Molecular diagnostic methods using genetic material probes have been employed in the health care field for the detection and quantitation of several species of microorganisms (2, 5). These methods are faster and more suitable than traditional culture methods. In addition, the possibility of the inclusion of difficult-to-culture, uncultivable, or uncharacterized species in the set of target species has led to a more comprehensive investigation of microorganism communities in oral infections (1, 7).
The checkerboard DNA-DNA hybridization method has been widely used in dentistry to detect pathogenic or nonpathogenic species harbored in the oral cavities of healthy and/or diseased patients (3, 4). This method enables the rapid and simultaneous identification of several microbial species (up to 45) in a large number (up to 28) of oral samples. The original method proposed by Socransky et al. (8) employs digoxigenin-labeled whole genomic DNA probes which are hybridized to DNA samples, followed by chemiluminescent detection, as a result of an antigen-antibody reaction. However, technical difficulties associated with this reaction, material costs, and mainly the processing time needed due to the higher number of membranes to be evaluated can limit the use of this protocol or make it difficult to use. Thus, in the present study, we describe a modified protocol which employs rapid direct labeling and detection of whole genomic DNA probes as an alternative form of the checkerboard DNA-DNA hybridization method.
Genomic DNA was extracted from isolates of the bacterial species according to a modification of the method originally described by Pitcher et al. (6). The whole genomic DNA probes from the 13 bacterial species were directly labeled with thermostable alkaline phosphatase enzyme using the AlkPhos Direct Labeling and Detection System (GE Healthcare UK). Briefly, 100 ng of denatured DNA was mixed with the labeling buffer and alkaline phosphatase enzyme. Formaldehyde was then added to covalently cross-link the enzyme to the probe. The resulting alkaline phosphatase-labeled probes were adjusted to a final concentration of 1 ng/μl.
Tests with different DNA probe concentrations were performed in order to optimize the amounts of labeled probe necessary to distinctively detect 104, 105, 106, and 107 cells with the lowest possible background. The labeled probes were hybridized against whole genomic DNA extracted from each bacterial species. After the calibration tests, the labeled DNA probes were tested against oral biofilm samples. Subgingival biofilm samples from 10 periodontally healthy or minimally diseased subjects were collected with sterile paper points. After harvesting, each sample was transferred to a microtube containing 150 μl of TE (10 mM Tris-HCl, 1 mM EDTA, pH 7.6), followed by the addition of 150 μl of 0.5 M NaOH. The samples were kept at −20°C until processing by the checkerboard DNA-DNA hybridization method. The samples were applied to the membranes according to Socransky et al. (8). The membranes were prehybridized at 60°C for 2 h in a hybridization buffer consisting of NaCl at 0.5 M and blocking reagent at 0.4% (wt/vol). Defined amounts of labeled whole genomic probes were applied to the membranes. Hybridization was performed overnight at 60°C under gentle agitation. On the following day, the membranes were washed twice at 65°C for 30 min in primary wash buffer (urea at 2 M, sodium dodecyl sulfate at 0.1%, NaH2PO4 at 50 mM [pH 7.0], NaCl at 150 mM, MgCl2 at 1 mM, blocking reagent at 0.2% [wt/vol]) and twice in secondary wash buffer (Tris base at 1 M, NaCl at 2 M, MgCl2 at 1 M) at room temperature for 15 min. After washing, the hybrids were direct detected by chemiluminescence using the Gene Images CDP-Star Reagent (GE Healthcare UK). Signals were detected by exposing the membrane to ECL Hyperfilm-MP (GE Healthcare UK) for 10 min. The images obtained on Hyperfilm were digitized and analyzed with the ImageQuant TL software (GE Healthcare UK). Based on the pixel intensities of the chemiluminescent signals that originated from the cell concentrations of each sample compared with the control lanes, the amounts of bacterial cells collected from the subgingival samples could be classified according to the following scores, as proposed by Socransky et al. (8): 0, no cells detected; 1, <105 cells; 2, ∼105 cells; 3, 105 to 106 cells; 4, ∼106 cells; 5, >106 cells.
Calibration of the probe concentrations allowed the detection of DNA amounts corresponding to 105 and 106 cells of the species tested for while maintaining enough sensitivity to discriminate among the six proposed scores. As in the original protocol, hybridization of labeled probes and DNA quantities equivalent to 104 cells did not result in either detectable or reproducible signals. On the other hand, quantities equivalent to 107 cells result in higher-intensity signals and higher background levels.
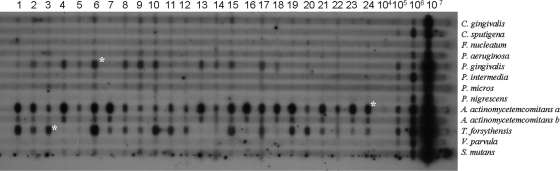
Figure 1 represents the hybridization of labeled probes against 24 subgingival biofilm samples from healthy subjects. The signals can be quantified by comparing the intensities of the signals from the dental samples to those of the control samples (lanes 105, 106, and 107) in the membrane. The results revealed that all of the species tested for were detected in most of the subgingival biofilm samples investigated. Aggregatibacter actinomycetemcomitans a was present in amounts of >105 and ≤106 cells in 14 of the samples tested. Prevotella nigrescens and Veillonella parvula were the species founded with a lower incidence. Our results revealed that this alternative protocol provides a shortened processing time with a sensitivity and specificity comparable to those of the original and that it is suitable for use in studies using the checkerboard DNA-DNA hybridization format.
FIG. 1.
Checkerboard DNA-DNA hybridization format. Shown is the membrane used to detect 13 bacterial species (Capnocytophaga gingivalis, Capnocytophaga sputigena, Fusobacterium nucleatum, Pseudomonas aeruginosa, Porphyromonas gingivalis, Prevotella intermedia, Peptostreptococcus micros, P. nigrescens, A. actinomycetemcomitans a and b, Tannerella forsythensis, V. parvula, and Streptococcus mutans) in 24 subgingival biofilm samples from healthy subjects. Vertical lanes 1 to 24 contained DNA extracted from the samples. The rightmost lanes are standards containing DNA amounts equivalent to 104, 105, 106, and 107 cells of each species tested for. The horizontal rows contained the indicated DNA probes diluted in hybridization buffer. An asterisk at the intersection of the horizontal rows and vertical lanes indicates the presence of a species. The intensity of the signals obtained is proportional to the amount of DNA immobilized in the membrane.
Footnotes
Published ahead of print on 16 June 2010.
REFERENCES
- 1.Alves, F. R., J. F. Siqueira, Jr., F. L. Carmo, A. L. Santos, R. S. Peixoto, I. N. Rocas, and A. S. Rosado. 2009. Bacterial community profiling of cryogenically ground samples from the apical and coronal root segments of teeth with apical periodontitis. J. Endod. 35:486-492. [DOI] [PubMed] [Google Scholar]
- 2.Baumgartner, S., T. Imfeld, O. Schicht, C. Rath, R. E. Persson, and G. R. Persson. 2009. The impact of the stone age diet on gingival conditions in the absence of oral hygiene. J. Periodontol. 80:759-768. [DOI] [PubMed] [Google Scholar]
- 3.De Souza, R. F., C. Nascimento, R. R. Regis, C. H. Silva-Lovato, and H. F. Paranhos. 2009. Effects of the domestic use of a disclosing solution on the denture biofilm: a preliminary study. J. Oral Rehabil. 36:491-497. [DOI] [PubMed] [Google Scholar]
- 4.do Nascimento, C., R. F. Albuquerque, Jr., J. P. M. Issa, I. Y. Ito, C. H. Lovato da Silva, H. F. Paranhos, and R. F. de Souza. 2009. Use of the DNA checkerboard hybridization method for detection and quantitation of Candida species in oral microbiota. Can. J. Microbiol. 55:622-626. [DOI] [PubMed] [Google Scholar]
- 5.Papaioannou, W., S. Gizani, A. D. Haffajee, M. Quirynen, E. Mamai-Homata, and L. Papagiannoulis. 2009. The microbiota on different oral surfaces in healthy children. Oral Microbiol. Immunol. 24:183-189. [DOI] [PubMed] [Google Scholar]
- 6.Pitcher, D. G., N. A. Sanders, and R. J. Owen. 1989. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 8:151-156. [Google Scholar]
- 7.Sakamoto, M., J. F. Siqueira, Jr., I. N. Rocas, and Y. Benno. 2009. Diversity of spirochetes in endodontic infections. J. Clin. Microbiol. 47:1352-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Socransky, S. S., C. Smith, L. Martin, B. J. Paster, F. E. Dewhirst, and A. E. Levin. 1994. “Checkerboard” DNA-DNA hybridization. Biotechniques 17:788-792. [PubMed] [Google Scholar]