Abstract
Noroviruses (NoVs) are recognized as the leading cause of epidemic and sporadic acute gastroenteritis. Early detection of NoV is crucial to control the spread of the disease. In this study, we evaluated the diagnostic accuracy, analytical sensitivity, and analytical reactivity of the IDEIA Norovirus assay (an enzyme immunoassay [EIA]) in a prospective and retrospective study design. A total of 557 prospectively collected fecal samples and a panel of 97 archived fecal samples, including 21 different GI and GII genotypes, were tested by conventional reverse transcription-PCR (RT-PCR)/bidirectional sequencing, real-time RT-PCR, and electron microscopy. The sensitivity and specificity of the EIA were 57.6% and 91.9%, respectively. The sensitivity for detecting NoV in fecal samples from outbreaks improved from 44.1% when three samples were tested to 76.9% when five samples per outbreak were tested. The EIA was able to detect strains from 7 GI and 11 GII genotypes. The analytical sensitivity of the EIA was 3.1 × 106 and 1.6 × 107 virus particles g−1 of fecal sample for NoV GI and GII strains, respectively. Most GII samples positive by EIA had a threshold cycle (CT) of <26.5, and 50% of the GII samples negative by EIA had a CT of >25.6, suggesting that, although strains from genotypes GI.8, GII.10, and GII.16 were not detected, the low sensitivity of the EIA is primarily caused by low virus concentration. In conclusion, the current EIA may be of use as a rapid screening test during a norovirus outbreak investigation when multiple fecal samples are available; however, sporadic samples should be tested by molecular methods.
Noroviruses (NoVs) cause approximately 23 million infections each year in the United States, accounting for 60% of the disease burden caused by known enteric pathogens (35). They are the leading cause of epidemics of gastroenteritis worldwide (39) and an important cause of sporadic gastroenteritis illnesses in children and adults (40). Major factors for the public health impact of NoVs include: (i) great strain diversity (five genogroups [GI to -V] and 32 genotypes) (33, 52, 53), (ii) low infectious dose (as low as 18 particles) (46), (iii) prolonged asymptomatic shedding (3), (iv) environmental stability (10), and (v) the lack of long-term immunity (38, 46). NoV outbreaks often occur in semiclosed environments that favor person-to-person transmission, including long-term care facilities, cruise ships, prisons, hospitals, and recreational facilities (6, 9, 20, 23, 24, 36, 41, 45, 49).
To date, only strains of GI, GII, and GIV have been detected in humans (53), and although several genotypes may cocirculate in the community, GII.4 strains have been responsible for the majority of the outbreaks since the mid 1990s (54). Recent studies demonstrated that GII.4 strains have evolved over time into new emerging variants that have replaced previously dominant variant strains (4, 32, 43, 53).
Experimental human infection with NoV has shown that virus shedding may precede clinical symptoms (GI.1 viral load peak range, 108 to 10 12 copies g−1 of stool) and continues for several weeks in the presence or absence of clinical symptoms (3). However, during outbreaks lower viral loads (GI, 104 to 108; GII, 104 to 1010) are typically found (37).
Early detection of NoV is crucial to control the spread of disease in outbreaks (15, 21). Since the discovery of Norwalk virus in 1972 by electron microscopy (EM), followed by its cloning and sequencing in 1990 (25), detection of NoV has evolved from a conventional reverse transcription-PCR (RT-PCR) assay in the mid 1990s (2, 51) to real-time RT-PCR (rRT-PCR) assays, which have become the method in most clinical laboratories for the diagnosis of NoV (26, 30, 47). These molecular techniques, combined with sequence analysis, have resulted in a better understanding of the role of NoV as the leading cause of epidemic gastroenteritis and an important cause of sporadic gastroenteritis (18, 40). Several enzyme immunoassays (EIAs) for rapid detection of NoV (5, 12, 13) have been developed recently and are commercially available in Europe, Japan, and Australia. These EIAs can be applied in large-scale clinical and epidemiological studies. To overcome the great antigenic diversity of NoV strains and to allow for detection of new strains, EIAs require antibodies that cross-react between NoV genogroups and/or genotypes. Current EIAs are based on polyclonal or monoclonal antibodies raised against a panel of different virus-like particles (VLPs). Previous studies have demonstrated that the sensitivity and specificity of NoV EIAs vary with the diagnostic goal (outbreak or sporadic cases) and test design (19), which raises questions about their usefulness for routine screening of samples.
In this study, we determined the diagnostic accuracy of a commercial NoV EIA and assessed its analytical sensitivity and reactivity by comparing the obtained results with rRT-PCR, EM, and conventional RT-PCR results. Prospective samples from three sites as well as samples from a comprehensive panel of archived NoV genotypes were evaluated.
MATERIALS AND METHODS
Study design.
To evaluate the EIA, the IDEIA Norovirus kit (Oxoid Ltd., Ely, United Kingdom), two separate studies were performed: (i) to evaluate the diagnostic accuracy and analytical sensitivity, a prospective study with fecal samples collected during outbreaks was performed; (ii) to evaluate the analytical reactivity, a retrospective study was performed by testing a panel of archived fecal samples representing strains of most circulating NoV genotypes, other enteric viral pathogens, and 10 negative fecal samples for known viral pathogens. This panel of 97 archived fecal samples had been collected from outbreaks of acute gastroenteritis in the United States from 2000 to 2007 and had been stored as whole stool at 4°C. These samples had been tested previously for GI and GII NoV by real-time TaqMan RT-PCR (rRT-PCR) (47) and genotyped by sequencing of region C and region D RT-PCR products (29, 50).
Specimen collection.
A total of 557 fecal samples (102 outbreaks [with three or more samples] and 136 sporadic cases) were collected between February and December 2008 from infants, children, and adults with symptoms of viral gastroenteritis (acute onset of nausea, vomiting, abdominal cramps, and diarrhea). The samples were collected within 72 h of onset of symptoms at the Commonwealth of Virginia Division of Consolidated Laboratory Services (VA; n = 251), Oregon State Public Health Laboratory (OR; n = 206), and Colchester Hospital in the United Kingdom (UK; n = 100).
IDEIA Norovirus kit (EIA).
The IDEIA Norovirus kit is a single-plate assay in which microwells are coated with monoclonal antibodies to both GI and GII NoV strains. Two different lot numbers of the kit were used during the study (594141 and 651103). Fecal samples were tested according to the manufacturer's instructions upon receipt. Briefly, 10% fecal sample suspensions (clinical samples or quality controls) were prepared by adding 0.1 g of raw fecal sample or 100 μl of liquid fecal sample to 1 ml of sample diluent provided with the EIA kit. After vortexing, solids were removed by centrifugation for 5 min at 6,000 × g. Positive controls (NoV virus-like particles) and negative controls (sample diluent) which were provided in the kit and quality control (QC) samples (see below) were included in each run. One hundred microliters of clarified fecal sample and 100 μl of conjugate were added to each well and incubated for 1 h at room temperature. After washing with freshly prepared wash buffer, 2 drops of substrate were added to each well and strips were incubated for 30 min in the dark. The reaction was stopped by adding 2 drops of Stop solution, and optical densities (ODs) were measured at 450/630 nm with an MRX Revelation spectrophotometer (Dynex Technologies, Inc., Chantilly, VA.). The cutoff was calculated as the average OD value for the negative controls plus 0.1. Samples with OD values greater than the cutoff value plus 0.01 were regarded as positive, and samples with OD values lower than the cutoff value minus 0.01 were regarded as negative. Samples with OD values between the cutoff value ± 0.01 were retested and removed from the study if they remained equivocal.
QCs.
Eight fecal samples (three NoV negative, two NoV GI positive, and three NoV GII positive) were provided by Oxoid Ltd. and were included in each EIA run. Clarified supernatants of the QC samples were stored at −20°C in 100-μl aliquots.
Inter- and intra-assay reproducibility.
To evaluate the interassay reproducibility of the IDEIA Norovirus kit, the eight QCs were tested in triplicate on the same day using two different kit lot numbers (594141 and 651103). We also evaluated the interassay reproducibility between runs for which the eight QC samples were included in each run throughout the study (10 months). The intra-assay reproducibility was evaluated by testing replicates of the eight QC samples in a single run.
Conventional RT-PCR.
RNA was extracted from 50 μl of a clarified 10% fecal suspension using the KingFisher instrument and MagMAX 96 viral RNA isolation kit (Ambion, Austin, TX), according to the manufacturer's instructions. RT-PCR was performed using GI- and GII-specific oligonucleotide primers (regions B, C, and D), currently used for NoV characterization at the CDC, and the Qiagen (Valencia, CA) one-step RT-PCR kit as previously described (1, 29, 34, 50). Amplified products were analyzed on ethidium bromide-stained 2% agarose gels.
Bidirectional sequencing.
RT-PCR products of appropriate sizes were gel purified (QIAquick PCR purification kit; Qiagen, Valencia, CA) and sequenced using the BigDye terminator cycle sequencing ready reaction kit (PE Applied Biosystems). Sequences were edited by using Sequencher (Gene Core Corporation, Ann Arbor, MI) and genotyped by local BLAST analysis against NoV reference sequence databases at CDC. Phylogenetic trees were generated using TreeCon software (48).
NoV GI/GII TaqMan rRT-PCR.
All rRT-PCRs were performed using GI- and GII-specific probe/oligonucleotide primer sets targeting the ORF1/2 junction region (47). The protocol was optimized for the Ag-Path one-step RT-PCR kit (Ambion, Austin, TX) and Applied Biosystems 7500 platform. Standard curves were generated using 10-fold serial dilutions of NoV GI.4 and GII.4 T7 RNA transcripts (17). The number of NoV RNA copies in a fecal sample was determined based on the CT value and corresponding number of RNA copies/μl extrapolated from the appropriate standard curve (GI or GII).
RT-PCR inhibitors.
rRT-PCR-negative samples were retested in the presence of 3 × 103 copies of GII.4 T7 RNA transcripts (17) to assess the presence of inhibitors. If inhibition was detected, the sample was removed from the study.
Electron microscopy.
Samples were tested according to validated laboratory standard operating procedures. Briefly, 20% fecal suspensions in phosphate-buffered saline (PBS) were clarified by centrifugation at 2,500 × g for 30 min. Supernatants were recovered, and virus was concentrated through 30% (wt/vol) sucrose by ultracentrifugation at 45,000 × g. Pellets were resuspended in PBS and clarified by centrifugation at 2,500 × g for 8 min, and 25 μl was transferred to a petri dish. A 300-mesh Formvar- and carbon-coated grid was floated on the drop for 15 to 60 min at room temperature. Any excess fluid was blotted, and 25 μl of 2% (wt/vol) phosphotungstic acid (pH 6.5) was added. The grid was air dried and analyzed under an electron microscope (50,000× magnification) (16).
Diagnostic accuracy.
To evaluate the diagnostic accuracy of the EIA, all 557 prospectively collected fecal samples were tested with the IDEIA Norovirus kit, conventional RT-PCR/bidirectional sequencing, and rRT-PCR. One hundred of these 557 samples were randomly selected and submitted for EM analysis.
The rRT-PCR and EIAs were performed at different sites but with identical protocols. All 251 fecal samples collected in VA were tested by rRT-PCR at the state laboratory and submitted to the CDC for further testing by EIA. All 206 fecal samples collected in OR were tested by rRT-PCR and EIA at the state laboratory and then submitted to the CDC. All 100 fecal samples from the United Kingdom were tested by EIA at the hospital and then submitted to the CDC for further testing by rRT-PCR. In addition, all 557 fecal samples were tested by conventional RT-PCR/bidirectional sequencing at the CDC. A total of 100 fecal samples (VA and OR, n = 75; UK, n = 25) were randomly selected and submitted to the Leeds Teaching Hospitals NHS Trust, United Kingdom, no later than 15 days after collection, and EM was performed within 31 days after collection. Fecal samples were stored at 4°C throughout the study.
Analytical reactivity.
To evaluate the ability of the EIA to detect different NoV genotypes, a panel of archived fecal samples was assembled, including 77 NoV GI- or GII-positive samples, 10 samples that tested negative for NoV GI and GII, and 10 samples that were positive for other enteric viruses (rotavirus, n = 3; sapovirus, n = 3; enterovirus n = 2; astrovirus, n = 2) but were negative for NoV (Table 1). All samples were tested by using the EIA kit, by conventional RT-PCR/bidirectional sequencing to confirm the genogroup/genotype, and by rRT-PCR to determine the CT value at the CDC.
TABLE 1.
Analytical reactivity study: human norovirus genotypes and subclusters included in the panel of archived fecal samplesa
| Genotype (subcluster) | GenBank accession no. | Reference strain ID | No. of specimens |
|---|---|---|---|
| I.1 | L23828 | Hu/GI.1/KY89/1989/JP | 2 |
| I.2 | L07418 | Hu/GI.2/Southampton/1991/UK | 2 |
| I.3 | U04469 | Hu/GI.3/Desert Shield 395/1990/SA | 1 |
| I.3b | AF145709 | Hu/GI.3b/Stavanger/1995/NO | 2 |
| I.4 | AB042808 | Hu/GI.4/Chiba/2000/JP | 3 |
| I.5 | AJ277614 | Hu/GI.5/Musgrove/1989/UK | 4 |
| I.6 | AF093797 | Hu/GI.6/Hesse/1998/DE | 2 |
| I.7 | AJ277609 | Hu/GI.7/Winchester/1994/UK | 2 |
| I.8 | AF538679 | Hu/GI.8/Boxer/2001/US | 2 |
| II.1 | U07611 | Hu/GII.1/Hawaii virus/1971/US | 3 |
| II.2 | X81879 | Hu/GII.2/Melksham/1989/UK | 4 |
| II.3 | U02030 | Hu/GII.3/Toronto/1993/CA | 10 |
| II.4 (95/96-US) | AJ004864 | Hu/GII.4/Grimsby/1995/UK | 1 |
| II.4 (DenHaag) | EF126965 | Hu/GII.4/DenHaag89/2006/NL | 7 |
| II.4 (Hunter) | DQ078814 | Hu/GII.4/Hunter 504D/2004/AU | 2 |
| II.4 (Yerseke) | EF126963 | Hu/GII.4/Yerseke38/2006/NL | 4 |
| II.4 (Osaka) | AB434770 | Hu/GII.4/OC07138/2007/JP | 2 |
| II.5 | AJ277607 | Hu/GII.5/Hillingdon/1994/UK | 5 |
| II.6 | AJ277620 | Hu/GII.6/Seacroft/1990/UK | 4 |
| II.7 | AJ277608 | Hu/GII.7/Leeds/1990/UK | 4 |
| II.10 | AF427118 | Hu/GII.10/Erfurt/546/2000/DE | 1 |
| II.12 | AJ277618 | Hu/GII.12/Wortley/1990/UK | 3 |
| II.13 | AY113106 | Hu/GII.13/Fayetteville/2002/US | 2 |
| II.14 | AY130761 | Hu/GII.14/M7/1999/US | 2 |
| II.16 | AY502010 | Hu/GII.16/Tiffin/1999/US | 1 |
| II.17 | AY502009 | Hu/GII.17/CS-E1/2002/US | 2 |
Analytical sensitivity.
To estimate the minimum number of viral particles required for a positive signal with the EIA kit (limit of detection), six fecal samples (three GI and three GII) were selected based on the following two parameters: (i) samples that tested positive by EIA and (ii) samples that had a low CT value (i.e., high copy number) by rRT-PCR. A 20% (wt/vol) suspension (0.5 g stool in 2.5 ml of buffer) and seven 10-fold serial dilutions were prepared and tested in triplicate by EM, EIA, and rRT-PCR.
Data analysis.
The presence/absence of NoV for each specimen (clinical diagnostic truth) was determined based on three different criteria or reference standards: (i) conventional RT-PCR/bidirectional sequencing; (ii) EM; (iii) a composite reference method algorithm, including EM, conventional RT-PCR/bidirectional sequencing, and rRT-PCR (Table 2). The median CT value and interquartile range were calculated for rRT-PCR-positive samples and EIA-positive and -negative samples; EIA-positive and -negative groups were compared by the rank sum test in SPSS 17.0 (44).
TABLE 2.
Clinical diagnostic truth, based on the composite reference methods algorithm to determine the presence or absence of NoV in specimens collected between February and December 2008
| Reference methoda |
Clinical diagnostic truth | ||
|---|---|---|---|
| rRT-PCR | Conventional RT-PCR/sequence | EM | |
| +/− | + | + | + |
| +/− | +/− | + | + |
| +/− | + | −/NA | + |
| − | − | − | − |
| + | − | − | Indeterminateb |
+/−, positive or negative by the indicated methodology; NA, not available.
If the EM testing result was negative, then it was counted against IDEIA test performance in the positive and negative agreement calculation regardless of the IDEIA norovirus kit test result, unless there was other information (e.g., epidemiologic information) available to assist in establishing diagnostic truth.
Inter- and intra-assay reproducibility.
Statistical analysis was performed using a two-sample t test between the different lots of the EIA kit (P < 0.05). Means, standard deviations and percent coefficient of variation were calculated to evaluate the inter- and intra-assay reproducibility (8).
Diagnostic accuracy.
The diagnostic accuracy of the IDEIA Norovirus kit was evaluated for individual fecal samples and for outbreaks. An outbreak was considered positive when two or more samples tested positive. Sensitivity, specificity, positive and negative predictive values (PPV and NPV, respectively), overall agreement, and 95% confidence intervals (CI) were calculated as previously described (22, 28). Sensitivity levels were compared using the χ2 test (P < 0.05).
Analytical sensitivity.
The number of particles per gram of fecal sample was calculated based on the highest dilution in which particles were detected by EM. The number of NoV RNA copies per gram of fecal sample was calculated for each sample by rRT-PCR. The limit of detection of the EIA was established as the lowest number of viral particles (based on EM) and NoV RNA copies g−1 (based on rRT-PCR) that tested positive in at least two of three replicates.
RESULTS
All EIA control samples (positive, negative, and QC) gave reproducible results in all three sites (CDC, OR, and United Kingdom). Inter- and intra-assay reproducibility studies did not show significant differences between lots, runs, or replicates within the same run. Seven of the 557 fecal samples, including 2 samples that tested equivocal by EIA and 5 samples that contained RT-PCR inhibitors, were removed from the study (data not shown).
Diagnostic accuracy of the EIA.
The sensitivity and specificity of the IDEIA Norovirus kit ranged from 57.4% to 77.8% and 79.7% to 91.9%, respectively, when the results were compared by different reference standards. With conventional RT-PCR/bidirectional sequencing as the reference standard, the 100 samples tested by EM gave sensitivities for EIA, EM, and rRT-PCR of 57.8% (95% CI, 50.4 to 61.9), 51.6% (95% CI, 44.4 to 55.0), and 100% (95% CI, 93.2 to 100.0), respectively, and specificities of 88.9% (95% CI, 75.5 to 96.2), 91.7% (95% CI, 78.9 to 97.8), and 97.2% (95% CI, 84.6 to 99.9), respectively. The sensitivity and specificity of the EM were 51.6% (95% CI, 44.4 to 55.0) and 91.7% (95% CI, 78.9 to 97.8), regardless of the reference standard. The sensitivity and specificity of the rRT-PCR were 96.1% (95% CI, 94.5 to 97.0) and 97.7% (95% CI, 95.3 to 99.1), respectively, with no significant difference when the RT-PCR/bidirectional sequencing or the algorithm were included as the reference standard. However, the sensitivity and specificity of the rRT-PCR decreased to 91.7% (95% CI, 79.0 to 97.8) and 50.0% (95% CI, 42.8 to 53.4), respectively, when the EM was used as reference standard. The decrease in the rRT-PCR specificity was caused by 32 samples that tested negative by EM but positive by rRT-PCR. However, 31 of these 32 samples were confirmed as NoV-positive by conventional RT-PCR/bidirectional sequencing (data not shown). Therefore, the algorithm (Table 2) was used as reference standard throughout the rest of the study.
Overall, the sensitivity and specificity of the IDEIA Norovirus kit were 57.6% (95% CI, 55.0 to 59.6) and 91.9% (95% CI, 87.9 to 94.8), respectively (Table 3). The sensitivity and specificity of the EIA for assessing the cause of an outbreak by detecting NoV in two or more samples were 58.7% (95% CI, 52.6 to 61.6) and 88.9% (95% CI, 72.2 to 97.0), respectively. The sensitivity and specificity for detecting NoV in specimens from sporadic cases of gastroenteritis were 59.0% (95% CI, 50.6 to 64.0) and 93.3% (95% CI, 86.5 to 97.4), respectively. The sensitivity of the EIA to detect NoV in two or more samples per outbreak was determined for outbreaks from which three to more than five samples were collected. A statistically significant increase in sensitivity was observed when 5 samples/outbreak were tested rather than 3 (z = ±2.014; P = 0.044).
TABLE 3.
Diagnostic accuracy parameters for the IDEIA Norovirus kit
| Sample source (n) | Diagnostic accuracy (95% CI) |
% Overall agreement | |||
|---|---|---|---|---|---|
| Sensitivity | Specificity | PPV | NPV | ||
| All samples (557) | 57.6 (55.0-59.6) | 91.9 (87.9-94.8) | 91.5 (87.2-94.6) | 59.0 (56.4-60.9) | 71.3 |
| Outbreaksa,b (102) | 58.7 (52.6-61.6) | 88.9 (72.2-97.0) | 93.6 (84.0-98.3) | 43.6 (35.4-47.6) | 66.7 |
| Sporadic cases (136) | 59.0 (50.6-64.0) | 93.3 (86.5-97.4) | 87.8 (75.3-95.2) | 73.7 (68.3-76.9) | 77.9 |
| Outbreaksb with indicated no. of samples collected | |||||
| 3 (46) | 44.1 (34.3-46.9) | 100.0 (71.8-100) | 93.8 (72.9-99.7) | 38.7 (27.9-41.8) | 57.4 |
| 4 (27) | 63.2 (47.9-68.1) | 100.0 (62.8-100.0) | 92.3 (70.0-99.6) | 53.3 (34.0-59.6) | 71.4 |
| 5 (17) | 76.9c (64.6-89.0) | 100 (45.4-100.0) | 83.3 (70.0-96.4) | 40.0 (7.9-71.3) | 76.4 |
| >5 (12) | 77.8c (55.8-88.3) | 100.0 (38.3-100.0) | 100.0 (59.6-97.9) | 60.0 (20.5-78.9) | 83.3 |
Outbreaks include three or more samples.
An outbreak was considered positive if two or more samples were positive.
Significantly different from outbreaks in which only three samples were collected (P < 0.05).
The PPV and NPV were 91.5% (95% CI, 87.2 to 94.6) and 59.0% (95% CI, 56.4 to 60.9), respectively, and the overall agreement of the results was 71.3% when the total data set was analyzed. When results from sporadic cases were analyzed, the PPV and NPV were 87.8% (95% CI, 75.3 to 95.2) and 73.7% (95% CI, 68.3 to 76.9), respectively, and 93.6% (95% CI, 84.0 to 98.3) and 43.6% (95% CI, 35.4 to 47.6), respectively, when results from outbreaks were analyzed (Table 3).
Analytical reactivity.
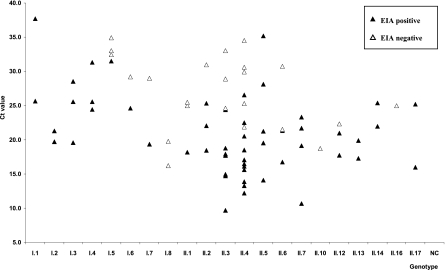
The IDEIA Norovirus kit uses pools of 4 and 10 monoclonal antibodies specific for different NoV GI and GII strains, respectively. The panel of archived fecal samples included 77 specimens representing 8 NoV GI, 13 NoV GII, 10 NoV-negative samples, and 10 samples positive for other enteric viruses (Table 1). Cross-reactivity with other enteric viruses or false-positives result were not detected (data not shown). Overall, the EIA detected 7/8 GI and 11/13 GII genotypes tested (Fig. 1). Results showed that GI.1 to GI.4 samples were detected regardless of the virus concentration (CT range, 19.6 to 37.7), whereas GI.8, showing higher virus concentration (CT range, 16.2 to 19.7) was not detected. Some, but not all, GI.5 to GI.7 strains were detected by EIA. Similarly, all GII.5, -7, -13, -14, and -17 samples (CT range, 10.7 to 35.2) were detected, whereas only some GII.1, GII.2, GII.3, GII.4, GII.6 and GII.12 samples (CT range, 9.7 to 34.5) were detected by EIA. None of the GII.10 and GII.16 samples (CT 18.7 and 24.9, respectively) were detected by EIA.
FIG. 1.
Analytical reactivity, based on the NoV genotypes (8 GI and 13 GII) detected by EIA and their CT values. A panel of 77 NoV strains was tested by EIA and rRT-PCR.
Detection of NoV genotypes in samples collected during the prospective study.
A total of 336 fecal samples were positive by conventional RT-PCR/bidirectional sequencing (Table 4). The NoV genotypes were determined based on CDC's reference database for regions B, C, and D. Thirty-one of 336 samples (9.2%) were classified as GI, and 305 of 336 (90.8%) were classified as GII. GII.4 was the most frequently (76.5%) detected genotype, followed by GII.3 (7.7%), GI.4 (5.1%), GI.8 (2.7%), and GII.1 (2.1%). The IDEIA Norovirus kit detected 58% (95% CI, 52.4 to 62.9) of the samples, including most but not all of the GII.4, GII.3, GI.4, and GII.1 samples. However, it failed to detect GI.7, GI.8, GI.14, GII.6, GII.9, and GII.14. Strains from four different GII.4 subclusters (GII.4 DenHaag, GII.4 Osaka, GII.4 Yerseke, and a novel GII.4 strain) were detected by RT-PCR/bidirectional sequencing. The percentage of strains of each GII.4 subcluster detected by the EIA ranged from 34.0% to 100%, although samples belonging to the Osaka subcluster were not detected by the IDEIA Norovirus kit.
TABLE 4.
Sensitivity of the IDEIA Norovirus kit by genotype for 336 NoV-positive samples by conventional RT-PCR/bidirectional sequencing
| Genotypea | Reference norovirus strain | GenBank accession no. | No. of sequenced specimens | Specimens detected by EIA |
|
|---|---|---|---|---|---|
| No. (%) | 95% CI | ||||
| I.4 | Hu/GI.4/Chiba 407/1987/JP | AB022679 | 1 | 1 (100) | 16.8-100.0 |
| Hu/GI.4/Queens Arms/1992/UK | AJ313030 | 3 | 3 (100) | 38.3-100.0 | |
| Hu/GI.4/Valetta/1995/MT | AJ277616 | 13 | 13 (100) | 73.4-100.0 | |
| I.6 | Hu/GI.6/Hesse/1998/DE | AB112109 | 1 | 1 (100) | 16.8-100.0 |
| I.7 | Hu/GI.7/Winchester/1994/UK | AJ277609 | 1 | 0 (0) | 0-83.3 |
| I.8 | Hu/GI.8/Boxer/2001/US | AF538679 | 9 | 0 (0) | 0-34.5 |
| I.14b | Hu/GI.14/Saitama T25GI/2001/JP | AB112100 | 3 | 0 (0) | 0-61.8 |
| II.1 | Hu/GII.1/Hawaii/1971/US | U07611 | 7 | 3 (43) | 15.8-75.0 |
| II.3 | Hu/GII.3/Arg320/1995/AR | AF190817 | 24 | 14 (58) | 38.8-75.6 |
| Hu/GII.3/Toronto/1991/CA | EF202567 | 2 | 1 (50) | 9.4-90.5 | |
| II.4 | Hu/GII.4/Yerseke38/2006/NL | EF126963 | 9 | 6 (67) | 35.1-88.3 |
| Hu/GII.4/DenHaag89/2006/NL | EF126965 | 237 | 139 (59) | 52.3-64.7 | |
| Hu/GII.4/OsakaCity07138/2007/JP | AB434770 | 1 | 0 (0) | 0-83.3 | |
| Hu/GII.4/Apeldoorn317/2007/NL | AB445395 | 10 | 7 (70) | 39.2-89.7 | |
| II.5 | Hu/GII.5/White river/290/1994/US | AF414423 | 3 | 3 (100) | 38.3-100.0 |
| II.6 | Hu/GII.6/Seacroft/1990/UK | AJ277620 | 2 | 0 (0) | 0-71.0 |
| II.7 | Hu/GII.7/Gwynedd/273/1994/US | AF414409 | 3 | 2 (67) | 20.2-94.4 |
| Hu/GII.7/Leeds/1990/UK | AJ277608 | 2 | 1 (50) | 9.4-90.5 | |
| II.9c | Hu/GII.9/ICB2162/1998/BR | DQ386959 | 3 | 0 (0) | 0-61.8 |
| II.14 | Hu/GII.14/M7/1999/US | AY130761 | 2 | 0 (0) | 0-71.0 |
| Total | 336 | 194 (58) | 52.4-62.9 | ||
Analytical sensitivity.
Three GI.4 and three GII.4 samples that previously tested positive by rRT-PCR and EIA were 10-fold serially diluted and tested in triplicate by EM, EIA, and rRT-PCR. The number of virus particles in the highest dilution positive by EM and the number of NoV RNA copies by rRT-PCR in the highest dilution positive by EIA were used to calculate the virus concentration per gram of fecal sample. The limit of detection of the EIA was different for strains of each genogroup (Table 5). For GI, the average lowest number of particles required for a positive signal by EIA was 3.1 × 106 particles g−1 of fecal sample (range, 2.1 × 106 to 4.9 × 106), which corresponds to 4.1 × 107 NoV RNA copies g−1 of fecal sample. For GII.4, the detection limit was 1.6 × 107 particles g−1 of fecal sample (range, 6.8 × 106 to 2.1 × 107), or 4.2 × 108 NoV RNA copies g−1 of fecal sample.
TABLE 5.
Analytical sensitivitya
| Sample (genotype) | Virus concnb (particles g−1c/RNA copies g−1d) | EIA | rRT-PCR (CT) | EM (no. of particles) | Minimum detectable virus concne |
|
|---|---|---|---|---|---|---|
| No. of particles g−1 | No. of RNA copies g−1 | |||||
| A (GII.4) | 2.09E+08/1.15E+10 | + | 21.3 | 52,557 | 2.10E+07 | 1.15E+09 |
| B (GII.4) | 6.80E+07/2.07E+09 | + | 23.9 | 16,969 | 6.80E+06 | 2.07E+08 |
| C (GII.4) | 2.10E+09/5.85E+09 | + | 25.6 | 52,321 | 2.09E+07 | 5.85E+07 |
| Avg for A, B, and C | 1.62E+07 | 4.20E+08 | ||||
| D (GI.4) | 2.12E+08/1.03E+10 | + | 23.6 | 5,303 | 2.12E+06 | 1.03E+08 |
| E (GI.4) | 4.94E+06/6.45E+06 | + | 27.9 | 12,370 | 4.94E+06 | 6.45E+06 |
| F (GI.4) | 2.15E+08/1.41E+09 | + | 25.3 | 53,735 | 2.15E+06 | 1.41E+07 |
| Avg for D, E, and F | 3.07E+06 | 4.12E+07 | ||||
Based on the minimum number of EM-detectable virus particles and NoV RNA copies detected by rRT-PCR required for a positive signal in the EIA (limit of detection).
The number of viral particles and number of RNA copies per g of fecal sample in the original (undiluted) specimen.
Particle counts were calculated based on the EM results.
The number of NoV RNA copies per g were calculated based on the number of copies/μl in a 20% suspension.
The number of viral particles or NoV RNA copies per g of fecal sample in the highest dilution positive by EIA.
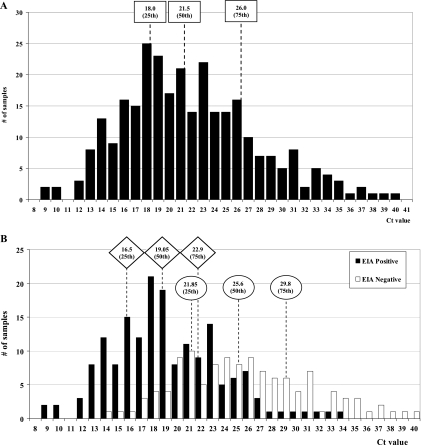
Distribution of CT values for GII-positive samples collected during the prospective study.
Because 291 (95.4%) of 305 RT-PCR/bidirectional sequencing-positive GII samples were also positive by rRT-PCR, we analyzed the distribution of their CT values and their outcome by EIA (Fig. 2A and B). Overall, the rRT-PCR GII-positive samples (n = 291) had a median CT value of 21.5 (interquartile range [IQR], 18.0 to 26.0) (Fig. 2A). The ability of the EIA to detect NoV in this subset of samples was also analyzed. Only 59.6% (174 of 291) of the rRT-PCR NoV GII-positive samples were detected by EIA. There was an overlap in the distribution of CT values between EIA-positive samples (CT range, 9.1 to 34.4) and negative samples (CT range, 14.4 to 40.5) (Fig. 2B). However, a significant difference in the median CT value of EIA-positive samples (median CT, 19.1; IQR, 16.5 to 22.9) compared to EIA-negative samples (median CT, 25.6; IQR, 21.9 to 29.8) was observed (P < 0.0001).
FIG. 2.
Distribution of CT values of GII-positive samples collected during the prospective study in 291 samples regardless of the EIA result (A) or positive by EIA (174/291) and negative by EIA (117/291) (B). The median CT value (50th) and interquartile range (25th to 75th) were calculated for rRT-PCR-positive samples (□) and EIA positive (▵) and negative (○) samples.
DISCUSSION
In this study, we evaluated the diagnostic accuracy, analytical sensitivity, and reactivity of the latest version of the IDEIA Norovirus assay. The assay showed excellent specificity (91.9%) and PPV (91.5%) but low sensitivity (57.6%) and NPV (59.0%). We demonstrated that the lack of sensitivity of the assay during the prospective study was primarily due to the low analytical sensitivity (or limit of detection) of the assay. The kit detects most of the NoV GI and GII genotypes circulating in humans (53), although some uncommon genotypes could not be detected.
Laboratory diagnosis of NoV outbreaks relies on the detection of virus particles by EM, detection of viral RNA by RT-PCR, or detection of viral antigens by EIA (12, 16, 39). A critical parameter for evaluating the performance of EIAs is the selection of the reference method. When using EM as the reference method we found that the sensitivity of the IDEIA Norovirus assay was 77.8% and the sensitivity of the rRT-PCR was 91.7%. The specificity of the rRT-PCR was only 50%, because 32 samples confirmed as NoV by sequence analysis tested negative by EM. Using conventional RT-PCR/bidirectional sequencing as the reference method, the sensitivity of the rRT-PCR increased to 100%, compared to 57.8% and 51.6% for the EIA and EM, respectively. Therefore, we chose an algorithm that combined conventional RT-PCR, rRT-PCR, and EM results as the reference method.
The low sensitivity of the IDEIA Norovirus assay in our study is similar or higher than previously reported results (5, 11, 12, 19, 42). There are several reasons that could explain the low sensitivity of the EIA. First, collection of samples at >72 h after onset of symptoms is directly correlated with the lack of detecting NoV by the EIA (12, 18). In addition, storage of fecal samples for long periods of time before testing may also compromise the level of virus, as proteolytic degradation may occur. In our study, fecal samples were collected and tested within 72 h after onset of viral gastroenteritis symptoms (12, 19, 42), while in other studies samples were tested by EIA after storage for 2 to 3 years (11). Second, differences in inclusion criteria for the fecal samples between different studies may influence the outcome. For example, in our study, samples were only included when the clinical symptoms were consistent with a NoV outbreak. Other studies tested all samples that were submitted to a public health laboratory (12, 42) or a subset of samples reflecting the strain diversity, but not necessarily the prevalence of NoVs (5). Finally, the antigenic diversity of the NoV strains may explain the discrepancy in sensitivity of the EIA kit found in different studies. The IDEIA Norovirus kit uses a cocktail of monoclonal and polyclonal antibodies generated against virus-like particles representing a selection of GI and GII genotypes, and certain genotypes may be missed (5). This was clearly shown in a study where the sensitivity, but not the specificity, dropped significantly when samples from 2002, when the pandemic strain GII.4 Farmington Hills emerged, were tested (11, 53), or when samples were selected to reflect the known NoV genotypes at that time (5).
The low sensitivity of the assay raises serious questions about the usefulness of this kit for routine screening for NoV. In our study, the sensitivity of the IDEIA Norovirus assay to diagnose a NoV outbreak compared to detecting NoV in a single fecal sample did not differ significantly (58.7% and 59.0%, respectively). However, the sensitivity of the assay to confirm a norovirus outbreak increased from 44.1% when three samples were tested to 76.9% when five samples were tested (19, 42). A previous study using statistical analysis demonstrated that when at least five fecal samples per outbreak are tested, finding two out of five samples positive was sufficient to diagnose an outbreak as NoV with at least 95% confidence, with a test sensitivity of 72% (14). However, in our study, 71.6% of the outbreaks had less than five samples, which increased the risk of false-negative outbreaks.
With the great genetic and antigenic diversities of human NoV strains (3 genogroups [GI, GII, and GIV] and at least 25 genotypes) (53) and the continuous emergence of new strains (43, 54), it is crucial that a diagnostic test detects the wide antigenic variety of contemporary NoV strains. The IDEIA Norovirus assay successfully detected strains from seven genotypes in fecal samples collected during the prospective arm of the study, but it failed to detect strains of another six genotypes (GI.7, GI.8, GI.14, GII.6, GII.9, and GII.14). However, in the retrospective study, 18 of the 21 genotypes were detected and only strains of GI.8, GII.10, and GII.16 were missed. The failure to detect specific NoV genotypes was also reported in previous studies (19), but unlike our study, no contemporary strains were included in those earlier studies. For GII.4 strains, the majority of the samples with a CT lower than 26.5 tested positive by the EIA kit, which suggests that although a few genotypes are not detected in the EIA kit (e.g., GI.8), the low sensitivity for samples collected during the prospective study is primarily caused by an insufficient amount of virus (analytical sensitivity).
Our study is the first to estimate the detection limit of a commercial norovirus EIA. The lowest number of GII.4 RNA copies that resulted in a positive signal by EIA was 4.20 × 108 copies g−1 of fecal sample, which equals a CT value of 25.6 based on the standard curve. Fifty percent of the GII samples that tested negative by EIA had a CT value higher than 26.5, suggesting that this is the approximate detection limit of the EIA. For GI.4, the lowest number of NoV RNA copies g−1 of fecal sample that resulted in a positive signal by EIA was 4.94 × 106, which equals a CT value of 27.9 based on the standard curve. During the first 48 h of a NoV outbreak, the viral load in fecal samples ranges from 107 to 108 copies g−1 (7, 31, 37), which suggests that samples that are collected later than 48 h after the onset of symptoms could be missed by the IDEIA Norovirus kit. However, additional studies are needed to evaluate the limit of detection of other genotypes.
In summary, the IDEIA Norovirus assay showed excellent specificity and PPV but low sensitivity and NPV both for outbreaks as well as for samples from sporadic cases. The low NPV indicates that EIA-negative results need to be confirmed by RT-PCR, but a positive result strongly correlates with the presence of NoV. The kit successfully detected 18 of the 21 genotypes evaluated and demonstrated that at least 107 virus particles g−1 of fecal sample are required for a positive signal. Taken together our data demonstrate that (i) the IDEIA Norovirus kit should only be used for samples collected during an outbreak; (ii) samples should be collected preferably during the first 48 h after onset of symptoms; and (iii) testing more than five samples per outbreak significantly increases the likelihood of a positive NoV diagnosis for the outbreak.
In conclusion, the IDEIA Norovirus kit may be useful for rapid screening of fecal samples collected during an outbreak of acute gastroenteritis. However, negative samples will have to be confirmed by a second technique, such as rRT-PCR.
Acknowledgments
We thank Richard Winetrobe for his technical assistance with the EM analysis. We thank the staff of the public health laboratories of Virginia and Oregon and from the Colchester Hospital in the United Kingdom for providing us with the fecal specimens. We thank Oxoid Ltd., Ely, United Kingdom for providing the IDEIA Norovirus kit for this study.
This study was supported by a grant from the Centers for Disease Control Foundation.
The findings and conclusions in this article are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention. This article did receive clearance through the appropriate channels at the CDC prior to submission.
Footnotes
Published ahead of print on 16 June 2010.
REFERENCES
- 1.Anderson, A. D., V. D. Garrett, J. Sobel, S. S. Monroe, R. L. Fankhauser, K. J. Schwab, J. S. Bresee, P. S. Mead, C. Higgins, J. Campana, and R. I. Glass. 2001. Multistate outbreak of Norwalk-like virus gastroenteritis associated with a common caterer. Am. J. Epidemiol. 154:1013-1019. [DOI] [PubMed] [Google Scholar]
- 2.Ando, T., Q. Jin, J. R. Gentsch, S. S. Monroe, J. S. Noel, S. F. Dowell, H. G. Cicirello, M. A. Kohn, and R. I. Glass. 1995. Epidemiologic applications of novel molecular methods to detect and differentiate small round structured viruses (Norwalk-like viruses). J. Med. Virol. 47:145-152. [DOI] [PubMed] [Google Scholar]
- 3.Atmar, R. L., A. R. Opekun, M. A. Gilger, M. K. Estes, S. E. Crawford, F. H. Neill, and D. Y. Graham. 2008. Norwalk virus shedding after experimental human infection. Emerg. Infect. Dis. 14:1553-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bok, K., E. J. Abente, M. Realpe-Quintero, T. Mitra, S. V. Sosnovtsev, A. Z. Kapikian, and K. Y. Green. 2009. Evolutionary dynamics of GII. 4 noroviruses over a 34-year period. J. Virol. 83:11890-11901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burton-MacLeod, J. A., E. M. Kane, R. S. Beard, L. A. Hadley, R. I. Glass, and T. Ando. 2004. Evaluation and comparison of two commercial enzyme-linked immunosorbent assay kits for detection of antigenically diverse human noroviruses in stool samples. J. Clin. Microbiol. 42:2587-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calderon-Margalit, R., R. Sheffer, T. Halperin, N. Orr, D. Cohen, and T. Shohat. 2005. A large-scale gastroenteritis outbreak associated with Norovirus in nursing homes. Epidemiol. Infect. 133:35-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan, M. C., J. J. Sung, R. K. Lam, P. K. Chan, N. L. Lee, R. W. Lai, and W. K. Leung. 2006. Fecal viral load and norovirus-associated gastroenteritis. Emerg. Infect. Dis. 12:1278-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. 2005. User verification of performance for precision and trueness; approved guideline, 2nd ed. CLSI document EP15-A2. Clinical and Laboratory Standards Institute, Wayne, PA.
- 9.Costas, L., A. Vilella, A. Llupia, J. Bosch, M. T. Jimenez de Anta, and A. Trilla. 2007. Outbreak of norovirus gastroenteritis among staff at a hospital in Barcelona, Spain, September 2007. Euro Surveill. 12:E071122.5. [DOI] [PubMed] [Google Scholar]
- 10.Dancer, S. J. 2009. The role of environmental cleaning in the control of hospital-acquired infection. J. Hosp. Infect. 73:378-385. [DOI] [PubMed] [Google Scholar]
- 11.de Bruin, E., E. Duizer, H. Vennema, and M. P. Koopmans. 2006. Diagnosis of norovirus outbreaks by commercial ELISA or RT-PCR. J. Virol. Methods 137:259-264. [DOI] [PubMed] [Google Scholar]
- 12.Dimitriadis, A., L. D. Bruggink, and J. A. Marshall. 2006. Evaluation of the Dako IDEIA norovirus EIA assay for detection of norovirus using faecal specimens from Australian gastroenteritis outbreaks. Pathology 38:157-165. [DOI] [PubMed] [Google Scholar]
- 13.Dimitriadis, A., and J. A. Marshall. 2005. Evaluation of a commercial enzyme immunoassay for detection of norovirus in outbreak specimens. Eur. J. Clin. Microbiol. Infect. Dis. 24:615-618. [DOI] [PubMed] [Google Scholar]
- 14.Duizer, E., A. Pielaat, H. Vennema, A. Kroneman, and M. Koopmans. 2007. Probabilities in norovirus outbreak diagnosis. J. Clin. Virol. 40:38-42. [DOI] [PubMed] [Google Scholar]
- 15.Friesema, I. H., H. Vennema, J. C. Heijne, C. M. de Jager, G. Morroy, J. H. van den Kerkhof, E. J. de Coster, B. A. Wolters, H. L. ter Waarbeek, E. B. Fanoy, P. F. Teunis, R. van der Linde, and Y. T. van Duynhoven. 2009. Norovirus outbreaks in nursing homes: the evaluation of infection control measures. Epidemiol. Infect. 137:1722-1733. [DOI] [PubMed] [Google Scholar]
- 16.Gallimore, C. I., J. Green, D. Lewis, A. F. Richards, B. A. Lopman, A. D. Hale, R. Eglin, J. J. Gray, and D. W. Brown. 2004. Diversity of noroviruses cocirculating in the north of England from 1998 to 2001. J. Clin. Microbiol. 42:1396-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gentry, J., J. Vinje, and E. K. Lipp. 2009. A rapid and efficient method for quantitation of genogroups I and II norovirus from oysters and application in other complex environmental samples. J. Virol. Methods 156:59-65. [DOI] [PubMed] [Google Scholar]
- 18.Glass, R. I., U. D. Parashar, and M. K. Estes. 2009. Norovirus gastroenteritis. N. Engl. J. Med. 361:1776-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gray, J. J., E. Kohli, F. M. Ruggeri, H. Vennema, A. Sanchez-Fauquier, E. Schreier, C. I. Gallimore, M. Iturriza-Gomara, H. Giraudon, P. Pothier, I. Di Bartolo, N. Inglese, E. de Bruin, B. van der Veer, S. Moreno, V. Montero, M. C. de Llano, M. Hohne, and S. M. Diedrich. 2007. European multicenter evaluation of commercial enzyme immunoassays for detecting norovirus antigen in fecal samples. Clin. Vaccine Immunol. 14:1349-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greig, J. D., and M. B. Lee. 2009. Enteric outbreaks in long-term care facilities and recommendations for prevention: a review. Epidemiol. Infect. 137:145-155. [DOI] [PubMed] [Google Scholar]
- 21.Harris, J. P., B. A. Lopman, and S. J. O'Brien. 2010. Infection control measures for norovirus: a systematic review of outbreaks in semi-enclosed settings. J. Hosp. Infect. 74:1-9. [DOI] [PubMed] [Google Scholar]
- 22.Health Decision Strategies, L.L.C. 2008. Epidemiology and lab statistics from study counts. www.healthstrategy.com/epiperl/epiperl.htm.
- 23.Honish, L., J. Talbot, D. Dragon, and D. Utgoff. 2008. Outbreak of norovirus gastroenteritis at a university student residence, Edmonton, Alberta, 2006. Can. Commun. Dis. Rep. 34:1-7. [PubMed] [Google Scholar]
- 24.Isakbaeva, E. T., M. A. Widdowson, R. S. Beard, S. N. Bulens, J. Mullins, S. S. Monroe, J. Bresee, P. Sassano, E. H. Cramer, and R. I. Glass. 2005. Norovirus transmission on cruise ship. Emerg. Infect. Dis. 11:154-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang, X., M. Wang, K. Wang, and M. K. Estes. 1993. Sequence and genomic organization of Norwalk virus. Virology 195:51-61. [DOI] [PubMed] [Google Scholar]
- 26.Kageyama, T., S. Kojima, M. Shinohara, K. Uchida, S. Fukushi, F. B. Hoshino, N. Takeda, and K. Katayama. 2003. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J. Clin. Microbiol. 41:1548-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kageyama, T., M. Shinohara, K. Uchida, S. Fukushi, F. B. Hoshino, S. Kojima, R. Takai, T. Oka, N. Takeda, and K. Katayama. 2004. Coexistence of multiple genotypes, including newly identified genotypes, in outbreaks of gastroenteritis due to;Norovirus in Japan. J. Clin. Microbiol. 42:2988-2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelsey, J. L., A. S. Whittemore, A. S. Evans, and W. D. Thompson. 1996. Methods in observational epidemiology, vol. 26. Oxford University Press, New York, NY.
- 29.Kojima, S., T. Kageyama, S. Fukushi, F. B. Hoshino, M. Shinohara, K. Uchida, K. Natori, N. Takeda, and K. Katayama. 2002. Genogroup-specific PCR primers for detection of Norwalk-like viruses. J. Virol. Methods 100:107-114. [DOI] [PubMed] [Google Scholar]
- 30.Koopmans, M. 2008. Progress in understanding norovirus epidemiology. Curr. Opin. Infect. Dis. 21:544-552. [DOI] [PubMed] [Google Scholar]
- 31.Lee, N., M. C. Chan, B. Wong, K. W. Choi, W. Sin, G. Lui, P. K. Chan, R. W. Lai, C. S. Cockram, J. J. Sung, and W. K. Leung. 2007. Fecal viral concentration and diarrhea in norovirus gastroenteritis. Emerg. Infect. Dis. 13:1399-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lindesmith, L. C., E. F. Donaldson, A. D. Lobue, J. L. Cannon, D. P. Zheng, J. Vinje, and R. S. Baric. 2008. Mechanisms of GII.4 norovirus persistence in human populations. PLoS Med. 5:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martella, V., M. Campolo, E. Lorusso, P. Cavicchio, M. Camero, A. L. Bellacicco, N. Decaro, G. Elia, G. Greco, M. Corrente, C. Desario, S. Arista, K. Banyai, M. Koopmans, and C. Buonavoglia. 2007. Norovirus in captive lion cub (Panthera leo). Emerg. Infect. Dis. 13:1071-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mattison, K., E. Grudeski, B. Auk, H. Charest, S. J. Drews, A. Fritzinger, N. Gregoricus, S. Hayward, A. Houde, B. E. Lee, X. L. Pang, J. Wong, T. F. Booth, and J. Vinje. 2009. Multicenter comparison of two norovirus ORF2-based genotyping protocols. J. Clin. Microbiol. 47:3927-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Navarro, G., R. M. Sala, F. Segura, C. Arias, E. Anton, P. Varela, P. Pena, T. Llovet, I. Sanfeliu, M. Canals, G. Serrate, and A. Nogueras. 2005. An outbreak of norovirus infection in a long-term-care unit in Spain. Infect. Control Hosp. Epidemiol. 26:259-262. [DOI] [PubMed] [Google Scholar]
- 37.Ozawa, K., T. Oka, N. Takeda, and G. S. Hansman. 2007. Norovirus infections in symptomatic and asymptomatic food handlers in Japan. J. Clin. Microbiol. 45:3996-4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parrino, T. A., D. S. Schreiber, J. S. Trier, A. Z. Kapikian, and N. R. Blacklow. 1977. Clinical immunity in acute gastroenteritis caused by Norwalk agent. N. Engl. J. Med. 297:86-89. [DOI] [PubMed] [Google Scholar]
- 39.Patel, M. M., A. J. Hall, J. Vinje, and U. D. Parashar. 2009. Noroviruses: a comprehensive review. J. Clin. Virol. 44:1-8. [DOI] [PubMed] [Google Scholar]
- 40.Patel, M. M., M. A. Widdowson, R. I. Glass, K. Akazawa, J. Vinje, and U. D. Parashar. 2008. Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerg. Infect. Dis. 14:1224-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Podewils, L. J., L. Zanardi Blevins, M. Hagenbuch, D. Itani, A. Burns, C. Otto, L. Blanton, S. Adams, S. S. Monroe, M. J. Beach, and M. Widdowson. 2007. Outbreak of norovirus illness associated with a swimming pool. Epidemiol. Infect. 135:827-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Richards, A. F., B. Lopman, A. Gunn, A. Curry, D. Ellis, H. Cotterill, S. Ratcliffe, M. Jenkins, H. Appleton, C. I. Gallimore, J. J. Gray, and D. W. Brown. 2003. Evaluation of a commercial ELISA for detecting Norwalk-like virus antigen in faeces. J. Clin. Virol. 26:109-115. [DOI] [PubMed] [Google Scholar]
- 43.Siebenga, J. J., H. Vennema, D. P. Zheng, J. Vinje, B. E. Lee, X. L. Pang, E. C. Ho, W. Lim, A. Choudekar, S. Broor, T. Halperin, N. B. Rasool, J. Hewitt, G. E. Greening, M. Jin, Z. J. Duan, Y. Lucero, M. O'Ryan, M. Hoehne, E. Schreier, R. M. Ratcliff, P. A. White, N. Iritani, G. Reuter, and M. Koopmans. 2009. Norovirus illness is a global problem: emergence and spread of norovirus GII.4 variants, 2001-2007. J. Infect. Dis. 200:802-812. [DOI] [PubMed] [Google Scholar]
- 44.SPSS Inc. 2008. SPSS statistics 17. SPSS Inc., Chicago, IL.
- 45.Takkinen, J. 2006. Recent norovirus outbreaks on river and seagoing cruise ships in Europe. Euro Surveill. 11:E060615.2. [DOI] [PubMed] [Google Scholar]
- 46.Teunis, P. F., C. L. Moe, P. Liu, S. E. Miller, L. Lindesmith, R. S. Baric, J. Le Pendu, and R. L. Calderon. 2008. Norwalk virus: how infectious is it? J. Med. Virol. 80:1468-1476. [DOI] [PubMed] [Google Scholar]
- 47.Trujillo, A. A., K. A. McCaustland, D. P. Zheng, L. A. Hadley, G. Vaughn, S. M. Adams, T. Ando, R. I. Glass, and S. S. Monroe. 2006. Use of TaqMan real-time reverse transcription-PCR for rapid detection, quantification, and typing of norovirus. J. Clin. Microbiol. 44:1405-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van de Peer, Y., and R. De Wachter. 1994. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10:569-570. [DOI] [PubMed] [Google Scholar]
- 49.van Duynhoven, Y. T., C. M. de Jager, L. M. Kortbeek, H. Vennema, M. P. Koopmans, F. van Leusden, W. H. van der Poel, and M. J. van den Broek. 2005. A one-year intensified study of outbreaks of gastroenteritis in The Netherlands. Epidemiol. Infect. 133:9-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vinje, J., R. A. Hamidjaja, and M. D. Sobsey. 2004. Development and application of a capsid VP1 (region D) based reverse transcription PCR assay for genotyping of genogroup I and II noroviruses. J. Virol. Methods 116:109-117. [DOI] [PubMed] [Google Scholar]
- 51.Vinje, J., and M. P. Koopmans. 1996. Molecular detection and epidemiology of small round-structured viruses in outbreaks of gastroenteritis in the Netherlands. J. Infect. Dis. 174:610-615. [DOI] [PubMed] [Google Scholar]
- 52.Wang, Q. H., M. G. Han, S. Cheetham, M. Souza, J. A. Funk, and L. J. Saif. 2005. Porcine noroviruses related to human noroviruses. Emerg. Infect. Dis. 11:1874-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng, D. P., T. Ando, R. L. Fankhauser, R. S. Beard, R. I. Glass, and S. S. Monroe. 2006. Norovirus classification and proposed strain nomenclature. Virology 346:312-323. [DOI] [PubMed] [Google Scholar]
- 54.Zheng, D. P., M. A. Widdowson, R. I. Glass, and J. Vinje. 2010. Molecular epidemiology of genogroup II-genotype 4 noroviruses in the United States between 1994 and 2006. J. Clin. Microbiol. 48:168-177. [DOI] [PMC free article] [PubMed] [Google Scholar]