Abstract
Recent evidence strongly suggests an association between the use of fluoroquinolones and Clostridium difficile infection (CDI). Resistance to fluoroquinolones has been described not only in the hypervirulent strain 027, but also in other important PCR ribotypes circulating in hospital settings. In a European prospective study conducted in 2005, strains resistant to moxifloxacin represented 37.5% of C. difficile clinical isolates. In this study, we investigated a sample of 147 toxigenic C. difficile isolates, collected in Italy from 1985 to 2008, for the presence of mutations in gyr genes that conferred resistance to fluoroquinolones based on a LightCycler assay. Results were confirmed by the determination of MICs for moxifloxacin. Strains resistant to moxifloxacin were also investigated for resistance to three other fluoroquinolones and for a possible association between fluoroquinolone and macrolide-lincosamide-streptogramin B resistance. C. difficile isolates were typed by PCR ribotyping. In total, 50 clinical isolates showed substitutions in gyr genes and were resistant to fluoroquinolones. Ninety-six percent of the C. difficile resistant isolates showed the substitution Thr82-to-Ile in GyrA, as already observed in the majority of resistant strains worldwide. A significant increase of resistance (P < 0.001) was observed in the period 2002 to 2008 (56% resistant) compared to the period 1985 to 2001 (10% resistant). Coresistance with erythromycin and/or clindamycin was found in 96% (48/50) of the isolates analyzed and, interestingly, 84% of resistant strains were erm(B) negative. The majority of the fluoroquinolone-resistant isolates belonged to PCR ribotype 126 or 018. PCR ribotype 126 was the most frequently found from 2002 to 2005, whereas PCR ribotype 018 was predominant in 2007 and 2008 and still represents the majority of strains typed in our laboratory. Overall, the results demonstrate an increasing number of C. difficile strains resistant to fluoroquinolones in Italy and changes in the prevalence and type of C. difficile isolates resistant to fluoroquinolones circulating over time.
Clostridium difficile is an anaerobic, Gram-positive, spore-forming bacillus that may cause a spectrum of diseases, ranging from uncomplicated mild diarrhea to pseudomembranous colitis, collectively known as C. difficile infections (CDIs) (3, 39). The risk of CDI appears to be greater with certain antimicrobial agents and increases if strains are resistant to administered antimicrobials (30).
Recently, the new hypervirulent C. difficile strain, typed as PCR ribotype 027, toxinotype III, pulsed-field gel electrophoresis pattern NAP1, has been associated with more severe and fatal cases in the United States, Canada, Japan, and Europe (4, 16, 18, 20, 21, 22). Strain 027 isolates are characterized by a hyperproduction of toxins A and B, production of binary toxin, and resistance to erythromycin and fluoroquinolones (12, 18, 34). Resistance to these antibiotics characterizes not only strain 027 but the majority of C. difficile strains circulating in hospital settings and responsible for disease (2, 34).
The use of macrolide-lincosamide-streptogramin B (MLSB) antibiotics has long been known to be one of the major risk factors for CDI (15, 25). With C. difficile, resistance to these antibiotics, in particular to erythromycin and clindamycin, is due to an erm(B) gene carried by Tn5398, a mobile element that shows heterogeneous genetic organization. Nevertheless, an increased number of C. difficile erm(B)-negative isolates resistant to MLSB has been described, including strains characterized as PCR ribotype 027 (1, 19).
Historically, fluoroquinolones were considered a low risk in C. difficile diseases, but recent evidence strongly suggests an association between their use and CDI (6, 24, 26). Resistance to fluoroquinolones has been described not only in the epidemic strain 027 but also in other important PCR ribotypes, and it is increasing, since recent data have shown that resistant strains represent 37.5% of C. difficile clinical isolates in Europe (2). Two main mechanisms of fluoroquinolone resistance have been described: alterations in the targets of antibiotics, DNA gyrase and topoisomerase IV, and decreased accumulation inside bacteria (13, 29). In C. difficile, as in many other bacterial species, resistance is determined by amino acid substitutions in the quinolone resistance-determining region (QRDR) of the target enzymes (27). Since this bacterium does not have genes for topoisomerase IV, these alterations are located in the QRDR of either GyrA or GyrB, the DNA gyrase subunits (11, 34). Recent studies indicated that the replacement of Thr82 with Ile in GyrA characterized 93% of European toxigenic C. difficile isolates resistant to fluoroquinolones, including the hypervirulent epidemic clone 027/NAP1/III (12, 34). The remaining 7% showed a substitution in position 426 (Asp to Asn or Val) of GyrB (34).
Few data on fluoroquinolone resistance of C. difficile clinical isolates are available for Italy (34) and, for this reason, we analyzed a convenient sample of toxigenic isolates, collected by the Istituto Superiore di Sanità (ISS) from 1985 to 2008, for substitutions in GyrA and GyrB and for their MIC values to moxifloxacin. All C. difficile resistant strains were also tested for their resistance to ciprofloxacin, gatifloxacin, and levofloxacin and investigated for a possible association between fluoroquinolone and MLSB resistance. Typing of resistant isolates was carried out using the already-described PCR ribotyping method (5).
MATERIALS AND METHODS
C. difficile clinical isolates.
A total of 147 toxigenic C. difficile isolates, selected from the ISS C. difficile national collection, were analyzed. These strains were isolated from sporadic cases and five different outbreaks between 1985 and 2008 and were sent to ISS by nine different Italian hospitals (arbitrarily denominated from A to I) located in the center and north of Italy for toxigenic assays and/or molecular analysis.
An outbreak was defined as two or more related cases over a locally defined period, whereas a sporadic case of CDI was defined as diarrhea (≥3 loose stools per day for at least 2 days) in a patient with a stool culture positive for C. difficile and/or fecal cytotoxin B (18).
C. difficile strains are voluntarily sent to ISS, so the number of strains received each year from the different hospitals is variable, and it is possible that each hospital is not always represented in the different years. At least 50% of strains received per year were selected and included in the study. Only two C. difficile strains per outbreak and only one strain per single patient were considered for the analysis, whereas strains from sporadic cases were randomly selected. Since the attention to CDI has increased during the last years in our country, there has been a rise in the number of C. difficile strains received by our institute. From 1985 to 1989, we only received C. difficile strains isolated from outbreaks, and the six strains selected for this study represent three different epidemics that occurred in three different hospitals (A, B, and C). From the beginning of the 1990s a higher number of strains, isolated from both outbreaks and sporadic cases, was sent to ISS by several hospitals. The 64 strains selected from 1991 to 2001 were isolated in five different hospitals (D, E, F, G, and H), whereas the 77 strains selected from 2002 to 2008 were isolated in four different hospitals (E, F, G, and I). Two different outbreaks occurred in 2002 and 2007 in hospitals H and G, respectively.
For convenience, we divided the strains into two groups for the analysis of the results: the first group included 70 isolates from 1985 to 2001, and the second group included 77 isolates from 2002 to 2008.
Two C. difficile strains, representing the predominant types circulating in Italy during the period 1987 to 2001 (denominated A and R) (32) and two international reference strains belonging to PCR ribotype 027 or 017, kindly supplied by Ed Kuijper (European Centre for Disease Prevention and Control [ECDC]), were used as control strains in PCR ribotyping.
LightCycler gyr mutation assay.
In the gyr mutation study, the presence of mutations conferring resistance in codon 82 of gyrA and codon 426 of gyrB was determined by using a LightCycler assay recently designed in our laboratory (35). Briefly, purified genomic DNA, used as template, was isolated using the NucleoBond buffer set III and the NucleoBond AXG 20 (Macherey-Nagel, Düren, Germany). Amplification of targets was performed by an initial denaturation at 95°C for 10 min and 30 cycles of 95°C for 10 s, 49°C for 20 s, and 72°C for 20 s. The melting curves for the annealing of the probes with the PCR product were determined from 42°C to 95°C, with a temperature transition rate of 0.1°C/s, by monitoring the reporter dye fluorescence emission at 640 nm (gyrB) or at 705 nm (gyrA). Data were analyzed with LightCycler software, version 5.32, according to the manufacturer's instructions. Different melting peaks characterized different nucleotide transitions.
The international reference strain, C. difficile 630, susceptible to fluorquinolones, was used as a representative of isolates showing gyr genes with a wild-type sequence.
Antibiotic susceptibilities.
MICs to moxifloxacin (MX), levofloxacin (LE), gatifloxacin (GA), ciprofloxacin (CI), erythromycin (EM), and clindamycin (CM) were determined using the Etest (AB Biodisk, Solna, Sweden) on brucella agar plates containing vitamin K1 (0.5 mg/liter), hemin (5 mg/liter), and 5% defibrinated sheep red blood cells, according to the manufacturer's instructions. The breakpoint used for all antibiotics tested in the study was 8 μg/ml, in accordance with the guidelines established by the Clinical and Laboratory Standards Institute (CLSI) (8). Bacteroides thetaiotaomicron ATCC 29741 was used as a control strain.
Detection of erm(B) genes.
Since erm(B) is known to be predominant in MLSB-resistant C. difficile, the presence of genes belonging to this class was investigated by PCR using primers E5 and E6, as previously described (32). Five microliters of crude DNA was denatured for 5 min at 94°C and amplified for 30 cycles consisting of 1 min at 94°C, 1 min at 50°C, and 1 min 30 s at 72°C. The expected size of the PCR fragment was 0.6 kb.
PCR ribotyping and phylogenetic analysis.
PCR ribotyping analyses were performed by using the method of Bidet et al. (5). The first isolate identified for each PCR ribotype was submitted to the Anaerobe Reference Laboratory, University Hospital of Wales, Cardiff, United Kingdom, to be assigned to a reference type of the United Kingdom collection.
The genomic DNA fingerprinting patterns produced by PCR ribotyping were compared and analyzed with GelCompar II v6.0 (Applied Maths, Sint-Martens-Latem, Belgium). Similarity analysis was performed by using Pearson's correlation with a 0.5% optimization. Clustering was performed by using the unweighted pair group mean association (UPGMA). C. difficile strains showing a percentage of similarity of ≥90% were considered closely related.
RESULTS
Detection of mutations in gyr genes and resistance to fluoroquinolones.
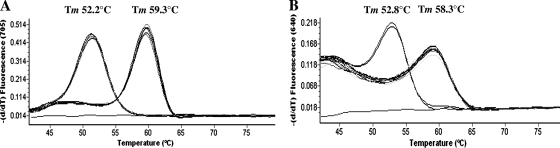
In total, 50 of the 147 (34%) isolates showed mutations in gyr genes. As far as the mutation in gyrA is concerned, we identified 48 strains with a Tm of about 52.2°C (Fig. 1 A). This Tm characterizes the transition of ACT (Thr) to ATT (Ile) in codon 82 of gyrA, whereas a melting peak at about 59.3°C characterized the wild-type gyrA sequence (35). gyrB analysis showed that two strains had a Tm at about 52.8°C, indicating the presence of the transition GAT (Asp) to AAT (Asn) in position 426 of the gene (Fig. 1B) (35). All the other strains had a Tm of about 58.3°C, which characterizes gyrB genes without mutations in position 426. The substitutions identified in this study have been previously described for C. difficile isolates (34).
FIG. 1.
gyr gene melting curve analysis in a representative number of C. difficile clinical isolates analyzed in this study. (A) For the gyrA gene, isolates showing the substitution Thr82 to Ile had a Tm of 52.2°C, whereas isolates showing the wild-type codon in position 82 had a Tm of 59.3°C. (B) For the gyrB gene, isolates showing the substitution Asp426 to Asn had a Tm of 52.8°C, whereas those with the wild-type gene had a Tm of 58.3°C.
In general, MIC values for MX ranged from 0.5 to ≥32 μg/ml. Etest values for MX confirmed the results obtained by real-time PCR; in fact, all strains without substitutions in gyr genes were susceptible, whereas the 50 isolates showing substitutions were resistant to MX, with MICs between 8 and ≥32 μg/ml. The MIC ranges of these strains for the other fluoroquinolones tested were as follows: 8 to ≥32 μg/ml for GA, 24 to ≥32 μg/ml for CI, and ≥32 μg/ml for LE. Resistant strains with different mutations did not show significant differences in MIC levels for the fluoroquinolones tested in this study. In the group of strains isolated from 1985 to 2001 we found 10% (7/70) of C. difficile isolates were resistant to fluoroquinolones, whereas in the second group, from 2002 to 2008, the percentage rose to 56% (43/77).
Association with MLSB resistance and detection of erm(B) genes.
Ninety-eight percent (49/50) of the C. difficile isolates resistant to fluoroquinolones were also resistant to EM and/or CM. MIC levels ranged between 1 and ≥256 μg/ml for EM and between 0.5 and ≥256 μg/ml for CM. In total, only eight C. difficile MLSB-resistant isolates (16%) showed an erm(B) gene. In particular, among the resistant strains isolated from 1985 to 2001 we found that 57% (4/6) had an erm(B) gene, whereas among those isolated from 2002 and 2008 the erm(B)-positive isolates only represented 9% (4/43). All isolates with an erm(B) gene were resistant to both EM and CM (MICs between 16 and 256 μg/ml and 12 and 256 μg/ml, respectively). C. difficile isolates that were erm(B) negative were resistant to EM (MICs, ≥256 μg/ml) and resistant (MICs between 8 and 16 μg/ml) or susceptible to CM, except for one isolate susceptible to EM and resistant to CM (MICs of 3 and 12 μg/ml, respectively).
PCR ribotyping and phylogenetic analysis.
In total, six different PCR ribotypes (001, 020, 012, 018, 078, and 126) were identified among C. difficile strains resistant to fluoroquinolones. No strains were identified as PCR ribotype 027 or 017. C. difficile strains isolated from 1985 to 2001 belonged to type 001 (two strains), type 126 (two strains), and types 012, 020, and 078 (one strain for each type). Among the strains collected between 2002 and 2008, 56% (24/43) belonged to type 126, 40% (17/43) to type 018, and 2% (1 of each type among the 43) to types 020 and 001, respectively. Interestingly, strains with PCR ribotype 126 were predominant between 2002 and 2005, whereas strains of PCR ribotype 018 were predominant between 2007 and 2008. C. difficile strains isolated from two different outbreaks that occurred in 2002 and 2007 belonged to PCR ribotypes 126 and 018, respectively.
The PCR ribotypes predominant in Italy during the period 1987 to 2001, denominated A and R (32), have been typed in this study as PCR ribotypes 012 and 078, respectively.
Two of the four C. difficile erm(B)-positive strains isolated between 1985 and 2001 belonged to PCR ribotype 001, one to 012, and one to 078, whereas two of those isolated between 2002 and 2008 were PCR ribotype 126, one was PCR ribotype 001, and one was PCR ribotype 020.
The 97 susceptible C. difficile strains analyzed in this study belonged to PCR ribotype 078 (26.5%), 012 (17.6%), 020 (16.2%), 002 (8.8%), 045 (4.4%), or other PCR ribotypes (26.6%).
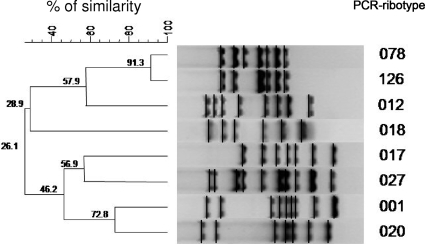
Phylogenetic analysis indicated that C. difficile strains resistant to fluoroquinolones belong to two distinct clusters (Fig. 2). The first one grouped PCR types 001 and 020 and the two control strains, typed as 027 and 017; the second cluster included PCR types 012, 018, 078, and 126. Interestingly, types 078 and 126 were closely related genetically, showing 91.3% similarity, whereas type 018 belonged to the more distant branch, showing only 57.9% similarity in comparison with the other types included in the same cluster.
FIG. 2.
Dendrogram obtained from the analysis of the representative C. difficile PCR ribotype patterns identified in this study. Similarity analysis was performed with Pearson's correlation and clustering by the unweighted pair group mean association (UPGMA) method. Two international reference strains belonging to PCR ribotypes 027 and 017 were used as control strains in the analysis.
DISCUSSION
Fluoroquinolones, in particular the broad-spectrum C-8-methoxyfluoroquinolones, are a class of antibiotics with a wide spectrum of activity that have been used extensively to treat a great variety of infections since the 1990s (13, 29). Fluoroquinolone resistance has recently increased in C. difficile clinical isolates, so the use of fluoroquinolones has emerged as an important risk factor for CDI (2, 6, 7, 24, 26). In Italy, the classes of drugs most affecting national expenditure for antibiotics are macrolides (23.8%) and quinolones/fluoroquinolones (21.3%) (31). The average consumption of macrolides in recent years has been constant, while there has been a substantial increase in the average consumption of quinolones/fluoroquinolones, with an increase of 4 million doses between 2000 and 2008. Even if international literature on antibiotic use in Italian hospitals is scarce, a study carried out in five hospitals located in the north of Italy from 2002 to 2004 indicated that the overall consumption of fluoroquinolones increased by 29%, from 11.3 defined daily doses (DDD)/100 bed days in 2002 to 14.57 DDD/100 bed days in 2004 (38). In particular, levofloxacin and ciprofloxacin together represented about 95% of the fluoroquinolones used in 2004.
The present study is the first characterization of fluoroquinolone-resistant C. difficile clinical isolates circulating in different Italian hospitals from 1985 to 2008. In total, we found 50 resistant isolates among the 147 analyzed. A significant increase of resistance (P < 0.001) was observed when comparing the period 1985 to 2001 (10% resistance) to the period 2002 to 2008 (56% resistance). The percentage of resistance observed in the latter period is higher than the European average (37.5%) (2) and indicates a large spread of fluoroquinolone-resistant C. difficile strains in hospital settings, coinciding with the increased use of fluoroquinolones over recent years.
Ninety-six percent of the C. difficile resistant isolates showed the substitution Thr82 to Ile in GyrA, as already observed in the majority of resistant C. difficile strains worldwide (34). Only two isolates had the substitution Asp426 to Asn in GyrB, which was previously described in some C. difficile strains (34). The results also confirmed the effectiveness of the LightCycler assay that we recently set up to detect mutations in codons 82 and 426 of gyrA and gyrB, respectively (35). In fact, all strains showing substitutions resulted in resistance to fluoroquinolones, and no discrepancies were detected. For this reason, the LightCycler assay can be considered a cost-effective, fast, and reproducible alternative to screen several C. difficile strains for fluoroquinolone resistance in comparison with DNA sequencing and susceptibility testing, which are more time-consuming. The limits of this method are that mutations occur in different codons and the resistance due to other mechanisms, such as efflux, that occurs even if these mechanisms seems to be absent in C. difficile.
As for erythromycin and clindamycin resistance, 58% (29/50) of the strains examined were resistant only to erythromycin, 38% (19/50) to both erythromycin and clindamycin, and 2% (1/50) only to clindamycin. Interestingly, 84% (41/49) of the C. difficile isolates resistant to erythromycin and/or clindamycin were erm(B) negative. In a previous study (32), we observed an increased number of erm(B)-negative clinical isolates in Italy from 1990 to 2001. The results obtained in the present study seem to confirm this trend. Since C. difficile erm(B)-negative strains are also negative for other important erm classes and efflux pumps (33), further studies will be necessary to identify the mechanism of resistance present in these strains.
The use of MLSB antibiotics and fluoroquinolones has been known to be one of the major risk factors for CDI (15, 25, 6, 24, 26). Overall, coresistance to moxifloxacin and erythromycin was found in 96% (48/50) of the C. difficile isolates analyzed in this study, confirming the high percentage of association also observed in other countries (1, 14). The accumulation and maintenance of different mechanisms of resistance, such as transferable genetic elements and nucleotide mutations, make C. difficile resistant to an increased number of antibiotics, limiting therapeutic options and management of severe infections.
The majority of the fluoroquinolone-resistant strains analyzed in this study belonged to PCR ribotype 126 or 018. Both PCR ribotypes were responsible either for sporadic cases or outbreaks. PCR ribotype 126 was the most frequent from 2002 to 2005, whereas PCR ribotype 018 was predominant in 2007 and 2008. Both PCR ribotypes have already been identified among human isolates (2, 9, 23, 28); furthermore, these PCR ribotypes have also been isolated from animals (10, 17). The two predominant types, A and R, found in our country between 1990 and 2001 (32) were assigned to PCR ribotypes 012 and 078, respectively, in this study. Phylogenetic analysis showed that isolates of PCR ribotypes 126 and 078 had a high percentage of similarity (92.3%). We found that 62% of the old Italian strains belonging to PCR ribotype 078 were resistant to erythromycin, erm(B) negative (32, 33), and susceptible to fluoroquinolones. Since isolates of PCR ribotypes 078 and 126 are closely related genetically, it is possible that the progenitor of resistant strains, PCR ribotype 126, is derived from an erm(B)-negative C. difficile PCR ribotype 078 isolate that was subsequently selected, by fluoroquinolone pressure, for its resistance to these antibiotics.
As far as PCR ribotype 018 is concerned, phylogenetic analysis indicates that it is not related to the types previously found in our country, so it seems to be a “new” PCR ribotype for Italy. Interestingly, this PCR ribotype represents 51% of the total number of strains analyzed in our institute since 2006 (data not shown). All Italian isolates belonging to this PCR ribotype were found to be resistant to fluoroquinolones, suggesting that the increased use of these antibiotics has played a determinant role in selection and spread of these strains. PCR ribotype 018 has rarely been identified in other countries (9, 28, 37) and, unfortunately, no data are available on their susceptibility to fluoroquinolones. C. difficile strains of PCR ribotype 018 do not present the typical characteristics of the current hypervirulent strains; in fact, they are typed as toxinotype 0, are binary toxin negative (36), and do not have significant changes in the regulatory toxins genes tcdD and tcdC, as demonstrated by our analysis on the pathogenicity locus (PaLoc) (data not shown). Besides resistance to fluoroquinolones, the capability to spread and cause disease for these strains may be due to other virulence factors, so a further characterization of isolates of PCR ribotype 018 is ongoing in our laboratory.
Overall, the results obtained in this study indicate an increasing number of C. difficile strains resistant to fluoroquinolones in our country and a change in the prevalence and type of the C. difficile isolates circulating over time. The results highlight the importance of typing of isolates and monitoring antimicrobial susceptibility for rapid detection of changes in C. difficile populations and rapid responses to the emergence of epidemic clones.
Acknowledgments
This work was partially supported by EC Project LSHE-CT-2006-037870, “European approach to combat outbreaks of Clostridium difficile associated diarrhea by development of new diagnostic tests.”
We are indebted to J. Brazier (Anaerobe Reference Laboratory, University Hospital of Wales, Cardiff, United Kingdom) for the PCR ribotyping of the representative C. difficile isolates. We thank Tonino Sofia for editing the manuscript.
Footnotes
Published ahead of print on 16 June 2010.
REFERENCES
- 1.Ackermann, G., A. Degner, H. Cohen, J. Silva, Jr., and A. C. Rodloff. 2003. Prevalence and association of macrolide-lincosamide-streptogramin B (MLSB) resistance with resistance to moxifloxacin in Clostridium difficile. J. Antimicrob. Chemother. 51:599-603. [DOI] [PubMed] [Google Scholar]
- 2.Barbut, F., P. Mastrantonio, M. Delmee, J. Brazier, E. Kuijper, and I. Poxton. 2007. Prospective study of Clostridium difficile infections in Europe with phenotypic and genotypic characterisation of the isolates. European Study Group on Clostridium difficile. Clin. Microbiol. Infect. 13:1048-1057. [DOI] [PubMed] [Google Scholar]
- 3.Bartlett, J. G. 2002. Clostridium difficile-associated enteric disease. Curr. Infect. Dis. Rep. 4:477-483. [DOI] [PubMed] [Google Scholar]
- 4.Bartlett, J. G. 2006. Narrative review: the new epidemic of Clostridium difficile-associated enteric disease. Ann. Intern. Med. 145:758-764. [DOI] [PubMed] [Google Scholar]
- 5.Bidet, P., F. Barbut, V. Lalande, B. Burghoffer, and J. C. Petit. 1999. Development of a new PCR-ribotyping method for Clostridium difficile based on ribosomal RNA gene sequencing. FEMS Microbiol. Lett. 175:261-266. [DOI] [PubMed] [Google Scholar]
- 6.Biller, P., B. Shank, L. Lind, M. Brennan, L. Tkatch, G. Killgore, A. Thompson, and L. C. McDonald. 2007. Moxifloxacin therapy as a risk factor for Clostridium difficile-associated disease during an outbreak: attempts to control a new epidemic strain. Infect. Control Hosp. Epidemiol. 28:198-201. [DOI] [PubMed] [Google Scholar]
- 7.Blondeau, J. M. 2009. What have we learned about antimicrobial use and the risks for Clostridium difficile-associated diarrhoea? J. Antimicrob. Chemother. 63:238-242. [DOI] [PubMed] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. 2007. Methods for antimicrobial susceptibility testing of anaerobic bacteria, 7th ed. Approved standard M11-A7. Clinical and Laboratory Standards Institute, Wayne, PA.
- 9.Coia, J. E. 2009. What is the role of antimicrobial resistance in the new epidemic of Clostridium difficile? Int. J. Antimicrob. Agents 33(Suppl. 1):S9-S12. [DOI] [PubMed] [Google Scholar]
- 10.Debast, S. B., L. A. van Leengoed, A. Goorhuis, C. Harmanus, E. J. Kuijper, and A. A. Bergwerff. 2009. Clostridium difficile PCR ribotype 078 toxinotype V found in diarrhoeal pigs identical to isolates from affected humans. Environ. Microbiol. 11:505-511. [DOI] [PubMed] [Google Scholar]
- 11.Dridi, L., J. Tankovic, B. Burghoffer, F. Barbut, and J. C. Petit. 2002. gyrA and gyrB mutations are implicated in cross-resistance to ciprofloxacin and moxifloxacin in Clostridium difficile. Antimicrob. Agents Chemother. 46:3418-3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drudy, D., L. Kyne, R. O'Mahony, and S. Fanning. 2007. gyrA mutations in fluoroquinolone-resistant Clostridium difficile PCR-027. Emerg. Infect. Dis. 13:504-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hooper, D. C. 2001. Emerging mechanisms of fluoroquinolone resistance. Emerg. Infect. Dis. 7:337-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang, H., S. Wu, M. Wang, Y. Zhang, H. Fang, A. C. Palmgren, A. Weintraub, and C. E. Nord. 2009. Clostridium difficile infections in a Shanghai hospital: antimicrobial resistance, toxin profiles and ribotypes. Int. J. Antimicrob. Agents 33:339-342. [DOI] [PubMed] [Google Scholar]
- 15.Johnson, S., M. H. Samore, K. A. Farrow, G. E. Killgore, F. C. Tenover, D. Lyras, J. I. Rood, P. DeGirolami, A. L. Baltch, M. E. Rafferty, S. M. Pear, and D. N. Gerding. 1999. Epidemics of diarrhea caused by a clindamycin-resistant strain of Clostridium difficile in four hospitals. N. Engl. J. Med. 341:1645-1651. [DOI] [PubMed] [Google Scholar]
- 16.Kato, H., Y. Ito, R. van den Berg, E. J. Kuijper, and Y. Arakawa. 2007. First isolation of Clostridium difficile 027 in Japan. Euro Surveill. 12:E070111.3. http://www.eurosurveillance.org/ew/2007/070111.asp#3. [DOI] [PubMed] [Google Scholar]
- 17.Keel, K., J. S. Brazier, K. W. Post, S. Weese, and J. G. Songer. 2007. Prevalence of PCR ribotypes among Clostridium difficile isolates from pigs, calves, and other species. J. Clin. Microbiol. 45:1963-1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuijper, E. J., B. Coignard, and P. Tüll. 2006. Emergence of Clostridium difficile-associated disease in North America and Europe. Clin. Microbiol. Infect. 12(Suppl. 6):2-18. [DOI] [PubMed] [Google Scholar]
- 19.Kuijper, E. J., R. J. van den Berg, S. Debast, C. E. Visser, D. Veenendaal, A. Troelstra, T. van der Kooi, S. van den Hof, S. and D. W. Notermans. 2006. Clostridium difficile ribotype 027, toxinotype III, the Netherlands. Emerg. Infect. Dis. 12:827-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuijper, E. J., B. Coignard, J. Brazier, C. Suetens, D. Drudy, C. Wiuff, H. Pituch, P. Reichert, F. Schneider, A. F. Widmer, K. E. Olsen, F. Allerberger, D. W. Notermans, F. Barbut, M. Delmée, M. Wilcox, P. Pearson, B. Patel, D. J. Brown, R. Frei, T. Akerlund, I. R. Poxton, and P. Tüll. 2007. Update of Clostridium difficile-associated disease due to PCR ribotype 027 in Europe. Euro Surveill. 12:714. http://www.eurosurveillance.org/em/v12n06/1206-221.asp. [DOI] [PubMed] [Google Scholar]
- 21.Labbé, A. C., L. Poirier, D. MacCannell, T. Louie, M. Savoie, C. Béliveau, M. Laverdière, and J. Pépin. 2008. Clostridium difficile infections (CDI) in a Canadian tertiary care hospital before and during a regional epidemic associated with the BI/NAP1/027 strain. Antimicrob. Agents Chemother. 52:3180-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loo, V. G., L. Poirier, M. A. Miller, M. Oughton, M. D. Libman, S. Michaud, A. M. Bourgault, T. Nguyen, C. Frenette, M. Kelly, A. Vibien, P. Brassard, S. Fenn, K. Dewar, T. J. Hudson, R. Horn, P. René, Y. Monczak, and A. Dascal. 2005. A predominantly clonal multiinstitutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N. Engl. J. Med. 353:2442-2449. [DOI] [PubMed] [Google Scholar]
- 23.Mutlu, E., A. J. Wroe, K. Sanchez-Hurtado, J. S. Brazier, and I. R. Poxton. 2007. Molecular characterization and antimicrobial susceptibility patterns of Clostridium difficile strains isolated from hospitals in south-east Scotland. J. Med. Microbiol. 56:921-929. [DOI] [PubMed] [Google Scholar]
- 24.Muto, C. A., M. Pokrywka, K. Shutt, A. B. Mendelsohn, K. Nouri, K. Posey, T. Roberts, K. Croyle, S. Krystofiak, S. Patel-Brown, A. W. Pasculle, D. L. Paterson, M. Saul, and L. H. Harrison. 2005. A large outbreak of Clostridium difficile-associated disease with an unexpected proportion of deaths and colectomies at a teaching hospital following increased fluoroquinolone use. Infect. Control Hosp. Epidemiol. 26:273-280. [DOI] [PubMed] [Google Scholar]
- 25.Owens R. C., Jr., C. J. Donskey, R. P. Gaynes, V. G. Loo, and C. A. Muto. 2008. Antimicrobial-associated risk factors for Clostridium difficile infection. Clin. Infect. Dis. 15(Suppl. 1):S19-S31. [DOI] [PubMed] [Google Scholar]
- 26.Pepin, J., N. Saheb, M. A. Coulombe, M. F. Alary, M. P. Corriveau, S. Authier, M. Leblanc, G. Rivard, M. Bettez, V. Primeau, M. Nguyen, C. E. Jacob, and L. Lanthier. 2005. Emergence of fluoroquinolones as the predominant risk factor for Clostridium difficile-associated diarrhea: a cohort study during an epidemic in Quebec. Clin. Infect. Dis. 41:1254-1260. [DOI] [PubMed] [Google Scholar]
- 27.Piddock, L. J. 1999. Mechanisms of fluoroquinolone resistance: an update, 1994-1998. Drugs 58(Suppl. 2):11-18. [DOI] [PubMed] [Google Scholar]
- 28.Pituch, H., J. S. Brazier, P. Obuch-Woszczatynski, D. Wultanska, F. Meisel-Mikolajczyk, and M. Luczak. 2006. Prevalence and association of PCR ribotypes of Clostridium difficile isolated from symptomatic patients from Warsaw with macrolide-lincosamide-streptogramin B (MLSB) type resistance. J. Med. Microbiol. 55:207-213. [DOI] [PubMed] [Google Scholar]
- 29.Ruiz, J. 2003. Mechanisms of resistance to quinolones: target alterations, decreased accumulation and DNA gyrase protection. J. Antimicrob. Chemother. 51:1109-1117. [DOI] [PubMed] [Google Scholar]
- 30.Rupnik, M., M. H. Wilcox, and D. N. Gerding. 2009. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat. Rev. Microbiol. 7:526-536. [DOI] [PubMed] [Google Scholar]
- 31.Russo, P. 2009. Uso dei farmaci antibiotici in Italia e in Europa, p. 15-17. In A. Pantosti and M. Del Grosso (ed.), Giornata europea degli antibiotici: uso responsabile per il controllo dell'antibiotico-resistenza. Rapporti ISTISAN 09/32. Istituto Superiore di Sanità, Rome, Italy.
- 32.Spigaglia, P., and P. Mastrantonio. 2004. Comparative analysis of Clostridium difficile clinical isolates belonging to different genetic lineages and time periods. J. Med. Microbiol. 53:1129-1136. [DOI] [PubMed] [Google Scholar]
- 33.Spigaglia, P., V. Carucci, F. Barbanti, and P. Mastrantonio. 2005. ErmB determinants and Tn916-like elements from clinical isolates of Clostridium difficile. Antimicrob. Agents Chemother. 49:2550-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spigaglia, P., F. Barbanti, P. Mastrantonio, J. S. Brazier, F. Barbut, M. Delmée, E. Kuijper, and I. R. Poxton. 2008. Fluoroquinolone resistance in Clostridium difficile isolates from a prospective study of C. difficile infections in Europe. European Study Group on Clostridium difficile. J. Med. Microbiol. 57:784-789. [DOI] [PubMed] [Google Scholar]
- 35.Spigaglia, P., A. Carattoli, F. Barbanti, and P. Mastrantonio. 21 October 2009, posting date. Detection of gyrA and gyrB mutations in Clostridium difficile isolates by real-time PCR. Mol. Cell Probes [Epub ahead of print.] doi: 10.1016/j.mcp.2009.10.002. [DOI] [PubMed]
- 36.Stubbs, S., M. Rupnik, M. Gibert, J. Brazier, B. Duerden, and M. Popoff. 2000. Production of actin-specific ADP-ribosyltransferase (binary toxin) by strains of Clostridium difficile. FEMS Microbiol. Lett. 186:307-312. [DOI] [PubMed] [Google Scholar]
- 37.Terhes, G., J. S. Brazier, E. Urbán, J. Sóki, and E. Nagy. 2006. Distribution of Clostridium difficile PCR ribotypes in regions of Hungary. J. Med. Microbiol. 55:279-282. [DOI] [PubMed] [Google Scholar]
- 38.Vaccheri, A., M. C. Silvani, L. Bersaglia, D. Motola, P. Strahinja, A. Vargiu, E. Poluzzi, and N. Montanaro. 2008. A 3 year survey on the use of antibacterial agents in five Italian hospitals. J. Antimicrob. Chemother. 61:953-958. [DOI] [PubMed] [Google Scholar]
- 39.Wilcox, M. H. 2003. Gastrointestinal disorders and the critically ill. Clostridium difficile infection and pseudomembranous colitis. Best Pract. Res. Clin. Gastroenterol. 17:475-493. [DOI] [PubMed] [Google Scholar]