X, Nanterre, France.
- 7.Cao, K., A. M. Moormann, K. E. Lyke, C. Masaberg, O. P. Sumba, O. K. Doumbo, D. Koech, A. Lancaster, M. Nelson, D. Meyer, R. Single, R. J. Hartzman, C. V. Plowe, J. Kazura, D. L. Mann, M. B. Sztein, G. Thomson, and M. A. Fernandez-Vina. 2004. Differentiation between African populations is evidenced by the diversity of alleles and haplotypes of HLA class I loci. Tissue Antigens 63:293-325. [DOI] [PubMed] [Google Scholar]
- 8.Carr, W. H., M. J. Pando, and P. Parham. 2005. KIR3DL1 polymorphisms that affect NK cell inhibition by HLA-Bw4 ligand. J. Immunol. 175:5222-5229. [DOI] [PubMed] [Google Scholar]
- 9.Carrington, M., M. P. Martin, and J. van Bergen. 2008. KIR-HLA intercourse in HIV disease. Trends Microbiol. 16:620-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carrington, M., and S. J. O'Brien. 2003. The influence of HLA genotype on AIDS. Annu. Rev. Med. 54:535-551. [DOI] [PubMed] [Google Scholar]
- 11.Clavel, F., D. Guetard, F. Brun-Vezinet, S. Chamaret, M. A. Rey, M. O. Santos-Ferreira, A. G. Laurent, C. Dauguet, C. Katlama, C. Rouzioux, et al. 1986. Isolation of a new human retrovirus from West African patients with AIDS. Science 233:343-346. [DOI] [PubMed] [Google Scholar]
- 12.De Cock, K. M., K. Odehouri, R. L. Colebunders, G. Adjorlolo, M. F. Lafontaine, A. Porter, E. Gnaore, L. Diaby, J. Moreau, W. L. Heyward, et al. 1990. A comparison of HIV-1 and HIV-2 infections in hospitalized patients in Abidjan, Cote d'Ivoire. AIDS 4:443-448. [DOI] [PubMed] [Google Scholar]
- 13.Denis, L., J. Sivula, P. A. Gourraud, N. Kerdudou, R. Chout, C. Ricard, J. P. Moisan, K. Gagne, J. Partanen, and J. D. Bignon. 2005. Genetic diversity of KIR natural killer cell markers in populations from France, Guadeloupe, Finland, Senegal, and Reunion. Tissue Antigens 66:267-276. [DOI] [PubMed] [Google Scholar]
- 14.Diouf, K., A. D. Sarr, G. Eisen, S. Popper, S. Mboup, and P. Kanki. 2002. Associations between MHC class I and susceptibility to HIV-2 disease progression. J. Hum. Virol. 5:1-7. [PubMed] [Google Scholar]
- 15.Duvall, M. G., A. Jaye, T. Dong, J. M. Brenchley, A. S. Alabi, D. J. Jeffries, M. van der Sande, T. O. Togun, S. J. McConkey, D. C. Douek, A. J. McMichael, H. C. Whittle, R. A. Koup, and S. L. Rowland-Jones. 2006. Maintenance of HIV-specific CD4+ T-cell help distinguishes HIV-2 from HIV-1 infection. J. Immunol. 176:6973-6981. [DOI] [PubMed] [Google Scholar]
- 16.Excoffier, L. G. L., and S. Schneider. 2005. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online 1:47-50. [PMC free article] [PubMed] [Google Scholar]
- 17.Fellay, J., K. V. Shianna, D. Ge, S. Colombo, B. Ledergerber, M. Weale, K. Zhang, C. Gumbs, A. Castagna, A. Cossarizza, A. Cozzi-Lepri, A. De Luca, P. Easterbrook, P. Francioli, S. Mallal, J. Martinez-Picado, J. M. Miro, N. Obel, J. P. Smith, J. Wyniger, P. Descombes, S. E. Antonarakis, N. L. Letvin, A. J. McMichael, B. F. Haynes, A. Telenti, and D. B. Goldstein. 2007. A whole-genome association study of major determinants for host control of HIV-1. Science 317:944-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frahm, N., P. Kiepiela, S. Adams, C. H. Linde, H. S. Hewitt, K. Sango, M. E. Feeney, M. M. Addo, M. Lichterfeld, M. P. Lahaie, E. Pae, A. G. Wurcel, T. Roach, M. A. St John, M. Altfeld, F. M. Marincola, C. Moore, S. Mallal, M. Carrington, D. Heckerman, T. M. Allen, J. I. Mullins, B. T. Korber, P. J. Goulder, B. D. Walker, and C. Brander. 2006. Control of human immunodeficiency virus replication by cytotoxic T lymphocytes targeting subdominant epitopes. Nat. Immunol. 7:173-178. [DOI] [PubMed] [Google Scholar]
- 19.Gao, X., G. W. Nelson, P. Karacki, M. P. Martin, J. Phair, R. Kaslow, J. J. Goedert, S. Buchbinder, K. Hoots, D. Vlahov, S. J. O'Brien, and M. Carrington. 2001. Effect of a single amino acid change in MHC class I molecules on the rate of progression to AIDS. N. Engl. J. Med. 344:1668-1675. [DOI] [PubMed] [Google Scholar]
- 20.Guyader, M., M. Emerman, P. Sonigo, F. Clavel, L. Montagnier, and M. Alizon. 1987. Genome organization and transactivation of the human immunodeficiency virus type 2. Nature 326:662-669. [DOI] [PubMed] [Google Scholar]
- 21.Holmgren, B., P. Aaby, H. Jensen, O. Larsen, Z. da Silva, and I. M. Lisse. 1999. Increased prevalence of retrovirus infections among older women in Africa. Scand. J. Infect. Dis. 31:459-466. [DOI] [PubMed] [Google Scholar]
- 22.Holmgren, B., Z. da Silva, O. Larsen, P. Vastrup, S. Andersson, and P. Aaby. 2003. Dual infections with HIV-1, HIV-2 and HTLV-1 are more common in older women than in men in Guinea-Bissau. AIDS 17:241-253. [DOI] [PubMed] [Google Scholar]
- 23.Jonsson, A. H., and W. M. Yokoyama. 2009. Natural killer cell tolerance licensing and other mechanisms. Adv. Immunol. 101:27-79. [DOI] [PubMed] [Google Scholar]
- 24.Kaslow, R. A., M. Carrington, R. Apple, L. Park, A. Munoz, A. J. Saah, J. J. Goedert, C. Winkler, S. J. O'Brien, C. Rinaldo, R. Detels, W. Blattner, J. Phair, H. Erlich, and D. L. Mann. 1996. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat. Med. 2:405-411. [DOI] [PubMed] [Google Scholar]
- 25.Kilmarx, P. H. 2009. Global epidemiology of HIV. Curr. Opin. HIV AIDS 4:240-246. [DOI] [PubMed] [Google Scholar]
- 26.Lanier, L. L. 2008. Up on the tightrope: natural killer cell activation and inhibition. Nat. Immunol. 9:495-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larsen, O., Z. da Silva, A. Sandstrom, P. K. Andersen, S. Andersson, A. G. Poulsen, M. Melbye, F. Dias, A. Naucler, and P. Aaby. 1998. Declining HIV-2 prevalence and incidence among men in a community study from Guinea-Bissau. AIDS 12:1707-1714. [DOI] [PubMed] [Google Scholar]
- 28.Leligdowicz, A., L. M. Yindom, C. Onyango, R. Sarge-Njie, A. Alabi, M. Cotten, T. Vincent, C. da Costa, P. Aaby, A. Jaye, T. Dong, A. McMichael, H. Whittle, and S. Rowland-Jones. 2007. Robust Gag-specific T-cell responses characterize viremia control in HIV-2 infection. J. Clin. Invest. 117:3067-3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacDonald, K. S., J. Embree, S. Njenga, N. J. Nagelkerke, I. Ngatia, Z. Mohammed, B. H. Barber, J. Ndinya-Achola, J. Bwayo, and F. A. Plummer. 1998. Mother-child class I HLA concordance increases perinatal human immunodeficiency virus type 1 transmission. J. Infect. Dis. 177:551-556. [DOI] [PubMed] [Google Scholar]
- 30.Mackelprang, R. D., G. John-Stewart, M. Carrington, B. Richardson, S. Rowland-Jones, X. Gao, D. Mbori-Ngacha, J. Mabuka, B. Lohman-Payne, and C. Farquhar. 2008. Maternal HLA homozygosity and mother-child HLA concordance increase the risk of vertical transmission of HIV-1. J. Infect. Dis. 197:1156-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin, M. P., and M. Carrington. 2008. KIR locus polymorphisms: genotyping and disease association analysis. Methods Mol. Biol. 415:49-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin, M. P., X. Gao, J. H. Lee, G. W. Nelson, R. Detels, J. J. Goedert, S. Buchbinder, K. Hoots, D. Vlahov, J. Trowsdale, M. Wilson, S. J. O'Brien, and M. Carrington. 2002. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat. Genet. 31:429-434. [DOI] [PubMed] [Google Scholar]
- 33.Middleton, D., L. Menchaca, H. Rood, and R. Komerofsky. 2003. New allele frequency database: http://www.allelefrequencies.net. Tissue Antigens 61:403-407. [DOI] [PubMed] [Google Scholar]
- 34.Onyango, C., A. Leligdowicz, A. Yokoyama, H. Sato, H. Song, E. E. Nakayama, T. Shiodad, T. de Silva, J. Townend, A. Jaye, H. Whittle, S. Rowland-Jones, and M. Cotten. 2009. HIV-2 capsids distinguish high and low virus load patients in a West African community cohort. Vaccine [Epub ahead of print.] doi: 10.1016/j.vaccine.2009.08.060. [DOI] [PubMed]
- 35.Ricard, D., A. Wilkins, P. T. N′Gum, R. Hayes, G. Morgan, A. P. Da Silva, and H. Whittle. 1994. The effects of HIV-2 infection in a rural area of Guinea-Bissau. AIDS 8:977-982. [DOI] [PubMed] [Google Scholar]
- 36.Rowland-Jones, S., J. Sutton, K. Ariyoshi, T. Dong, F. Gotch, S. McAdam, D. Whitby, S. Sabally, A. Gallimore, T. Corrah, et al. 1995. HIV-specific cytotoxic T cells in HIV-exposed but uninfected Gambian women. Nat. Med. 1:59-64. [DOI] [PubMed] [Google Scholar]
- 37.Rowland-Jones, S. L., and H. C. Whittle. 2007. Out of Africa: what can we learn from HIV-2 about protective immunity to HIV-1? Nat. Immunol. 8:329-331. [DOI] [PubMed] [Google Scholar]
- 38.Sanchez-Mazas, A., Q. G. Steiner, C. Grundschober, and J. M. Tiercy. 2000. The molecular determination of HLA-Cw alleles in the Mandenka (West Africa) reveals a close genetic relationship between Africans and Europeans. Tissue Antigens 56:303-312. [DOI] [PubMed] [Google Scholar]
- 39.Schim van der Loeff, M. F., and P. Aaby. 1999. Towards a better understanding of the epidemiology of HIV-2. Aids 13(Suppl. A):S69-S84. [PubMed] [Google Scholar]
- 40.Schmidt, W. P., M. S. Van Der Loeff, P. Aaby, H. Whittle, R. Bakker, M. Buckner, F. Dias, and R. G. White. 2008. Behavioor change and competitive exclusion can explain the diverging HIV-1 and HIV-2 prevalence trends in Guinea-Bissau. Epidemiol. Infect. 136:551-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Serwanga, J., L. A. Shafer, E. Pimego, B. Auma, C. Watera, S. Rowland, D. Yirrell, P. Pala, H. Grosskurth, J. Whitworth, F. Gotch, and P. Kaleebu. 2009. Host HLA B*allele-associated multi-clade Gag T-cell recognition correlates with slow HIV-1 disease progression in antiretroviral therapy-naive Ugandans. PLoS One 4:e4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Single, R. M., M. P. Martin, X. Gao, D. Meyer, M. Yeager, J. R. Kidd, K. K. Kidd, and M. Carrington. 2007. Global diversity and evidence for coevolution of KIR and HLA. Nat. Genet. 39:1114-1119. [DOI] [PubMed] [Google Scholar]
- 43.Stewart, C. A., F. Laugier-Anfossi, F. Vely, X. Saulquin, J. Riedmuller, A. Tisserant, L. Gauthier, F. Romagne, G. Ferracci, F. A. Arosa, A. Moretta, P. D. Sun, S. Ugolini, and E. Vivier. 2005. Recognition of peptide-MHC class I complexes by activating killer immunoglobulin-like receptors. Proc. Natl. Acad. Sci. U. S. A. 102:13224-13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trachtenberg, E., B. Korber, C. Sollars, T. B. Kepler, P. T. Hraber, E. Hayes, R. Funkhouser, M. Fugate, J. Theiler, Y. S. Hsu, K. Kunstman, S. Wu, J. Phair, H. Erlich, and S. Wolinsky. 2003. Advantage of rare HLA supertype in HIV disease progression. Nat. Med. 9:928-935. [DOI] [PubMed] [Google Scholar]
- 45.Wilkins, A., D. Ricard, J. Todd, H. Whittle, F. Dias, and A. Paulo Da Silva. 1993. The epidemiology of HIV infection in a rural area of Guinea-Bissau. AIDS 7:1119-1122. [DOI] [PubMed] [Google Scholar]