Abstract
Human respiratory syncytial virus (HRSV) fusion (F) protein is an essential component of the virus envelope that mediates fusion of the viral and cell membranes, and, therefore, it is an attractive target for drug and vaccine development. Our aim was to analyze the neutralizing mechanism of anti-F antibodies in comparison with other low-molecular-weight compounds targeted against the F molecule. It was found that neutralization by anti-F antibodies is related to epitope specificity. Thus, neutralizing and nonneutralizing antibodies could bind equally well to virions and remained bound after ultracentrifugation of the virus, but only the former inhibited virus infectivity. Neutralization by antibodies correlated with inhibition of cell-cell fusion in a syncytium formation assay, but not with inhibition of virus binding to cells. In contrast, a peptide (residues 478 to 516 of F protein [F478-516]) derived from the F protein heptad repeat B (HRB) or the organic compound BMS-433771 did not interfere with virus infectivity if incubated with virus before ultracentrifugation or during adsorption of virus to cells at 4°C. These inhibitors must be present during virus entry to effect HRSV neutralization. These results are best interpreted by asserting that neutralizing antibodies bind to the F protein in virions interfering with its activation for fusion. Binding of nonneutralizing antibodies is not enough to block this step. In contrast, the peptide F478-516 or BMS-433771 must bind to F protein intermediates generated during virus-cell membrane fusion, blocking further development of this process.
Human respiratory syncytial virus (HRSV), a member of the Pneumovirus genus of the Paramyxoviridae family, is the main cause of severe lower respiratory tract infections in very young children (36), and it is a pathogen of considerable importance in the elderly (24, 26) and in immunocompromised adults (22). Currently, there is no effective vaccine against the virus although it is known that passive administration of neutralizing antibodies to individuals at high risk is an effective immunoprophylaxis (37, 38).
The HRSV genome is a single-stranded negative-sense RNA molecule of approximately 15 kb that encodes 11 proteins (16, 53). Two of these proteins are the main surface glycoproteins of the virion. These are (i) the attachment (G) protein, which mediates virus binding to cells (44), and (ii) the fusion (F) protein, which promotes both fusion of the viral and cell membranes at the initial stages of the infectious cycle and fusion of the membrane of infected cells with those of adjacent cells to form characteristic syncytia (72). These two glycoproteins are the only targets of neutralizing antibodies either induced in animal models (19, 63, 65, 70) or present in human sera (62).
The G protein is a highly variable type II glycoprotein that shares neither sequence identity nor structural features with the attachment protein of other paramyxoviruses (75). It is synthesized as a precursor of about 300 amino acids (depending on the strain) that is modified posttranslationally by the addition of a large number of N- and O-linked oligosaccharides and is also palmitoylated (17). The G protein is oligomeric (probably a homotetramer) (23) and promotes binding of HRSV to cell surface proteoglycans (35, 40, 49, 67). Whether this is the only interaction of G with cell surface components is presently unknown.
The F protein is a type I glycoprotein that is synthesized as an inactive precursor of 574 amino acids (F0) which is cleaved by furin during transport to the cell surface to yield two disulfide-linked polypeptides, F2 from the N terminus and F1 from the C terminus (18). Like other viral type I fusion proteins, the mature F protein is a homotrimer which is in a prefusion, metastable, conformation in the virus particle. After fusion, the F protein adopts a highly stable postfusion conformation. Stability of the postfusion conformation is determined to great extent by two heptad repeat (HR) sequences, HRA and HRB, present in the F1 chain. Mixtures of HRA and HRB peptides form spontaneously heterotrimeric complexes (43, 51) that assemble in six-helix bundles (6HB), consisting of an internal core of three HRA helices surrounded by three antiparallel HRB helices, as determined by X-ray crystallography (79).
The three-dimensional (3D) structure of the HRSV F protein has not been solved yet. Nevertheless, the structures of the pre- and postfusion forms of two paramyxovirus F proteins have revealed substantial conformational differences between the pre- and postfusion conformations (77, 78). The present hypothesis about the mechanism of membrane fusion mediated by paramyxovirus F proteins proposes that, following binding of the virus to the cell surface, the prefusion form of the F glycoprotein is activated, and membrane fusion is triggered. The F protein experiences then a series of conformational changes which include the exposure of a hydrophobic region, called the fusion peptide, and its insertion into the target membrane. Subsequent refolding of this intermediate leads to formation of the HRA and HRB six-helix bundle, concomitant with approximation of the viral and cell membranes that finally fuse, placing the fusion peptide and the transmembrane domain in the same membrane (4, 20). The formation of the 6HB and the associated free energy change are tightly linked to the merger of the viral and cellular membranes (60).
Antibodies play a major role in protection against HRSV. Animal studies have demonstrated that immunization with either F or G glycoproteins induces neutralizing antibodies and protects against a viral challenge (19, 63, 70). Furthermore, transfer of these antibodies (31, 56) or of anti-F or anti-G monoclonal antibodies (MAbs) protects mice, cotton rats, or calves against either a human or bovine RSV challenge, respectively (65, 68, 73). Likewise, infants at high risk of severe HRSV disease are protected by the prophylactic administration of immunoglobulins with high anti-HRSV neutralizing titers (33). Finally, a positive correlation was found between high titers of serum neutralizing antibodies and protection in adult volunteers challenged with HRSV (34, 74), while an inverse correlation was found between high titers of neutralizing antibodies and risk of infection in children (29) and in the elderly (25).
Whereas all the anti-G monoclonal antibodies reported to date are poorly neutralizing (1, 28, 48, 71), some anti-F monoclonal antibodies have strong neutralization activity (1, 3, 5, 28, 46). It is believed that HRSV neutralization by anti-G antibodies requires simultaneous binding of several antibodies to different epitopes, leading to steric hindrance for interaction of the G glycoprotein with the cell surface. Indeed, it has been shown that neutralization is enhanced by mixtures of anti-G monoclonal antibodies (1, 50), mimicking the effect of polyclonal anti-G antibodies. In contrast, highly neutralizing anti-F monoclonal antibodies do not require cooperation by other antibodies to block HRSV infectivity efficiently (1).
In addition to neutralizing antibodies, other low-molecular-weight compounds directed against the F protein are potent inhibitors of HRSV infectivity. Synthetic peptides that reproduce sequences of heptad repeat B inhibit both membrane fusion promoted by the F protein and HRSV infectivity (42). Also, other small molecules obtained by chemical synthesis have been shown to interact with F protein and inhibit HRSV infectivity. These HRSV entry inhibitors have been the topic of intense research in recent years (55).
This study explores the mechanisms of HRSV neutralization by different inhibitors of membrane fusion, including anti-F monoclonal antibodies, an HRB peptide, and the synthetic compound BMS-433771 (13-15). The results obtained indicate that antibodies and low-molecular-weight compounds block membrane fusion at different stages during virus entry.
MATERIALS AND METHODS
Viruses, cells, protein, and plasmid.
The Long strain of HRSV was propagated in HEp-2 cells grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 2.5% fetal calf serum (FCS).
A vaccinia virus recombinant that expresses a soluble form of the HRSV F glycoprotein (FTM−) has been reported (7, 8). The F soluble form (FTM−) was engineered by introducing a premature stop codon in the F gene that eliminated the transmembrane (TM) region and the cytoplasmic tail (7). The FTM− is secreted to the supernatants of cells infected with the corresponding vaccinia recombinant.
BSR-T7/5 cells (a BHK-derived cell line that constitutively expresses the T7 RNA polymerase; kindly provided by K.-K. Conzelmann) (11) were maintained in DMEM with 10% FCS. Alternate passages of BSR cells were supplemented with 1 mg/ml G418 sulfate (Sigma-Aldrich) in order to select for T7 polymerase expression.
A pTM1-derived plasmid (21), carrying a full-length cDNA copy of the HRSV F protein gene under transcriptional control of the T7 promoter (pTM1-F), has been described previously (30).
Monoclonal antibodies, peptide, and antiviral compound.
MAbs 2F, 47F, and 101F specific for F protein of HRSV and MAb 63G against G protein have been previously described (27, 28, 45, 76). They were purified from ascitic fluids by protein A-Sepharose chromatography (28). The MAb 1B.C11 against p72 capsid protein of African swine pest virus (ASPV) was used as a negative control (61).
The HRB-derived peptide F protein consisting of residues 478 to 516 (F478-516), YDPLVFPSDEFDASISQVNEKINQSLAFIRKSDELLHNV, was synthesized by solid-phase methods in an ABI 433A synthesizer (Applied Biosystems), using Fmoc (9-fluorenylmethoxy carbonyl) chemistry and 0.1-mmol-scale FastMoc protocols on Fmoc-Rink-amide (MBHA) resin, with 10-fold excess of Fmoc-protected l-amino acids and HBTU/HOBt (2-[H-benzotriazole-1-yl]-1.13.3-tetramethyluronium hexafluorophosphate/1-hydroxybenzotriazole) or HATU [2-(1-H-9-azobenzyltriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate] coupling chemistries. The protected peptide resin was N-deblocked with piperidine prior to full deprotection and cleavage with trifluoroacetic acid (TFA)-water-triisopropylsilane (95:2.5:2.5, vol/vol/vol; 90 min at room temperature). The peptide was precipitated by addition of chilled methyl tert-butyl ether, taken up in aqueous acetic acid ([HOAc] 10%, vol/vol) and lyophilized. Purification by preparative high-performance liquid chromatography (HPLC) was performed on a C18 column (Phenomenex Luna C8; 21.2 by 250 mm; particle size, 10 μm), using a linear gradient of solvent B into A for elution (solvent A, 0.045% TFA in H2O; solvent B, 0.036% in acetonitrile), at a flow rate of 25 ml/min. Fractions of adequate HPLC purity and with the expected mass by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry were combined and lyophilized.
BMS-433771, is a low-molecular-weight compound with antiviral properties (13-15). It was synthesized according to Provencal et al. (57), and it was a generous gift of Arrow Therapeutics (London, United Kingdom).
Preparation of Fab fragments.
Purified monoclonal antibodies were digested with papain (Roche Biochemicals), for 4 h at 37°C, at a mass ratio of 100:1 (MAb to papain) in the presence of the reducing agent 4.2 mM cysteine and 0.8 mM EDTA. The reaction was stopped by adding 300 mM iodoacetamide in phosphate-buffered saline (PBS) and incubating for 15 min at room temperature. Undigested antibody and Fc fragments were removed using protein A-Sepharose chromatography with Fab fragments collected in the column flowthrough. Fab fragments were further purified by size exclusion chromatography using a Superdex 200 10/30 GL column (GE Healthcare). Their purity was assessed by SDS-polyacrylamide gel electrophoresis (PAGE), and their reactivity with the F protein was determined by enzyme-linked immunosorbent assay (ELISA).
ELISAs. (i) Direct method.
Purified FTM− (6) was diluted in PBS and used to coat 96-well microtiter plates. After overnight incubation at 4°C, the wells were blocked with 5% pig serum in 0.05% Tween-20 in PBS. Dilutions of antibodies in blocking solution were added to the wells that were incubated for 1 h at 37°C. After plates were washed with water, antibodies bound to the wells were revealed with horseradish peroxidase-labeled anti-mouse Ig and O-phenyl-diamine (OPD) as a substrate, following the manufacturer's instructions (GE Healthcare). Negative controls consisting of blocking solution but no antigen were tested in parallel.
ELISAs done with Fab fragments were revealed with biotinylated anti-mouse kappa chain (GE Healthcare) for 1 h at 37°C, followed by horseradish peroxidase-labeled streptavidin (GE Healthcare) for 30 min at room temperature and OPD as a substrate.
(ii) Indirect method.
Purified anti-FTM− antibodies from rabbit serum were diluted in PBS and used to coat 96-well microtiter plates. After overnight incubation at 4°C, the wells were blocked with 5% pig serum in 0.05% Tween-20 in PBS. Dilutions of ultracentrifuged virus-antibody mixtures (see below) were made in blocking solution and added to the wells that were incubated for 2 h at 37°C. After plates were washed with water, biotin-labeled anti-mouse antibodies (GE Healthcare) or a pool of biotin-labeled monoclonal antibodies (2F, 47F and 101F) specific for the F protein, in blocking solution, were added and incubated for 1 h at 37°C, respectively. After plates were washed again, antibodies bound to the wells were revealed with horseradish peroxidase-labeled streptavidin (GE Healthcare) and OPD as a substrate, following the manufacturer's instructions (GE Healthcare).
Virus neutralization.
Inhibition of HRSV infectivity was assessed by two different methods. For the first method, HEp-2 cells were grown in 96-well plates in DMEM with 10% FCS. Dilutions of antibodies, peptide, or antiviral compound, done in DMEM with 2.5% inactivated FCS, were incubated with HRSV for 30 min at 37°C as indicated in figure legends. These mixtures were used to infect HEp-2 cells in DMEM with 2.5% inactivated FCS. After 1 h of adsorption at 37°C, DMEM with 2.5% inactivated FCS was added, and the cells were incubated for 72 h at 37°C. Then, the plates were washed three times with 0.05% Tween-20 in PBS and fixed with 80% cold acetone in PBS. Antigen production in the fixed monolayers was measured by ELISA, using a pool of monoclonal antibodies specific for the F and G glycoproteins.
For the second method, virus-inhibitor mixtures prepared as indicated in the previous paragraph, were incubated for 30 min at 37°C. Then these mixtures were loaded onto 25% glycerol cushions in PBS and ultracentrifuged at 125,812 × g for 2 h at 4°C in a Beckman SW60 rotor. The pellets were resuspended in DMEM with 2.5% inactivated FCS and used to infect HEp-2 cells. Viral antigen produced after 72 h was quantified as described in the previous paragraph.
Syncytium formation assay.
The syncytium formation assay was performed in either (i) HRSV-infected cells or (ii) transfected cells.
(i) HRSV-infected cells.
HEp-2 cells were grown in microchamber culture slides up to 90 to 100% confluence and infected with HRSV in DMEM-2.5% FCS for 90 min at 37°C, as indicated in the figure legends. Then, the inoculum was removed, and after cells were washed, they were incubated in DMEM-2.5% FCS for 5 h at 37°C, when the antibodies under test were added to the cultures. Incubation was continued until 48 h postinfection, and the cells were fixed with cold methanol for 5 min, followed by cold acetone for 30 s. Fixed cells were subsequently immunostained using anti-F and anti-G MAbs, followed by incubation with anti-mouse fluorescein-linked antibody (GE Healthcare), and examined for syncytia using a Zeiss microscope equipped with an AxioCam HRC digital camera and Axiovision, version 3.1, software.
(ii) Transfected cells.
BSR-T7/5 cells were grown in microchamber culture slides up to 90 to 100% confluence and then transfected in DMEM-2.5% FCS with 0.5 μg of plasmid DNA (pTM1-F) using FuGENE HD (Roche Biochemicals) at a 2:1 ratio (μl of FuGENE to μg of DNA). The transfection mixture was removed at 7 h posttransfection. Then, the cells were washed twice with DMEM-2.5% FCS, and antibodies were added to cultures in DMEM-2.5% FCS. Incubation was continued until 48 h posttransfection, when the cells were fixed and syncytium formation assessed as described in the paragraph above.
Flow cytometry.
Virus binding to cells and concurrent HRSV infection were assessed by flow cytometry as follows. Virus-antibody mixtures were incubated for 30 min at 37°C before being added to a suspension of HEp-2 cells in DMEM with 2% FCS for 1 h at 4°C. Unbound virus was removed by washing the cells twice with PBS-5% pig serum, and the cells were split in two tubes. One of the tubes was incubated for 30 min at 4°C with a purified anti-F antibody conjugated with the fluorochrome Cy5. After the cells were pelleted and washed twice with PBS-5% pig serum, they were fixed in 1.4% paraformaldehyde. The fluorescence corresponding to bound virus was measured on a FACSCanto flow cytometer (BD Biosciences, San Jose, CA) and analyzed using FlowJo, version 7.5.2, software (TreeStar, Inc.).
The other half of the cells were plated and cultured at 37°C for 48 h in DMEM with 2% FCS. Then the cells were washed with PBS and detached using PBS-10 mM EDTA. HRSV infection was quantified by measuring the level of F protein expression on the surface of infected cells by flow cytometry with an anti-F antibody conjugated with the fluorochrome Cy5 as described in the previous paragraph.
RESULTS
MAbs directed against different antigenic sites of the F protein differ in neutralization activity.
Different antigenic sites have been identified previously in the HRSV F protein by either loss of monoclonal antibody reactivity in escape mutants, pairwise competition of antibodies for virus binding, or reactivity of antibodies with synthetic peptides or protein fragments (3, 5, 9, 28, 45). To compare the neutralizing properties of antibodies specific for different F protein epitopes, three MAbs were selected for this study. Location of their respective epitopes in the F protein primary structure is illustrated in Fig. 1A. MAb 2F recognizes an epitope of antigenic site I. Sequence changes in residue P389 abolish reactivity with this antibody (45). MAb 47F recognizes an epitope of antigenic site II. Mutants selected with this antibody have changes at position N262, N268, K272, or S275 (3, 28, 45). Finally, MAb 101F, whose epitope was mapped in antigenic site IV, does not recognize escape mutants with the change K433T (76).
FIG. 1.
Antigen binding and virus neutralization with MAbs directed against the F protein of HRSV. (A) Diagram of the F protein primary structure, showing antigenic sites I, II and IV; the hydrophobic regions (▪) (the signal peptide [SP], the fusion peptide [FP], and the transmembrane region [TM]); HRA and HRB; and the sites of proteolytic processing (red arrow, site I; black arrow, site II). Shown below the diagram are the MAbs used in this study and the sequence changes in escape mutants that ablate reactivity with each MAb. (B) Serial dilutions of purified anti-F MAbs were tested in a direct ELISA for binding to a soluble form of the F protein (FTM−), as described in Materials and Methods. (C) Long virus (6.5 × 103 PFU) was incubated with different amounts of MAbs for 30 min at 37°C before being used to infect HEp-2 cells. Production of viral antigen was quantified by ELISA 72 h later, as described in Materials and Methods, and results are presented as a percentage of the value for control cells infected in the absence of antibody. MAb 1P (against HRSV phosphoprotein) was used as a negative control in neutralization. Data represent the mean and standard deviation from three independent experiments. OD, optical density.
The three purified MAbs displayed similar reactivities with a soluble form of the F protein (FTM−) in a direct ELISA (Fig. 1B). However, significant differences were observed between MAbs when their abilities to block virus infectivity were compared in a neutralization assay based on a reduction of viral antigen production (Fig. 1C). While MAb 47F (antigenic site II) and MAb 101F (antigenic site IV) inhibited completely the production of viral antigens in a dose-dependent manner, MAb 2F (antigenic site I) at low concentrations reduced antigen production by 50% compared with a negative control (MAb 1P), but this reduction did not increase even at the highest concentration tested. Thus, the neutralizing activity of the three anti-F antibodies was related to epitope specificity. Whereas MAbs 47F and 101F behaved as expected for truly neutralizing antibodies, the partial inhibition exhibited by MAb 2F may reflect some indirect effect of this antibody on virus infectivity, such as virus aggregation or cross-linking (see below).
MAbs 47F and 101F inhibit virus infectivity before viral adsorption to cells.
It is thought that, following binding of HRSV to the target membrane, the F protein is activated and drives membrane fusion by an irreversible change in conformation from the initial, metastable, prefusion form to a lower-free energy postfusion state. Therefore, it was of interest to determine whether neutralizing MAbs were able to inhibit viral infectivity before contact of the virus with the target cell, i.e., before the F protein underwent activation for membrane fusion. To this end, virus aliquots were incubated with no antibody or with a large excess of either 2F, 47F, 101F, or control antibody before being subjected to ultracentrifugation to pellet the virus and remove any antibody excess not bound to viral particles. Then, the pellets were resuspended and used to infect HEp-2 cells. Antigen production was measured 72 h later by ELISA. As shown in Fig. 2A, MAbs 47F and 101F efficiently inhibited viral infectivity in this type of assay, compared with no antibody or an irrelevant antibody (1B.C11) used as negative control. In contrast, MAb 2F did not reduce antigen production.
FIG. 2.
Neutralization with anti-F MAbs added before (A) or during (B) HRSV infection. (A) Long virus (8.2 × 105 PFU) was incubated in the absence or presence of 400 μg of the anti-F MAbs indicated on the figure. Virus-antibody mixtures were ultracentrifuged (125,812 × g for 2 h in a Beckman SW60 rotor), and the pellets were resuspended and used to infect HEp-2 cells. After 72 h of incubation, production of viral antigen was quantified by ELISA, as described in Materials and Methods, and results are presented as a percentage of the value for control cells infected in the absence of antibody. MAb 1B.C11 against a capsid protein of ASPV was used as a negative control (C−) in neutralization. (B) Long virus (1 × 105 PFU) ultracentrifuged as before in the absence of antibody was used to infect HEp-2 cells in the absence or presence of 3 μg of the antibodies indicated on the figure. Production of viral antigen was quantified as described for panel A. Data represent the mean and standard deviation from three independent experiments.
To confirm that the ultracentrifuged virus was still susceptible of neutralization by antibodies, aliquots of a virus pelleted in the absence of antibodies were mixed with either no antibody or with MAb 2F, 47F, 101F, or a control. The virus-antibody mixtures were then added to HEp-2 cultures, and antigen production was measured 72 h later by ELISA, as mentioned before. Again, MAbs 47F and 101F were able to inhibit essentially all the infectivity, but MAb 2F reduced viral antigen production by only around 50% (Fig. 2B). Thus, the conditions used to pellet the virus did not have a major impact either on virus infectivity or on its susceptibility to antibody neutralization (compare Fig. 1C and 2B).
The results shown in Fig. 2 demonstrate, therefore, that MAbs 47F and 101F can block viral infectivity if added to the virus before it contacts the cells, i.e., before the F protein is activated for fusion.
MAbs 2F, 47F, and 101F remain bound to ultracentrifuged virus.
The inactivation of HRSV infectivity by 47F and 101F antibodies described in the previous section could be the result of a stable interaction of these antibodies with the F protein. Alternatively, binding of antibodies to the F protein could induce irreversible changes in F by a hit-and-run mechanism, leading to virus inactivation (10). Of course, there are intermediates to these alternatives, such as induction of irreversible changes by antibodies that remain bound to the inactivated antigen.
To investigate if antibodies were still bound to virus particles after ultracentrifugation, virus-antibody mixtures prepared as before were ultracentrifuged, and the amount of antibody present in the pellets was quantified in an indirect ELISA. The same pellets were used in a similar assay to check if the same amount of virus had been sedimented, irrespective of the antibody used in the preincubation with the virus. Finally, aliquots of the pellets were used to infect HEp-2 cells, and virus antigen production was assessed 48 h later by flow cytometry.
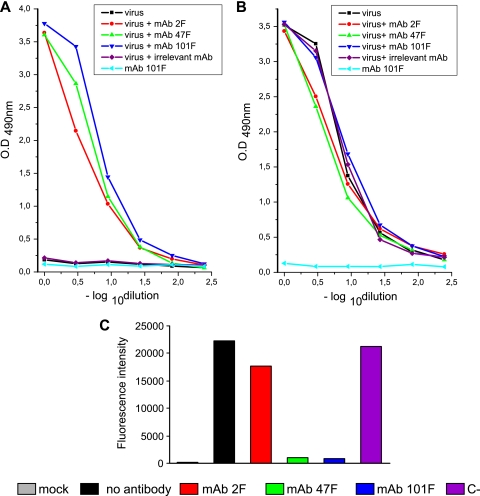
Despite differences in the neutralizing activity of the three MAbs (Fig. 1C and 2), similar amounts of each MAb tested (2F, 47F, and 101F) remained bound to the virus particles after sedimentation, as shown in Fig. 3A. The antibody present in the pellet required specific binding to HRSV since an irrelevant antibody directed against a capsid protein of ASPV was not trapped in the pellet. Also, antibody 101F was not found in the pellet unless it had been mixed with the virus before ultracentrifugation.
FIG. 3.
Quantification of antibodies, virus, and infectivity in pellets after centrifugation. Serial dilutions of pelleted virus-antibody mixtures, ultracentrifuged as described in the legend of Fig. 2A, were incubated with purified rabbit anti-FTM− antibodies bound to microtiter plates. After samples were washed, the amount of antibody bound to the captured virus was quantified with biotin-labeled anti-mouse antibodies (A), and the amount of captured virus was determined with a pool of biotin-labeled anti-F (2F, 47F, and 101F) antibodies (B), as detailed in Materials and Methods. MAb 1B.C11 against a capsid protein of ASPV was used as a negative control (C−). Controls of antibody (101F) without virus and virus without antibody were also ultracentrifuged and tested. (C) HEp-2 cells were infected with the virus-antibody mixtures present in the different pellets. After 48 h, the cells were detached, incubated with a Cy5-labeled anti-F antibody, and analyzed by flow cytometry. Mean fluorescence intensity values are shown in ordinates.
The amounts of virus present in the pellets were also similar, irrespective of the antibody mixed with the virus, and similar to the amount of virus sedimented without antibody (Fig. 3B). Therefore, antibodies had no effect on sedimentation of the virus under the conditions of the experiment shown in Fig. 3. However, as in previous experiments, the pellet infectivity was practically ablated, compared to results with an irrelevant antibody (1B.C11) or no antibody, if HRSV was incubated with 47F or 101F before ultracentrifugation (Fig. 3C). Antibody 2F only marginally reduced virus infectivity in this experiment. It is worth stressing that the effect of MAbs 2F, 47F, and 101F on virus infectivity, shown in Fig. 3C, faithfully reproduces the results presented in Fig. 1 and 2, even though different methodologies to assess viral antigen production were used.
In summary, a stable interaction of antibodies with HRSV virions seems to be required for neutralization of virus infectivity. However, as illustrated by MAb 2F, antibody binding is not sufficient to block virus infectivity, which emphasizes the relevance of antibody specificity for virus neutralization.
MAbs 47F and 101F do not inhibit binding of virus to cells.
Although HRSV binding to cells is mediated mainly by interactions of the G glycoprotein with cell surface proteoglycans, deletion mutants lacking the G protein gene can still infect cells in culture, presumably because the F protein can supersede, to some extent, virus binding to proteoglycans (66, 67). Therefore, it was of interest to test if anti-F neutralizing antibodies had an effect on HRSV binding to cells. To this end, HRSV was incubated with different antibodies before being added to suspensions of HEp-2 cells at 4°C to allow virus adsorption but not membrane fusion. The amount of virus bound to cells was then quantified by flow cytometry using Cy5-labeled anti-F antibodies.
As shown in Fig. 4A, preincubation of virus with 2F, 47F, or 101F antibody had minimal effect on the amount of HRSV bound to HEp-2 cells, in comparison with either an irrelevant antibody (1B.C11) or no antibody. In contrast, preincubation of virus with MAb 63G, directed against HRSV G glycoprotein (28), reduced fluorescence intensity to almost background levels (uninfected cells).
FIG. 4.
Quantification of virus binding to cells and infectivity in the presence of MAbs. (A) Long virus (6.5 × 105 PFU) preincubated in the absence or presence of 30 μg of the antibodies indicated on the figure, was mixed with a suspension of HEp-2 cells for 1 h at 4°C, and the virus bound to cells was detected by flow cytometry using an anti-F Cy5 antibody. (B) Aliquots of the virus-antibody-cell mixtures were replated. Forty-eight hours postinfection, cells were resuspended and labeled with an anti-F Cy5 antibody. Mock-infected cells or cells incubated with the MAb 63G against the attachment (G) glycoprotein or with the MAb 1B.C11 (C−) against a capsid protein of ASPV were used as controls in the experiment.
In parallel, aliquots of the same virus-antibody-cell mixtures were replated to evaluate infectivity 48 h later by flow cytometry with Cy5-labeled anti-F antibody (Fig. 4B). Again, 2F antibody had almost no effect on antigen production since fluorescence values were almost identical to those of negative controls (1B.C11 antibody or no antibody). In contrast, the amount of virus antigen expressed on the surface of cells incubated with either 47F, 101F, or 63G antibody was reduced almost to the values of a mock-infected control. Thus, whereas the 47F and 101F antibodies did not prevent binding of the virus to cells, they efficiently inhibited HRSV infection and therefore antigen production in this type of assay.
MAbs 47F and 101F also inhibit syncytium formation.
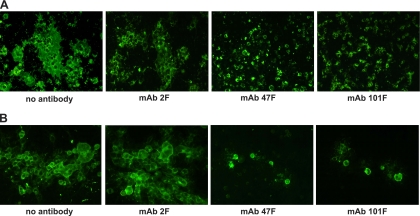
As mentioned in the introduction, the F protein also mediates fusion of the membrane of infected cells to those of adjacent cells to form characteristic syncytia (multinuclear, giant cells). To test if the anti-F MAbs were capable of inhibiting cell-cell fusion, they were tested in a syncytium formation assay done with either transfected cells, expressing the F protein as the only viral protein, or HRSV-infected cells. As shown in Fig. 5A, large multinucleated syncytia were visible by immunofluorescence in BSR-T7/5 cells 48 h after transfection with the plasmid pTM1-F. The presence and size of the syncytia were unaltered if MAb 2F was added to the medium of the transfected cells. In contrast, when neutralizing MAb 47F or 101F was added to the culture, only individual cells were visible, and syncytia were not detected.
FIG. 5.
Inhibition of syncytium formation by anti-F MAbs. (A) BSR-T7/5 cells were transfected with 500 ng of pTM1 plasmid encoding the full-length F gene. The transfection mixture was removed 7 h later, and the MAbs (40 μg/ml) indicated below each panel were added to the culture. Formation of syncytium was evaluated after incubation for 48 h, as described in Materials and Methods. (B) HEp-2 cells were infected with the Long strain of HRSV (multiplicity of infection, 0.1 PFU/cell). The inoculum was removed at 90 min postinfection, and cells were maintained with medium. Five hours later, MAbs were added to the culture. Syncytium formation was examined as described for panel A. The results shown in this figure are representative of three independent experiments.
Similar results were obtained with HEp-2 cells infected with HRSV (Fig. 5B). In this case, the antibodies were added to the culture medium after virus internalization. Large syncytia were seen 48 h after infection in either the absence or presence of MAb 2F. However, addition of MAb 47F or 101F inhibited not only the formation of multinucleated cells but also spreading of the virus to neighboring cells; consequently, only a few infected cells were observed by immunofluorescence when 47F or 101F was added to the cultures. In summary, a strict correlation was found between neutralization and cell-cell fusion inhibition with the antibodies used in this study.
MAb 101F neutralizes viral infectivity and inhibits syncytia even if monovalent.
To further investigate whether bivalent antibody binding was required to neutralize HRSV infectivity, the Fab fragment of the highly neutralizing MAb 101F was prepared (several attempts to prepare 47F Fab were unsuccessful since antigen binding was lost after papain treatment). To test whether bivalency was also required for the partial reduction of antigen production observed with MAb 2F and shown in Fig. 1C, the Fab fragment of this antibody was also prepared.
As shown in Fig. 6A, both MAb 2F and 101F and their Fab fragments bound to a soluble form of the F protein (FTM−) in a direct ELISA. The differences observed in the reactivity of MAbs and Fabs are likely due to differences in their avidities for the F protein.
FIG. 6.
Antigen binding, virus neutralization, and inhibition of syncytium formation with Fab fragments of anti-F MAbs. (A) Serial dilutions of MAbs and Fab fragments were tested for binding to a soluble form of the F protein (FTM−) in a direct ELISA, as described in Materials and Methods. (B) Long virus (6.5 × 103 PFU) was incubated with different amounts of the indicated Fab fragments for 30 min at 37°C. Virus-Fab mixtures were then used to infect HEp-2 cells. After a 72-h incubation, production of viral antigen was quantified by ELISA, as described in Materials and Methods, and results are presented as a percentage of the value for control cells infected in the absence of antibody. Data represent the mean and standard deviation from three independent experiments. (C) HEp-2 cells were infected with the Long strain of HRSV (multiplicity of infection, 0.1 PFU/cell). The inoculum was removed at 90 min postinfection, and cells were maintained with medium. Five hours later, MAbs (40 μg/ml) or Fab fragments (1 mg/ml) were added to the culture. Formation of syncytium was evaluated after incubation for 48 h as described in Materials and Methods. The results are representative of three independent experiments.
Then, the neutralization capacities of 101F and 2F Fabs were tested in the type of assay shown Fig. 1C. The Fab fragment of the MAb 101F (Fab 101F) completely inhibited antigen production if added together with the virus to HEp-2 cells (Fig. 6B) although it was around 50-fold less active, on a molar basis, than the intact antibody (compare Fig. 1C and 6B) (50% infectious dose [IC50]of 0.34 nM for Fab 101F versus 0.006 nM for MAb 101F). It is likely that these quantitative differences in neutralization, as in the case of the ELISA (Fig. 6A), are related to different avidities of MAb and Fab 101F for the F protein. However, the Fab fragment of MAb 2F (Fab 2F) had no significant effect on HRSV infectivity and consequently did not reduce antigen production, even at the highest concentration tested.
Since MAbs 2F and 101F also differed in their capacities to inhibit cell-cell fusion, both Fab 2F and Fab 101F were tested in a syncytium formation assay, in comparison with their respective antibodies. As shown in Fig. 6C, large syncytia were seen in either the absence (data not shown) or presence of MAb 2F or Fab 2F when either was added to infected cells after virus internalization. In contrast, when either MAb 101F or Fab 101F was added to the cell cultures, only individual cells but not syncytia were observed. As seen before in Fig. 5B, inhibition of syncytium formation also restrained spreading of the virus in the cultures.
Since monovalent Fab 101F inhibited viral infectivity and syncytia formation, the results presented here indicate that bivalent binding of that antibody to the F protein is not necessary for neutralization and cell-cell fusion inhibition. In contrast, the partial inhibition of antigen production in infected cells by MAb 2F is not observed when Fab 2F is used in the neutralization test, suggesting that bivalency is required for the partial neutralization by 2F antibody, perhaps by a mechanism related to aggregation of F protein or viral particles.
An HRB F-derived peptide and the organic compound BMS-433771 have to be present during viral adsorption to neutralize HRSV infectivity.
A number of low-molecular-weight compounds, like heptad repeat B (HRB)-derived peptides (42) and benzimidazolone small molecules (15), have been shown to be potent inhibitors of viral fusion. It was of interest to compare neutralizing activity by small molecular compounds with MAbs.
Previously, to confirm the reported neutralizing activity, peptide F478-516 containing a partial sequence of HRB (Fig. 7A) and BMS-433771 were tested in the antigen production assay. Production of viral antigen was totally inhibited by F478-516 (HRB) in the low micromolar range (Fig. 7B). The small molecule BMS-433771 was even more potent than the HRB peptide, on a molar basis, in blocking the production of viral antigen (Fig. 7B).
FIG. 7.
Neutralization of HRSV by HRB F478-516 peptide and BMS-433771. (A) Schematic diagram of the F protein primary structure. The expanded region shows the amino acid sequence of the HRB F-peptide used in the study. (B) Long virus (6.5 × 103 PFU) was incubated with increasing amounts of either HRB F478-516-derived peptide or with BMS-433771 for 30 min at 37°C before being used to infect HEp-2 cells. Production of viral antigen was quantified by ELISA 72 h later, as described in Materials and Methods, and results are presented as a percentage of the value for control cells infected in the absence of inhibitor. Data represent the mean and standard deviation from three independent experiments.
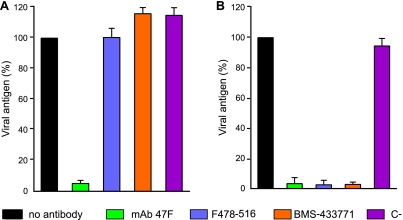
To obtain further insights into the inhibitory action of the HRB peptide and BMS-433771, an excess of these compounds was mixed with a fixed amount of virus, and the virus-inhibitor mixtures were ultracentrifuged in order to remove the excess of inhibitor. After resuspension, the pellets were used to infect cells, and antigen production was measured 72 h later. As seen in Fig. 8A, preincubation of the virus with either the peptide F478-516 (HRB) or BMS-433771, followed by ultracentrifugation, did not block viral infectivity. For comparison, and as shown before (Fig. 2A), the neutralizing MAb 47F ablated viral infectivity if preincubated with the virus before ultracentrifugation (Fig. 8A). However, the ultracentrifuged virus was still sensitive to neutralization by all the inhibitors tested when they were added during viral adsorption to cells (Fig. 8B).
FIG. 8.
Neutralization with MAb 47F, peptide F478-516, and BMS-433771 added before (A) or during (B) HRSV infection. (A) Long virus (8.2 × 105 PFU) was incubated in the absence or presence of 400 μg of MAb 47F, 925 μM F478-516, or 250 μM BMS-433771. Virus-inhibitor mixtures were ultracentrifuged (125,812 × g for 2 h in a Beckman SW60 rotor), and the pellets were resuspended and used to infect HEp-2 cells. After a 72-h incubation, production of viral antigen was quantified by ELISA as described in Materials and Methods, and results are presented as the percentage of values of infected controls without added inhibitor. MAb 1B.C11 against a capsid protein of ASPV was used as a negative control (C−) in neutralization. (B) Long virus (1 × 105 PFU), ultracentrifuged as before in the absence of inhibitor, was used to infect HEp-2 cells in the absence or presence of 3 μg of MAb 47F, 7.4 μM F478-516, or 2 μM BMS-433771. Production of viral antigen was quantified as described for panel A. Data represent the mean and standard deviation from three independent experiments.
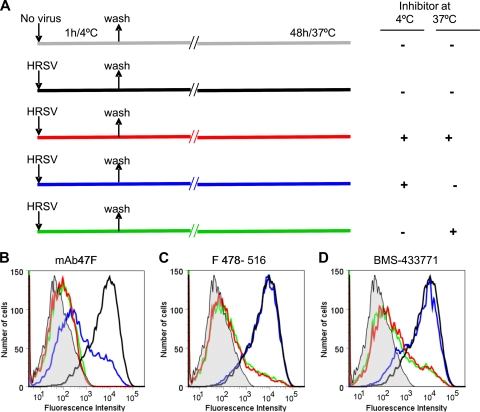
An alternative test for the step at which MAbs and small molecules inhibit virus infectivity is illustrated in Fig. 9A. In this case, virus was allowed to bind to cells for 1 h at 4°C in either the absence or presence of inhibitors. Under these conditions virus can adsorb to cells, but membrane fusion and virus entry are blocked since the F protein is not activated at low temperatures (59). After cultures were washed, they were shifted to 37°C in order to allow membrane fusion to proceed. Virus infectivity, reflected by virus antigen expression at the cell surface, was measured 48 h later by flow cytometry.
FIG. 9.
Effect of MAb 47F, peptide F418-516, and BMS-433771 during adsorption of HRSV to cells at 4°C. The experimental design is shown in panel A. Cultures of HEp-2 cells (90% confluent) were either mock infected (gray line) or infected with the Long strain of HRSV (multiplicity of infection, 0.2 PFU/cell) in the absence or presence of 80 μg of MAb 47F (B), 55 μM peptide F 478-516 (C), or 3 μM BMS-433771 (D) and incubated for 1 h at 4°C. The inoculum was subsequently removed, and after cultures were washed with DMEM-2.5% FCS, they were shifted up to 37°C either in the absence or presence of the same amounts of the corresponding inhibitor for another 48 h. At this time, cells were resuspended, and the amount of virus antigen present at the cell surface was quantified by flow cytometry as detailed in the legend of Fig. 4B.
As seen in Fig. 9B, addition of MAb 47F to cultures during virus binding at 4°C was enough to reduce antigen production essentially to the background level of mock-infected cells. This inhibition was slightly more effective if the MAb was added back to the medium after the adsorption period (Fig. 9B, red line) than if it was removed after the incubation at 4°C (blue line). Inhibition was also effective if MAb 47F was not present during virus binding but was added to the cultures when the incubation temperature shifted to 37°C (Fig. 9B, green line). Thus, the presence of MAb 47F during virus binding at 4°C prevents HRSV infection at higher temperatures even though the antibody does not inhibit virus binding at 4°C, as shown in Fig. 4A. Therefore, and in agreement with the results of previous sections, the inhibitory effect of MAb 47F is exerted on the virus before activation of the F protein for fusion.
In clear contrast with these results, addition of either peptide F478-516 or BMS-433771 to the cultures during virus binding at 4°C had minimal effect on virus infectivity (Fig. 9C and D, blue lines). However, these two low-molecular-weight inhibitors substantially reduced infectivity if present after shifting of the cultures to 37°C, irrespective of having been present (red lines) or absent (green lines) during the adsorption period. Therefore, the peptide F478-516 and the compound BMS-433771 need to be present during the active process of virus entry to exhibit their inhibitory activity. Thus, the results shown in Fig. 8 and 9 suggest that the low-molecular-weight inhibitors do not act on the prefusion F protein but probably bind and block some of the F protein intermediates that are formed during the process of membrane fusion.
DISCUSSION
Inhibitors of the HRSV F glycoprotein, such as palivizumab (Synagis), a humanized MAb (32), and low-molecular-weight compounds (2, 15, 52), have been the subject of intense research in recent years. Knowledge of their mode of action should contribute to improve their effectiveness and clinical use.
Although differences in the neutralizing capacities of anti-F antibodies have been reported previously (5, 28), the reason for such differences remained unknown. The fact that antibodies binding to epitopes which are altered in the same escape mutants differed in their neutralizing activity suggested that subtle differences in antigen binding may have a major impact in antibody-mediated neutralization (45). For instance, it was reported that virus binding of MAbs 47F and 49F was affected similarly by amino acid changes selected in mutants resistant to 47F. Both antibodies also competed for simultaneous binding to virus preparations, suggesting that they recognized overlapping epitopes, yet whereas MAb 47F neutralized HRSV efficiently, MAb 49F had no effect on virus infectivity (28). The results reported here emphasize the idea of epitope specificity for HRSV neutralization. While MAbs 2F, 47F, and 101F bound similarly to a soluble form of the F protein (FTM−) and comparable amounts of the three antibodies remained bound to the Long virus after ultracentrifugation, only 47F and 101F antibodies inactivated virus infectivity. This contrasts with the “coating theory” of virus neutralization which proposes that antibody coating of virions is sufficient to inhibit infectivity (54). Instead, the interaction of antibodies with specific residues of the F protein seems to be the mechanism for effective neutralization.
It is known that MAbs 2F, 47F, and 101F bind to different regions of the cone-shaped molecules of an anchorless form of the F protein (FTM−), probably folded in the postfusion conformation (12; also data not shown). It is conceivable that these antibodies could also bind to the prefusion form of the F protein which is present in virions. Figure 10 shows a model of the HRSV F prefusion structure built with the atomic coordinates of the structure determined for the parainfluenza virus 5 (PIV5) F homologue (78). In this model, the only known residue that affects MAb 2F binding (Fig. 10, red sphere) sits at the base of the globular head, whereas residues that alter reactivity with either MAb 47F (green spheres) or MAb 101F (blue spheres) are placed on the side of the head, not far from the fusion peptide (shown in magenta) and the two cleavage sites (red arrow, site I; black arrow, site II; shown in one monomer). Residues of antigenic site II (Fig. 10, green spheres) are also proximal in this model to sequences (shown in black) that form the central α-helical coil of the 6HB in the postfusion conformation of the F protein. Therefore, and with this model in mind, it is feasible to envisage that binding of MAbs to antigenic sites II or IV may have a greater impact on inhibition of some of the changes related to F protein activation (proteolytic cleavage, exposure of the fusion peptide, or formation of the HRA α-helix) than binding of MAbs to antigenic site I. In other words, binding of MAb 47F or 101F, but not 2F, to prefusion F would interfere with activation of this protein when the virus attaches to the target cell surface, where activation occurs (41). Although our data favor this mechanism of virus neutralization by anti-F antibodies, cone-shaped F protein spikes have been observed abundantly in purified particles of PIV5 (47). Therefore, it cannot be excluded that binding of neutralizing MAbs to F spikes in virions may have an indirect effect on activation of the prefusion form in the event that they coexist in the same virus particle. It will be difficult to distinguish between these two possibilities until a purified prefusion form of the F protein is available to test antibody binding.
FIG. 10.
Three-dimensional model of the HRSV F protein prefusion conformation. The model represents the proposed prefusion conformation of the HRSV F trimer, built using the SWISS-MODEL server facilities (http://swissmodel.expasy.org/) and the atomic coordinates of the prefusion structure of the PIV5 F protein (Protein Data Bank code, 2B9B) as a template (78). The backbone structure of the three monomers is shown in gray. Fusion peptide sequences of one monomer are shown in pink, and those of HRA are shown in black. Residues that are changed in virus isolates or in escape mutants selected with monoclonal antibodies, whose epitopes map in different antigenic sites of the F protein, are shown as colored spheres (antigenic site I, amino acid 389; antigenic site II, amino acids 262, 268, 272, and 275; and antigenic site IV, amino acids 429, 432, 433, 436, and 447). The two proteolytic cleavage sites are indicated with arrows in one of the monomers, colored as described in the legend of Fig. 1.
In any case, the result of binding of neutralizing anti-F antibodies to virus particles is inactivation of virus infectivity even though interaction of the virus-antibody complexes with cells is not altered (Fig. 4). Neutralization of virus infectivity correlated with inhibition of cell-cell fusion, as manifested in a syncytium formation assay (Fig. 5). Antibody bivalency was not required for the inhibitory activity of 101F, thus stressing the relevance of specific interactions between F and antibodies for neutralization. In contrast, the partial neutralization exhibited by MAb 2F when added to the culture medium (Fig. 1) required bivalency and was not reflected in the syncytium formation assay. Antibody 2F, as many other nonneutralizing anti-F antibodies (not shown), may reduce virus infectivity by cross-linking of F molecules in the same virus particle or by aggregation of virions (58). This inhibitory effect would be lost with monovalent Fab fragments and may not be observed in a cell-cell fusion assay due to different topologies of F molecules in the surfaces of virions and in infected/transfected cells.
Although most neutralizing antibodies targeted against enveloped viruses interfere with binding of virus to specific receptors, antibodies inhibiting virus-cell membrane fusion have also been found. For instance, two laboratories reported recently the isolation of cross-neutralizing antibodies directed against the influenza virus hemagglutinin (HA) (64, 69). While most neutralizing anti-HA antibodies interfered with receptor binding (39) and neutralized a restricted set of viral strains, the cross-neutralizing anti-HA antibodies block infection by binding to a conserved pocket near the fusion peptide (64), thus preventing membrane fusion. Whether antibodies 47F and 101F operate in a similar manner is an appealing possibility that cannot be tested until a detailed structure of the HRSV F protein is available.
The mode of action of neutralizing anti-F antibodies contrasts with that of the HRB peptide (F478-516) or the BMS-433771 compound. Incubation of virus with these low-molecular-weight inhibitors before ultracentrifugation (Fig. 8) or during the step of virus binding to cells at 4°C (Fig. 9) had no effect on infectivity. It has been reported that an HRB peptide of the PIV5 fusion protein can bind to a prehairpin intermediate of F, preventing the formation of the six-helix bundle and completion of membrane fusion (59). Consistent with this mode of action, the peptide F478-516 was ineffective against HRSV unless it was present during membrane fusion, presumably when the prehairpin intermediate was available.
Regarding BMS-433771, Cianci et al. (14) probed the interaction of a photoaffinity analog of this compound with HRSV virions, showing that it could be linked covalently to Tyr198. This residue is part of a hydrophobic cavity in the six-helix bundle of the postfusion F protein core structure (14). The results shown in Fig. 8 and 9 are in apparent contradiction with those of Cianci et al., (14) since incubation of HRSV with BMS-433771 before ultracentrifugation or during virus adsorption to cells at 4°C had no effect on infectivity. Resolution of this apparent disagreement requires the availability of homogeneous preparations of the prefusion HRSV F protein to test binding of BMS-433771 without the ambiguity of having mixtures of pre- and postfusion conformations in the same virus preparation. Nevertheless, our results favor the idea that, to be effective, BMS-433771 has to interact with an F intermediate before completion of membrane fusion.
In summary, neutralizing antibodies and low-molecular-weight compounds inhibit virus entry by acting at different steps of the membrane fusion process. In addition to expanding our view of their mode of action, the results presented here offer the possibility of testing a combined therapy of HRSV infections, increasing its effectiveness, and reducing the risk of selecting viruses resistant to monotherapy.
Acknowledgments
We thank K. Chappell for critical reading of the manuscript and English corrections and O. Cano for preparing the purified FTM− protein. We also thank Biomol-Informatics SL (Madrid, Spain) for bioinformatics advice.
This work was supported by grant SAF2009-11632 (to J.A.M.) from Ministerio de Ciencia e Innovación and BIO2008-04487-CO3-02 (to D.A.). The Biología Viral laboratory is part of the VIRHOST consortium funded by Comunidad de Madrid.
Footnotes
Published ahead of print on 9 June 2010.
REFERENCES
- 1.Anderson, L. J., P. Bingham, and J. C. Hierholzer. 1988. Neutralization of respiratory syncytial virus by individual and mixtures of F and G protein monoclonal antibodies. J. Virol. 62:4232-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andries, K., M. Moeremans, T. Gevers, R. Willebrords, C. Sommen, J. Lacrampe, F. Janssens, and P. R. Wyde. 2003. Substituted benzimidazoles with nanomolar activity against respiratory syncytial virus. Antiviral Res. 60:209-219. [DOI] [PubMed] [Google Scholar]
- 3.Arbiza, J., G. Taylor, J. A. Lopez, J. Furze, S. Wyld, P. Whyte, E. J. Stott, G. Wertz, W. Sullender, M. Trudel, et al. 1992. Characterization of two antigenic sites recognized by neutralizing monoclonal antibodies directed against the fusion glycoprotein of human respiratory syncytial virus. J. Gen. Virol. 73:2225-2234. [DOI] [PubMed] [Google Scholar]
- 4.Baker, K. A., R. E. Dutch, R. A. Lamb, and T. S. Jardetzky. 1999. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell 3:309-319. [DOI] [PubMed] [Google Scholar]
- 5.Beeler, J. A., and K. van Wyke Coelingh. 1989. Neutralization epitopes of the F glycoprotein of respiratory syncytial virus: effect of mutation upon fusion function. J. Virol. 63:2941-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Begoña Ruiz-Arguello, M., L. Gonzalez-Reyes, L. J. Calder, C. Palomo, D. Martin, M. J. Saiz, B. Garcia-Barreno, J. J. Skehel, and J. A. Melero. 2002. Effect of proteolytic processing at two distinct sites on shape and aggregation of an anchorless fusion protein of human respiratory syncytial virus and fate of the intervening segment. Virology 298:317-326. [DOI] [PubMed] [Google Scholar]
- 7.Bembridge, G. P., J. A. Lopez, R. Bustos, J. A. Melero, R. Cook, H. Mason, and G. Taylor. 1999. Priming with a secreted form of the fusion protein of respiratory syncytial virus (RSV) promotes interleukin-4 (IL-4) and IL-5 production but not pulmonary eosinophilia following RSV challenge. J. Virol. 73:10086-10094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bembridge, G. P., J. A. Lopez, R. Cook, J. A. Melero, and G. Taylor. 1998. Recombinant vaccinia virus coexpressing the F protein of respiratory syncytial virus (RSV) and interleukin-4 (IL-4) does not inhibit the development of RSV-specific memory cytotoxic T lymphocytes, whereas priming is diminished in the presence of high levels of IL-2 or gamma interferon. J. Virol. 72:4080-4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bourgeois, C., C. Corvaisier, J. B. Bour, E. Kohli, and P. Pothier. 1991. Use of synthetic peptides to locate neutralizing antigenic domains on the fusion protein of respiratory syncytial virus. J. Gen. Virol. 72:1051-1058. [DOI] [PubMed] [Google Scholar]
- 10.Brioen, P., B. Rombaut, and A. Boeye. 1985. Hit-and-run neutralization of poliovirus. J. Gen. Virol. 66:2495-2499. [DOI] [PubMed] [Google Scholar]
- 11.Buchholz, U. J., S. Finke, and K. K. Conzelmann. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calder, L. J., L. Gonzalez-Reyes, B. Garcia-Barreno, S. A. Wharton, J. J. Skehel, D. C. Wiley, and J. A. Melero. 2000. Electron microscopy of the human respiratory syncytial virus fusion protein and complexes that it forms with monoclonal antibodies. Virology 271:122-131. [DOI] [PubMed] [Google Scholar]
- 13.Cianci, C., E. V. Genovesi, L. Lamb, I. Medina, Z. Yang, L. Zadjura, H. Yang, C. D'Arienzo, N. Sin, K. L. Yu, K. Combrink, Z. Li, R. Colonno, N. Meanwell, J. Clark, and M. Krystal. 2004. Oral efficacy of a respiratory syncytial virus inhibitor in rodent models of infection. Antimicrob. Agents Chemother. 48:2448-2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cianci, C., D. R. Langley, D. D. Dischino, Y. Sun, K. L. Yu, A. Stanley, J. Roach, Z. Li, R. Dalterio, R. Colonno, N. A. Meanwell, and M. Krystal. 2004. Targeting a binding pocket within the trimer-of-hairpins: small-molecule inhibition of viral fusion. Proc. Natl. Acad. Sci. U. S. A. 101:15046-15051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cianci, C., K. L. Yu, K. Combrink, N. Sin, B. Pearce, A. Wang, R. Civiello, S. Voss, G. Luo, K. Kadow, E. V. Genovesi, B. Venables, H. Gulgeze, A. Trehan, J. James, L. Lamb, I. Medina, J. Roach, Z. Yang, L. Zadjura, R. Colonno, J. Clark, N. Meanwell, and M. Krystal. 2004. Orally active fusion inhibitor of respiratory syncytial virus. Antimicrob. Agents Chemother. 48:413-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collins, P. L., and J. E. Crowe. 2007. Respiratory syncytial virus and metapneumovirus, p. 1601-1646. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 17.Collins, P. L., and G. Mottet. 1992. Oligomerization and post-translational processing of glycoprotein G of human respiratory syncytial virus: altered O-glycosylation in the presence of brefeldin A. J. Gen. Virol. 73:849-863. [DOI] [PubMed] [Google Scholar]
- 18.Collins, P. L., and G. Mottet. 1991. Post-translational processing and oligomerization of the fusion glycoprotein of human respiratory syncytial virus. J. Gen. Virol. 72:3095-3101. [DOI] [PubMed] [Google Scholar]
- 19.Connors, M., P. L. Collins, C. Y. Firestone, and B. R. Murphy. 1991. Respiratory syncytial virus (RSV) F, G, M2 (22K), and N proteins each induce resistance to RSV challenge, but resistance induced by M2 and N proteins is relatively short-lived. J. Virol. 65:1634-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 21.Elroy-Stein, O., T. R. Fuerst, and B. Moss. 1989. Cap-independent translation of mRNA conferred by encephalomyocarditis virus 5′ sequence improves the performance of the vaccinia virus/bacteriophage T7 hybrid expression system. Proc. Natl. Acad. Sci. U. S. A. 86:6126-6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Englund, J. A., L. J. Anderson, and F. S. Rhame. 1991. Nosocomial transmission of respiratory syncytial virus in immunocompromised adults. J. Clin. Microbiol. 29:115-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Escribano-Romero, E., J. Rawling, B. Garcia-Barreno, and J. A. Melero. 2004. The soluble form of human respiratory syncytial virus attachment protein differs from the membrane-bound form in its oligomeric state but is still capable of binding to cell surface proteoglycans. J. Virol. 78:3524-3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Falsey, A. R., P. A. Hennessey, M. A. Formica, C. Cox, and E. E. Walsh. 2005. Respiratory syncytial virus infection in elderly and high-risk adults. N. Engl. J. Med. 352:1749-1759. [DOI] [PubMed] [Google Scholar]
- 25.Falsey, A. R., and E. E. Walsh. 1998. Relationship of serum antibody to risk of respiratory syncytial virus infection in elderly adults. J. Infect. Dis. 177:463-466. [DOI] [PubMed] [Google Scholar]
- 26.Falsey, A. R., and E. E. Walsh. 2000. Respiratory syncytial virus infection in adults. Clin. Microbiol. Rev. 13:371-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garcia-Barreno, B., T. Delgado, B. Akerlind-Stopner, E. Norrby, and J. A. Melero. 1992. Location of the epitope recognized by monoclonal antibody 63G on the primary structure of human respiratory syncytial virus G glycoprotein and the ability of synthetic peptides containing this epitope to induce neutralizing antibodies. J. Gen. Virol. 73:2625-2630. [DOI] [PubMed] [Google Scholar]
- 28.Garcia-Barreno, B., C. Palomo, C. Peñas, T. Delgado, P. Perez-Breña, and J. A. Melero. 1989. Marked differences in the antigenic structure of human respiratory syncytial virus F and G glycoproteins. J. Virol. 63:925-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glezen, W. P., L. H. Taber, A. L. Frank, and J. A. Kasel. 1986. Risk of primary infection and reinfection with respiratory syncytial virus. Am. J. Dis. Child. 140:543-546. [DOI] [PubMed] [Google Scholar]
- 30.Gonzalez-Reyes, L., M. B. Ruiz-Arguello, B. Garcia-Barreno, L. Calder, J. A. Lopez, J. P. Albar, J. J. Skehel, D. C. Wiley, and J. A. Melero. 2001. Cleavage of the human respiratory syncytial virus fusion protein at two distinct sites is required for activation of membrane fusion. Proc. Natl. Acad. Sci. U. S. A. 98:9859-9864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Graham, B. S., T. H. Davis, Y. W. Tang, and W. C. Gruber. 1993. Immunoprophylaxis and immunotherapy of respiratory syncytial virus-infected mice with respiratory syncytial virus-specific immune serum. Pediatr. Res. 34:167-172. [DOI] [PubMed] [Google Scholar]
- 32.Groothuis, J. R., and H. Nishida. 2002. Prevention of respiratory syncytial virus infections in high-risk infants by monoclonal antibody (palivizumab). Pediatr. Int. 44:235-241. [DOI] [PubMed] [Google Scholar]
- 33.Groothuis, J. R., E. A. Simoes, M. J. Levin, C. B. Hall, C. E. Long, W. J. Rodriguez, J. Arrobio, H. C. Meissner, D. R. Fulton, R. C. Welliver, et al. 1993. Prophylactic administration of respiratory syncytial virus immune globulin to high-risk infants and young children. The Respiratory Syncytial Virus Immune Globulin Study Group. N. Engl. J. Med. 329:1524-1530. [DOI] [PubMed] [Google Scholar]
- 34.Hall, C. B., E. E. Walsh, C. E. Long, and K. C. Schnabel. 1991. Immunity to and frequency of reinfection with respiratory syncytial virus. J. Infect. Dis. 163:693-698. [DOI] [PubMed] [Google Scholar]
- 35.Hallak, L. K., D. Spillmann, P. L. Collins, and M. E. Peeples. 2000. Glycosaminoglycan sulfation requirements for respiratory syncytial virus infection. J. Virol. 74:10508-10513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henderson, F. W., W. A. Clyde, Jr., A. M. Collier, F. W. Denny, R. J. Senior, C. I. Sheaffer, W. G. Conley III, and R. M. Christian. 1979. The etiologic and epidemiologic spectrum of bronchiolitis in pediatric practice. J. Pediatr. 95:183-190. [DOI] [PubMed] [Google Scholar]
- 37.IMpact-RSV Study Group. 1998. Palivizumab, a humanized respiratory syncytial virus monoclonal antibody, reduces hospitalization from respiratory syncytial virus infection in high-risk infants. Pediatrics 102:531-537. [PubMed] [Google Scholar]
- 38.Johnson, S., C. Oliver, G. A. Prince, V. G. Hemming, D. S. Pfarr, S. C. Wang, M. Dormitzer, J. O'Grady, S. Koenig, J. K. Tamura, R. Woods, G. Bansal, D. Couchenour, E. Tsao, W. C. Hall, and J. F. Young. 1997. Development of a humanized monoclonal antibody (MEDI-493) with potent in vitro and in vivo activity against respiratory syncytial virus. J. Infect. Dis. 176:1215-1224. [DOI] [PubMed] [Google Scholar]
- 39.Knossow, M., and J. J. Skehel. 2006. Variation and infectivity neutralization in influenza. Immunology 119:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krusat, T., and H. J. Streckert. 1997. Heparin-dependent attachment of respiratory syncytial virus (RSV) to host cells. Arch. Virol. 142:1247-1254. [DOI] [PubMed] [Google Scholar]
- 41.Lamb, R. A. 1993. Paramyxovirus fusion: a hypothesis for changes. Virology 197:1-11. [DOI] [PubMed] [Google Scholar]
- 42.Lambert, D. M., S. Barney, A. L. Lambert, K. Guthrie, R. Medinas, D. E. Davis, T. Bucy, J. Erickson, G. Merutka, and S. R. Petteway, Jr. 1996. Peptides from conserved regions of paramyxovirus fusion (F) proteins are potent inhibitors of viral fusion. Proc. Natl. Acad. Sci. U. S. A. 93:2186-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lawless-Delmedico, M. K., P. Sista, R. Sen, N. C. Moore, J. B. Antczak, J. M. White, R. J. Greene, K. C. Leanza, T. J. Matthews, and D. M. Lambert. 2000. Heptad-repeat regions of respiratory syncytial virus F1 protein form a six-membered coiled-coil complex. Biochemistry 39:11684-11695. [DOI] [PubMed] [Google Scholar]
- 44.Levine, S., R. Klaiber-Franco, and P. R. Paradiso. 1987. Demonstration that glycoprotein G is the attachment protein of respiratory syncytial virus. J. Gen. Virol. 68:2521-2524. [DOI] [PubMed] [Google Scholar]
- 45.Lopez, J. A., R. Bustos, C. Orvell, M. Berois, J. Arbiza, B. Garcia-Barreno, and J. A. Melero. 1998. Antigenic structure of human respiratory syncytial virus fusion glycoprotein. J. Virol. 72:6922-6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lopez, J. A., C. Peñas, B. Garcia-Barreno, J. A. Melero, and A. Portela. 1990. Location of a highly conserved neutralizing epitope in the F glycoprotein of human respiratory syncytial virus. J. Virol. 64:927-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ludwig, K., B. Schade, C. Bottcher, T. Korte, N. Ohlwein, B. Baljinnyam, M. Veit, and A. Herrmann. 2008. Electron cryomicroscopy reveals different F1+F2 protein states in intact parainfluenza virions. J. Virol. 82:3775-3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinez, I., J. Dopazo, and J. A. Melero. 1997. Antigenic structure of the human respiratory syncytial virus G glycoprotein and relevance of hypermutation events for the generation of antigenic variants. J. Gen. Virol. 78:2419-2429. [DOI] [PubMed] [Google Scholar]
- 49.Martinez, I., and J. A. Melero. 2000. Binding of human respiratory syncytial virus to cells: implication of sulfated cell surface proteoglycans. J. Gen. Virol. 81:2715-2722. [DOI] [PubMed] [Google Scholar]
- 50.Martinez, I., and J. A. Melero. 1998. Enhanced neutralization of human respiratory syncytial virus by mixtures of monoclonal antibodies to the attachment (G) glycoprotein. J. Gen. Virol. 79:2215-2220. [DOI] [PubMed] [Google Scholar]
- 51.Matthews, J. M., T. F. Young, S. P. Tucker, and J. P. Mackay. 2000. The core of the respiratory syncytial virus fusion protein is a trimeric coiled coil. J. Virol. 74:5911-5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McKimm-Breschkin, J. 2000. VP-14637 ViroPharma. Curr. Opin. Investig. Drugs 1:425-427. [PubMed] [Google Scholar]
- 53.Melero, J. A. 2007. Molecular biology of human respiratory syncytial virus, p. 1-42. In P. A. Cane (ed.), Respiratory syncytial virus: perspectives in medical virology. Elsevier, Amsterdam, Netherlands.
- 54.Parren, P. W., I. Mondor, D. Naniche, H. J. Ditzel, P. J. Klasse, D. R. Burton, and Q. J. Sattentau. 1998. Neutralization of human immunodeficiency virus type 1 by antibody to gp120 is determined primarily by occupancy of sites on the virion irrespective of epitope specificity. J. Virol. 72:3512-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Powell, K. L., and D. Alber. 2007. Development of antivirals against respiratory syncytial virus, p. 279-297. In P. A. Cane (ed.), Respiratory syncytial virus: perspectives in medical virology. Elsevier, Amsterdam, Netherlands.
- 56.Prince, G. A., V. G. Hemming, R. L. Horswood, and R. M. Chanock. 1985. Immunoprophylaxis and immunotherapy of respiratory syncytial virus infection in the cotton rat. Virus Res. 3:193-206. [DOI] [PubMed] [Google Scholar]
- 57.Provencal, D. P., K. D. Gesenberg, H. Wang, C. Escobar, H. Wong, M. A. Brown, A. J. Staab, and Y. R. Pendri. 2004. Development of an efficient and scalable process of a respiratory syncytial virus inhibitor. Org. Process Res. Dev. 8:903-908. [Google Scholar]
- 58.Reading, S. A., and N. J. Dimmock. 2007. Neutralization of animal virus infectivity by antibody. Arch. Virol. 152:1047-1059. [DOI] [PubMed] [Google Scholar]
- 59.Russell, C. J., T. S. Jardetzky, and R. A. Lamb. 2001. Membrane fusion machines of paramyxoviruses: capture of intermediates of fusion. EMBO J. 20:4024-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Russell, C. J., and L. E. Luque. 2006. The structural basis of paramyxovirus invasion. Trends Microbiol. 14:243-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sanz, A., B. Garcia-Barreno, M. L. Nogal, E. Viñuela, and L. Enjuanes. 1985. Monoclonal antibodies specific for African swine fever virus proteins. J. Virol. 54:199-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sastre, P., J. A. Melero, B. Garcia-Barreno, and C. Palomo. 2005. Comparison of affinity chromatography and adsorption to vaccinia virus recombinant infected cells for depletion of antibodies directed against respiratory syncytial virus glycoproteins present in a human immunoglobulin preparation. J. Med. Virol. 76:248-255. [DOI] [PubMed] [Google Scholar]
- 63.Stott, E. J., G. Taylor, L. A. Ball, K. Anderson, K. K. Young, A. M. King, and G. W. Wertz. 1987. Immune and histopathological responses in animals vaccinated with recombinant vaccinia viruses that express individual genes of human respiratory syncytial virus. J. Virol. 61:3855-3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sui, J., W. C. Hwang, S. Perez, G. Wei, D. Aird, L. M. Chen, E. Santelli, B. Stec, G. Cadwell, M. Ali, H. Wan, A. Murakami, A. Yammanuru, T. Han, N. J. Cox, L. A. Bankston, R. O. Donis, R. C. Liddington, and W. A. Marasco. 2009. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct. Mol. Biol. 16:265-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Taylor, G., E. J. Stott, M. Bew, B. F. Fernie, P. J. Cote, A. P. Collins, M. Hughes, and J. Jebbett. 1984. Monoclonal antibodies protect against respiratory syncytial virus infection in mice. Immunology 52:137-142. [PMC free article] [PubMed] [Google Scholar]
- 66.Techaarpornkul, S., N. Barretto, and M. E. Peeples. 2001. Functional analysis of recombinant respiratory syncytial virus deletion mutants lacking the small hydrophobic and/or attachment glycoprotein gene. J. Virol. 75:6825-6834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Techaarpornkul, S., P. L. Collins, and M. E. Peeples. 2002. Respiratory syncytial virus with the fusion protein as its only viral glycoprotein is less dependent on cellular glycosaminoglycans for attachment than complete virus. Virology 294:296-304. [DOI] [PubMed] [Google Scholar]
- 68.Thomas, L. H., R. S. Cook, S. G. Wyld, J. M. Furze, and G. Taylor. 1998. Passive protection of gnotobiotic calves using monoclonal antibodies directed at different epitopes on the fusion protein of bovine respiratory syncytial virus. J. Infect. Dis. 177:874-880. [DOI] [PubMed] [Google Scholar]
- 69.Throsby, M., E. van den Brink, M. Jongeneelen, L. L. Poon, P. Alard, L. Cornelissen, A. Bakker, F. Cox, E. van Deventer, Y. Guan, J. Cinatl, J. ter Meulen, I. Lasters, R. Carsetti, M. Peiris, J. de Kruif, and J. Goudsmit. 2008. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS One 3:e3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Walsh, E. E., C. B. Hall, M. Briselli, M. W. Brandriss, and J. J. Schlesinger. 1987. Immunization with glycoprotein subunits of respiratory syncytial virus to protect cotton rats against viral infection. J. Infect. Dis. 155:1198-1204. [DOI] [PubMed] [Google Scholar]
- 71.Walsh, E. E., C. B. Hall, J. J. Schlesinger, M. W. Brandriss, S. Hildreth, and P. Paradiso. 1989. Comparison of antigenic sites of subtype-specific respiratory syncytial virus attachment proteins. J. Gen. Virol. 70:2953-2961. [DOI] [PubMed] [Google Scholar]
- 72.Walsh, E. E., and J. Hruska. 1983. Monoclonal antibodies to respiratory syncytial virus proteins: identification of the fusion protein. J. Virol. 47:171-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Walsh, E. E., J. J. Schlesinger, and M. W. Brandriss. 1984. Protection from respiratory syncytial virus infection in cotton rats by passive transfer of monoclonal antibodies. Infect. Immun. 43:756-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Watt, P. J., B. S. Robinson, C. R. Pringle, and D. A. Tyrrell. 1990. Determinants of susceptibility to challenge and the antibody response of adult volunteers given experimental respiratory syncytial virus vaccines. Vaccine 8:231-236. [DOI] [PubMed] [Google Scholar]
- 75.Wertz, G. W., P. L. Collins, Y. Huang, C. Gruber, S. Levine, and L. A. Ball. 1985. Nucleotide sequence of the G protein gene of human respiratory syncytial virus reveals an unusual type of viral membrane protein. Proc. Natl. Acad. Sci. U. S. A. 82:4075-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu, S. J., A. Schmidt, E. J. Beil, N. D. Day, P. J. Branigan, C. Liu, L. L. Gutshall, C. Palomo, J. Furze, G. Taylor, J. A. Melero, P. Tsui, A. M. Del Vecchio, and M. Kruszynski. 2007. Characterization of the epitope for anti-human respiratory syncytial virus F protein monoclonal antibody 101F using synthetic peptides and genetic approaches. J. Gen. Virol. 88:2719-2723. [DOI] [PubMed] [Google Scholar]
- 77.Yin, H. S., R. G. Paterson, X. Wen, R. A. Lamb, and T. S. Jardetzky. 2005. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc. Natl. Acad. Sci. U. S. A. 102:9288-9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yin, H. S., X. Wen, R. G. Paterson, R. A. Lamb, and T. S. Jardetzky. 2006. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature 439:38-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhao, X., M. Singh, V. N. Malashkevich, and P. S. Kim. 2000. Structural characterization of the human respiratory syncytial virus fusion protein core. Proc. Natl. Acad. Sci. U. S. A. 97:14172-14177. [DOI] [PMC free article] [PubMed] [Google Scholar]