Abstract
The MTBDRsl assay (Hain Lifescience GmbH, Germany) is a new line probe assay for the detection of extensively drug-resistant tuberculosis (XDR TB). The test simultaneously detects resistance to ethambutol, aminoglycosides/cyclic peptides, and fluoroquinolones through detection of mutations in the relevant genes. The assay format is identical to the MTBDR Hain assay. The assay was evaluated for the detection of second-line-drug resistance in Vietnamese isolates using two sample sets from the microbiology department of Pham Ngoc Thach Hospital, Ho Chi Minh City, Viet Nam, with existing conventional phenotypic drug susceptibility results for second-line drugs: 41 consecutive fluoroquinolone-resistant isolates and 21 consecutive multidrug-resistant but fluoroquinolone-sensitive isolates. The sensitivity for detection of fluoroquinolone resistance was 75.6% (31/41) (95% confidence interval [95% CI], 59.7% to 87.6%), and for kanamycin resistance, the sensitivity was 100% (5/5) (95% CI, 47.8% to 100%). The sensitivity of the test for detection of ethambutol resistance was low, consistent with previous reports, at 64.2% (34/53) (95% CI, 49.8% to 76.9%). The specificity of the test was 100% for all three drugs. These data suggest that the MTBDRsl assay is a rapid, specific test for the detection of XDR TB but should not be used exclusively to “rule out” second-line-drug resistance. Further operational evaluation is required and should be integrated with evaluations of the MTBDR test.
The World Health Organization (WHO) has estimated that 5% of all tuberculosis (TB) cases globally are now multidrug-resistant tuberculosis (MDRTB) (resistance to at least rifampin [RIF] and isoniazid [INH]), based on data acquired since 2000 from more than 100 countries (14). Every year, an estimated 490,000 new cases of MDRTB occur, causing more than 130,000 deaths (14). In 2006, the documentation of a rapidly fatal TB outbreak among hospitalized HIV patients in Kwa Zulu Natal, South Africa (5) led to the definition of extensively drug-resistant tuberculosis (XDR TB) as TB resistant to a fluoroquinolone and injectable second-line drug (amikacin, capreomycin, or kanamycin) in addition to isoniazid and rifampin. XDR TB has subsequently been reported from over 50 countries by WHO (14). It is likely that the majority of XDR TB cases worldwide remain undetected due to the lack of second-line-drug testing in most high-burden settings. There are an estimated 40,000 new cases of extensively drug-resistant tuberculosis each year (15).
The recognition of XDR TB worldwide has made timely identification of XDR TB cases to achieve effective disease management and to prevent their spread a priority (3, 8).
Significant challenges exist; although standard protocols exist for second-line-drug susceptibility testing, strong evidence is lacking on many factors, such as the reproducibility and reliability of results, applicability of MIC to clinical outcomes, and intermethod variability. Proficiency testing for second-line-drug susceptibility testing has only recently been integrated into the supranational reference laboratory panel in an effort to improve standardization of second-line drugs across the WHO reference laboratory network.
Conventional drug resistance testing takes more than 2 weeks to return a result even after a positive culture has been isolated. Rapid commercial liquid-based culture systems, such as Bactec MGIT 960 testing (Becton Dickinson), for second-line drugs are not yet formally FDA/WHO approved but are reported to be accurate, widely used in developed settings, and reduce turnaround times to approximately 8 days (7). In recent years, many second-line-drug susceptibility testing methods have been developed. The most rapid results are achieved by direct testing of patient specimens by molecular methods; however, in addition to the high cost of such tests, the sensitivity remains suboptimal, and rigorous contamination control is required to maintain accuracy. The majority of high-burden settings currently lack the resources to implement such tests effectively. Two commercial DNA strip assays, INNO-LiPA RifTB (Innogenetics, Zwijndrecht, Belgium) and MTBDRplus (Hain Lifescience GmbH, Germany), targeting the rpoB plus katG and inhA genes have been extensively evaluated for use with Mycobacterium tuberculosis culture and directly on sputum to identify MDR TB cases (2, 10). The Foundation for Innovative Diagnostics (FIND) demonstration projects in South Africa of the GenoType MTBDRplus assay resulted in the recommendation of commercial line probe assays for use in high-burden settings by WHO (16). This assay is based on a multiplex PCR in combination with reverse hybridization. Either the absence of wild-type bands or the appearance of bands targeting specific mutations indicates the presence of a resistant strain. MDR TB cases can be detected within 1 or 2 days of sputum sampling using this assay.
In order to rapidly detect second-line-drug resistance, the MTBDRsl test (Hain Lifescience GmbH, Germany) has been developed. This assay can detect mutations in gyrA, rrs, and embB genes, detecting resistance to the fluoroquinolones (FQ), aminoglycosides/cyclic peptides, and ethambutol (EMB), respectively, with a single assay. A previous evaluation study in Germany showed that this assay has a high accuracy for FQ and amikacin-capreomycin resistance testing in clinical strains and sputum samples (6). EMB detection was specific (100%), but its sensitivity (69.2%) was low.
The MTBDRsl assay contains 22 probes, including 16 probes for gene mutation detection and 6 probes for the control of the test procedure. The six control probes include a conjugate control (CC), an amplification control (AC), a Mycobacterium tuberculosis complex control (TUB), and three locus probes (gyrA, rrs, and embB) for gene amplification control. The remaining probes detect FQ resistance (gyrA), amikacin/capreomycin resistance (rrs), and ethambutol resistance (embB). The probes contained in the assay do not detect all mutations in these genes but are targeted to the most commonly occurring mutations.
The aim of the present study was to determine the accuracy of this assay for detection of FQ, kanamycin, and ethambutol resistance against conventional phenotypic testing as the gold standard on Vietnamese isolates of M. tuberculosis.
MATERIALS AND METHODS
Samples.
Two sample sets with a high prevalence of second-line-drug resistance were used for evaluation. A total of 41 consecutive FQ-resistant isolates from individual patients were collected from Pham Ngoc Thach Hospital in Viet Nam between July 2005 and July 2006 (12). A set of 21 consecutive pulmonary MDR, FQ-sensitive isolates from individual patients collected at Pham Ngoc Thach Hospital between January and March 2005 was used to assess specificity of FQ resistance detection (4). Drug susceptibility testing (DST) is not routine in the Vietnamese national TB program and is done only on request from the treating clinician, usually following treatment failure or relapse. The outcome and follow-up data were not available for any of the patients.
Drug susceptibility testing.
Conventional 1% proportion phenotypic drug susceptibility testing (DST) on Lowenstein Jensen (LJ) medium was performed at Pham Ngoc Thach Hospital (PNT) Laboratory. All samples were tested for resistance to isoniazid (0.2 μg/ml), rifampin (40 μg/ml), streptomycin (STR) (4 μg/ml), ofloxacin (2 μg/ml), ethambutol (EMB) (2 μg/ml), kanamycin (20 μg/ml), thioacetone (10 μg/ml), pyrazinamide (PZA) (200 μg/ml), ethionamide (40 μg/ml), cycloserine (30 g/ml), and para-aminosalicylic acid (PAS) (0.5 g/ml). Pham Ngoc Thach Laboratory is the national reference laboratory for southern Vietnam and participates in WHO supranational proficiency testing for DST for all first-line drugs, FQ, and kanamycin. The performance of this laboratory is consistently higher than the minimum requirement.
MTBDRsl assay.
PCR and hybridization were performed according to the manufacturer's instructions. For amplification, 35 μl of primer nucleotide mix (PNM) (provided in the kit), 5 μl of 10× amplification buffer (Qiagen), 1.2 μl of 2.5 M MgCl2 (Qiagen), 5 μl of DNA (15-ng/μl) and water was added to a final volume of 50 μl. The PCR products were denatured by denaturing solution at room temperature, followed by hybridization with hybridization buffer at 45°C for 30 min in a shaking water bath. After stringent washing of the PCR products, hybridization was detected by colorimetric reaction. To control for cross contamination, water in lieu of DNA template was added to one negative control included in each run.
gyrA sequencing.
The quinolone resistance-determining region (QRDR) in gyrA was sequenced in all isolates to evaluate discrepancies between the MDRTBsl assay and phenotypic testing. All samples were cultured on LJ medium, and DNA was extracted for sequencing using standard methods (12). Primers GYRATBF (5′-CAG CTA CAT CGA CTA TGC GA-3′) and GYRATBR (5′-GGG CTT CGG TGT ACC TCA T-3′) were used to amplify a 320-bp fragment of the gyrA gene (12). Sequencing of PCR product was carried out with CEQ 8000 genetic analysis system (Beckman Coulter), and the results were analyzed with CEQ8000 software.
Statistics.
Data were analyzed with Stata 9 (Statacorp). McNemar's test was used to estimate agreement between the tests. The sensitivity, specificity and 95% confidence intervals (95% CIs) were calculated for each drug.
RESULTS
Phenotypic DST results.
Table 1 summarizes drug susceptibility patterns of FQ-resistant and FQ-sensitive isolates included in the study. Of the 41 FQ-resistant isolates, 43.9% (18/41) were resistant to INH, RIF, STR, EMB, and PZA but sensitive to all second-line drugs. Three isolates (7.3%) were MDR strains resistant to kanamycin in addition to ofloxacin and therefore XDR TB. One of these XDR isolates showed resistance to all second-line drugs tested except PAS.
TABLE 1.
Phenotypic drug susceptibility profiles of 41 isolates resistant to ofloxacin and 21 ofloxacin-sensitive control isolates
| Pattern | Drug susceptibility patterna |
No. (%) of ofloxacin-resistant isolates | No. (%) of ofloxacin-sensitive isolates | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| INH | RIF | STR | EMB | PZA | KANA | ETHIO | CYCLO | TB1 | PAS | |||
| 1 | R | R | R | R | S | S | S | R | S | S | 0 | 3 (14.3) |
| 2 | R | R | R | R | S | R | R | S | S | S | 0 | 2 (9.5) |
| 3 | R | R | R | R | S | S | R | R | R | S | 0 | 2 (9.5) |
| 4 | R | R | R | R | S | S | R | S | S | R | 0 | 1 (4.8) |
| 5 | R | R | R | R | S | S | S | S | R | S | 0 | 1 (4.8) |
| 6 | R | R | R | R | S | S | S | S | S | S | 6 (14.6) | 12 (57.1) |
| 7 | R | R | R | R | R | S | S | S | S | S | 18 (43.9) | 0 |
| 8 | R | R | R | S | R | S | S | S | S | S | 3 (7.3) | 0 |
| 9 | R | R | R | S | S | S | S | S | S | S | 2 (4.9) | 0 |
| 10 | R | S | R | R | R | S | S | S | S | S | 2 (4.9) | 0 |
| 11 | R | R | R | R | R | R | R | R | R | S | 1 (2.4) | 0 |
| 12 | R | R | R | R | R | R | S | S | S | R | 1 (2.4) | 0 |
| 13 | R | R | R | R | R | S | R | S | R | S | 1 (2.4) | 0 |
| 14 | R | R | R | R | R | S | R | S | S | S | 1 (2.4) | 0 |
| 15 | R | R | R | R | R | S | S | S | S | R | 1 (2.4) | 0 |
| 16 | R | R | R | S | S | R | S | S | S | S | 1 (2.4) | 0 |
| 17 | R | R | R | S | S | S | R | S | S | S | 1 (2.4) | 0 |
| 18 | R | R | S | R | S | S | S | S | S | R | 1 (2.4) | 0 |
| 19 | S | S | R | S | S | S | S | S | S | S | 1 (2.4) | 0 |
| 20 | S | S | S | S | S | S | S | S | S | S | 1 (2.4) | 0 |
| Total | 41 (100) | 21 (100) | ||||||||||
Abbreviations: INH, isoniazid; RIF, rifampin; STR, streptomycin; EMB, ethambutol; PZA, pyrazinamide; KANA, kanamycin; ETHIO, ethionamide; CYCLO, cycloserine; TB1, thioacetazone; PAS, para-aminosalicylic acid; R, resistant; S, sensitive.
Among the FQ-sensitive isolates, all isolates were resistant to INH, RIF, STR, and EMB. Specifically, 57.1% (12/21) isolates were resistant to INH, RIF, STR, and EMB but sensitive to PZA and all second-line drugs tested. The remaining isolates showed a range of second-line DST profiles.
MDRTBsl assay.
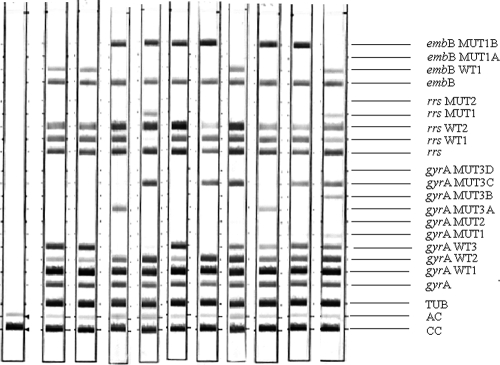
Representative hybridization patterns obtained are shown in Fig. 1. Interpretable results were obtained for all cultures tested.
FIG. 1.
Representative patterns obtained by the MTBDRsl assay. Lane 1, negative control; lanes 2 and 3, FQ-, EMB-, and KAN-susceptible strains; lanes 4, 5, 6, and 7, resistant strains (lane 4, resistant to FQ and EMB; lane 5, resistant to FQ, AMP/CMP, and EMB; lane 6, resistant to EMB; lane 7, resistant to FQ and EMB); lanes 8, 9, 10, and 11, strains containing wild-type and FQ-resistant populations. Specific probes are as follows: CC, conjugate control; AC, amplification control; TUB, Mycobacterium tuberculosis complex control; gyrA, gyrA gene amplification control; gyrA WT1 to WT3, gyrA wild-type probes; gyrA MUT1, gyrA MUT2, and gyrA MUT3A to MUT3D, gyrA mutant probes for A90V, S91P, D94A, D94N, D94Y-D94G, and D94H, respectively; rrs, rrs amplification control; rrs WT1 and WT2, rrs wild-type probes; rrs MUT1 and MUT2, rrs mutant probes for A1401G and G1484T, respectively; embB, embB amplification control; embB WT1, embB wild-type probe I codon 306; embB MUT1A and embB MUT1B, embB mutant probes for M306I and M306V, respectively.
FQ resistance.
The concordance between phenotypic testing and the MDRTBsl assay was 83.9% (52/62) for detecting FQ resistance (Table 2) (P = 0.002). The sensitivity of the MTBDRsl assay for detecting FQ resistance was 75.6% (31/41) (95% CI, 59.7 to 87.6). The specificity was 100% (95% CI, 83.9% to 100%).
TABLE 2.
Concordance between the MTBDRsl assay and phenotypic testing (gold standard) for ethambutol, ofloxacin, and kanamycin resistance
| Drug and phenotypic result (no. of isolates) | No. of isolates (%) with the following MTBDRsl assay result: |
|
|---|---|---|
| Resistant | Sensitive | |
| Ethambutol | ||
| Resistant (n = 53) | 34 (64.2) | 19 (35.8) |
| Sensitive (n = 9) | 0 | 9 (100) |
| Total (n = 62) | 34 (54.8) | 28 (45.2) |
| Ofloxacin | ||
| Resistant (n = 41) | 31 (75.6) | 10 (24.4) |
| Sensitive (n = 21) | 0 | 21 (100) |
| Total (n = 62) | 31 (50) | 31 (50) |
| Kanamycin | ||
| Resistant (n = 5) | 5 (100) | 0 |
| Sensitive (n = 57) | 0 | 57 (100) |
| Total (n = 62) | 5 (8.8) | 57 (91.9) |
The gyrA gene in the isolates were sequenced to evaluate discrepancies. Of 41 isolates, 33 (80.4%) had identical results by sequencing and the MTBDRsl assay (Table 3). Of those isolates with discrepant sequencing/MTBDRsl results, all but one isolate (FQ1319) had the same resistant/sensitive classification by both tests. This isolate had no mutation detected by sequencing but showed the presence of D94N gyrA mutation (MUT3B) by the MTBDRsl assay. For the seven remaining discrepant isolates, one sample (FQ1150) hybridized to the D94G mutation probe (MUT3C) by the MDRTBsl assay, but sequencing showed a D94N mutation, which should hybridize with probe MUT3B by the MTBDRsl assay. One isolate (FQ1155) was recorded as resistant by the MTBDRsl assay, because no hybridization was detected for the WT3 probe (D94X), but this isolate did not hybridize to any mutant probe. Mutation D94Y was shown by sequencing, which should have given hybridization for probe MUT3B by the MTBDRsl assay.
TABLE 3.
MTBDRsl assay and sequencing results for gyrA mutation of 42 phenotypically FQ-resistant strains included in the study
| Strain | MTBDRsl assay results |
GyrA sequencing result (mutation detected)c | Concordanced | ||
|---|---|---|---|---|---|
| FQ resulta | Hybridization patternb | Mutation detected | |||
| FQ1091 | R | WT + MUT3C | D94G | D94G* | Yes |
| FQ1092 | R | ΔWT3 + MUT3A | D94A | D94A | Yes |
| FQ1093 | R | ΔWT3 + MUT3C | D94G | D94G | Yes |
| FQ1094 | R | ΔWT3 + MUT3C | D94G | D94G | Yes |
| FQ1095 | R | WT + MUT3A | D94A | D94A* | Yes |
| FQ1150 | R | WT + MUT3C | D94G | D94N | No |
| FQ1151 | R | ΔWT3 + MUT3C | D94G | D94G | Yes |
| FQ1152 | R | WT + MUT3C | D94G | D94G* | Yes |
| FQ1153 | R | ΔWT2 + MUT1 | A90V | A90V | Yes |
| FQ1154 | S | WT | None (WT) | None | Yes |
| FQ1155 | R | ΔWT3 | D94X | D94Y | Partial |
| FQ1156 | R | ΔWT2 + MUT1 | A90V | A90V | Yes |
| FQ1157 | S | WT | None (WT) | None | Yes |
| FQ1158 | R | ΔWT2 + MUT1 | A90V | A90V | Yes |
| FQ1159 | R | ΔWT3 + MUT3A | D94A | D94A | Yes |
| FQ1160 | S | WT | None (WT) | None | Yes |
| FQ1161 | S | WT | None (WT) | None | Yes |
| FQ1162 | R | ΔWT3 + MUT3C | D94G | A90V*/D94G* | Partial |
| FQ1281 | R | WT + MUT3C | D94G | A90V*/D94G* | Partial |
| FQ1283 | R | WT + MUT1 + MUT3B + MUT3C | A90V/D94N/D84Y and D94G | A90V*/D94G* | Partial |
| FQ1284 | S | WT | None (WT) | None | Yes |
| FQ1286 | R | ΔWT3 + MUT3C | D94G | D94G | Yes |
| FQ1288 | S | WT | None (WT) | None | Yes |
| FQ1289 | R | WT+ MUT1 + MUT3A | A90V/D94A | A90V*/D94A* | Yes |
| FQ1290 | R | ΔWT3 + MUT3A | D94A | D94A | Yes |
| FQ1291 | R | ΔWT3 + MUT3C | D94G | D94G | Yes |
| FQ1292 | R | ΔWT3 + MUT3A + MUT3C | D94A/D94G | D94G*/D94A* | Yes |
| FQ1293 | R | ΔWT3 + MUT3A | D94A | D94A | Yes |
| FQ1294 | S | WT | None (WT) | None | Yes |
| FQ1295 | R | WT + MUT3C | D94G | D94G* | Yes |
| FQ1296 | R | ΔWT3 + MUT3C | D94G | D94G | Yes |
| FQ1297 | R | ΔWT3 + MUT3B + MUT3C | D94N/D94Y and D94G | D94G* | Partial |
| FQ1298 | R | ΔWT3 + MUT3B + MUT3C | D94N/D94Y and D94G | D94G* | Partial |
| FQ1318 | S | WT | None (WT) | None | Yes |
| FQ1319 | R | WT + MUT3B | D94N/D94Y | None | No |
| FQ1320 | R | ΔWT2 + MUT1 | A90V | A90V | Yes |
| FQ1321 | R | ΔWT2 + MUT1 | A90V | A90V | Yes |
| FQ1322 | R | ΔWT3 + MUT3C | D94G | D94G | Yes |
| FQ1323 | R | ΔWT2 + MUT1 | A90V | A90V | Yes |
| FQ1324 | S | WT | None (WT) | None | Yes |
| FQ1325 | S | WT | None (WT) | None | Yes |
Abbreviations: R, resistant; S, susceptible (result for FQ DST by MTBDRsl assay).
The hybridization pattern shows the probes the sample hybridized to. WT, wild type; ΔWT indicates lack of hybridization to the wild-type probe.
Asterisks indicate that the mutation was found in different peaks.
Partial concordance means that at least one identical mutation was detected by both tests.
Two samples (FQ1162 and FQ1281) had D94G mutations only by the MTBDRsl assay, but both D94G and A90V mutations were detected by sequencing; however, this was probably a consequence of a mixed wild-type-resistant population, as detected by sequencing. In both cases, the A90V mutant peak was only 30% of the wild-type height, indicating that the wild-type population appeared to be predominant.
The remaining three isolates (FQ1283, FQ1297, and FQ1298) showed multiple mutations with the MTBDRsl assay which were not all detected by sequencing. All of these samples recorded a resistant result by both tests. These discrepant results may have been due to cross contamination, but the seven discrepant isolates were tested again by the MTBDRsl assay and gave identical results, and no contamination was observed in negative controls or in FQ-sensitive isolates. The difference may therefore be due to differential amplification of targets by the PCRs for MTBDRsl and sequencing.
Aminoglycoside resistance.
The sensitivity (5/5) (95% CI, 47.8% to 100%), specificity (57/57) (95% CI, 93.7% to 100%), and concordance were all 100% for kanamycin resistance (P = 1). Mutations detected in rrs by the MTBDRsl assay were A1401G (4/5 [80%]) and G1484T (1/5 [20%]) (Table 4).
TABLE 4.
MTBDRsl pattern of 53 phenotypically ethambutol-resistant and 5 phenotypically kanamycin-resistant isolates
| Drug (gene) and MTBDRsl hybridization patterna | Codon mutation | No. (%) of isolates with MTBDRsl hybridization pattern |
|---|---|---|
| Ethambutol (embB) | ||
| ΔWT | ATG306ATC/ATT | 5 (9.4) |
| ΔWT + MUT1A | M306I | 5 (9.4) |
| ΔWT + MUT1B | M306V | 21 (39.6) |
| WT + MUT1A | None (WT) + M306I | 1 (1.9) |
| WT + MUT1B | None (WT) + M306V | 2 (3.8) |
| WT + MUT1A + MUT1B | None (WT) + M306I + M306V | 1 (1.9) |
| WT | None (WT) | 18 (34.0) |
| Total | 53 (100) | |
| Kanamycin (rrs) | ||
| ΔWT1 + WT2 + MUT1 | A1401G | 4 (44.4) |
| WT1 + WT2 + MUT1 | None (WT) + G1484T | 1 (11.1) |
| Total | 5 (100) |
Ethambutol resistance.
The concordance for detection of EMB resistance was 69.4% (43/62) (P < 0.001). The sensitivity for detection of EMB resistance was 64.2% (34/53) (95% CI, 49.8% to 76.9%). The specificity was 100% (9/9) (95% CI, 66.4% to 100%). The most common mutation detected in embB by the MTBDRsl assay was M306V in 45.3% (24/53) of EMB-resistant isolates. Mutations at ATG306ATC/ATT (5/53 [9.4%]) and M306I (7/53 [13.2%]) were also detected. Two isolates with mutation M306I and one isolate with mutation M306I were mixed wild-type-resistant populations, and one isolate had both M306I and M306V mutations in addition to the wild-type codon (Table 4).
DISCUSSION
The MTBDRsl assay is a specific test for the detection of resistance to fluoroquinolones, ethambutol, and kanamycin in M. tuberculosis. The sensitivity of this assay is variable for different drugs; therefore, this test should not be used to “rule out” resistance to second-line drugs, but it is an accurate rapid screening test for the identification of second-line drug resistance, especially in suspected cases of XDR TB. Detection of ethambutol (64.2%) resistance in Vietnamese isolates is similar to that previously reported by Hilleman et al. (6) for isolates from Germany and Brossier et al. from France (3a) at 69.2% and 57.0%, respectively. Similarly for kanamycin resistance, the sensitivity was 100% in this study and 86.7% and 77% in the other studies (6). Only a small number of kanamycin-resistant isolates were available for testing in all studies (n = 5, n = 8, and n = 13), and testing of a larger number of aminoglycoside-resistant isolates is required to define sensitivity accurately. Detection of FQ resistance was lower in this study (75.6%) than for German (90.6%) and French (87.0%) isolates, due to a larger percentage of FQ-resistant isolates without mutations in the gyrA region. We encountered similar rates of heteroresistance—the presence of mixed wild-type/resistant populations, as detected by different peaks by sequencing. This has now been reported from several settings, including Germany, Russia, and Vietnam (6, 9, 12). The reasons why mixed populations of fluoroquinolone-resistant/sensitive populations are frequently detected whereas they are not for the other TB drugs remains unclear but may be related to their over-the-counter availability and widespread short-course use as broad-spectrum antibiotics for the treatment of respiratory infections, allowing resistant populations to emerge but not be fully selected.
The low detection rate of ethambutol resistance is consistent with reports from other settings, with embB mutations accounting for between 30 and 70% of ethambutol-resistant isolates (1, 18). This underlines the need to identify other mutations for ethambutol resistance in order to improve the sensitivity of molecular tests, including line probe assays, for ethambutol resistance detection.
It was previously thought that cross-resistance among the amikacin and kanamycin aminoglycosides was total; however, substantial recent data have demonstrated that this is not the case (11, 13, 17). Specific mutations appear to confer complete cross-resistance, while others do not. For example, eis promoter mutations confer low-level resistance to kanamycin but not amikacin (17).
Line probe assays may offer the further advantage of identifying specific mutations conferring resistance which may allow the tailoring of optimized individual treatment regimens for patients based on MIC/cross-resistance patterns which would be a particular advantage in XDR TB patients where effective treatment options are severely restricted. However, further research is required to elucidate the nature of such cross-resistance patterns before clinical recommendations can be made.
The WHO recommends the use of commercial line probe assays (INNO-LiPA and Hain) for the rapid detection of MDR TB within national TB programs with the capacity to implement appropriate laboratory infrastructure, staff training, contamination control, external quality assessment schemes, standardized operating procedures, rapid reporting of results, and availability of quality-assured second-line drugs. Preliminary economic analysis suggests that line probe assays may offer cost savings over conventional practices: the cost per valid MTBDRplus assay result in South Africa was $19.56 when performed directly on primary specimens compared to $31.12 for MGIT culture and conventional DST (7H11 medium). Although this cost assessment did not include the costs of establishing laboratory infrastructure and staff training for molecular assays and will vary in different settings, these data suggest that commercial line probe assays are economically viable in some high-burden settings (16).
This study evaluated the performance of the MTBDRsl assay on DNA extracted from cultural isolates and did not evaluate performance of the test directly on sputum samples. Hilleman et al. (6) evaluated 64 sputum samples and achieved good sensitivity, but further data are required on performance directly on sputum. However, initial evaluations of the MTBDRsl test suggest that it may achieve performance characteristics similar to those of the line probe assays currently available for the detection of MDR TB and could be implemented for the timely identification of XDR TB cases in settings already performing the MDR TB assays. The rapid identification of XDR TB cases will allow early initiation of appropriate therapy and infection control and prevent further amplification of drug resistance. Operational studies should be conducted to evaluate the performance of the test in high-burden settings and to determine the optimal algorithms for implementation of the test by national TB programs in settings of high drug resistance.
Footnotes
Published ahead of print on 23 June 2010.
REFERENCES
- 1.Alcaide, F., G. E. Pfyffer, and A. Telenti. 1997. Role of embB in natural and acquired resistance to ethambutol in mycobacteria. Antimicrob. Agents Chemother. 41:2270-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnard, M., H. Albert, G. Coetzee, R. O'Brien, and M. E. Bosman. 2008. Rapid molecular screening for multidrug-resistant tuberculosis in a high-volume public health laboratory in South Africa. Am. J. Respir. Crit. Care Med. 177:787-792. [DOI] [PubMed] [Google Scholar]
- 3.Basu, S., G. H. Friedland, J. Medlock, J. R. Andrews, N. S. Shah, N. R. Gandhi, A. Moll, P. Moodley, A. W. Sturm, and A. P. Galvani. 2009. Averting epidemics of extensively drug-resistant tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 106:7672-7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Brossier, F., N. Veziris, A. Aubry, V. Jarlier, and W. Sougakoff. 2010. Detection by GenoType MTBDRs/test of complex mechanisms of resistance to second-line drugs and ethambutol in multidrug-resistant Mycobacterium tuberculosis complex isolates. J. Clin. Microbiol. 48:1683-1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caws, M., P. M. Duy, D. Q. Tho, N. T. Lan, D. V. Hoa, and J. Farrar. 2006. Mutations prevalent among rifampin- and isoniazid-resistant Mycobacterium tuberculosis isolates from a hospital in Vietnam. J. Clin. Microbiol. 44:2333-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gandhi, N. R., A. Moll, A. W. Sturm, R. Pawinski, T. Govender, U. Lalloo, K. Zeller, J. Andrews, and G. Friedland. 2006. Extensively drug-resistant tuberculosis as a cause of death in patients co-infected with tuberculosis and HIV in a rural area of South Africa. Lancet 368:1575-1580. [DOI] [PubMed] [Google Scholar]
- 5a.Hain Lifescience. 2009. Genotype MTBDRsl version 1.0 manual. Hain Lifescience, Nehren, Germany.
- 6.Hillemann, D., S. Rusch-Gerdes, and E. Richter. 2009. Feasibility of the GenoType MTBDRsl assay for fluoroquinolone, amikacin-capreomycin, and ethambutol resistance testing of Mycobacterium tuberculosis strains and clinical specimens. J. Clin. Microbiol. 47:1767-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin, S. Y., E. Desmond, D. Bonato, W. Gross, and S. Siddiqi. 2009. Multicenter evaluation of Bactec MGIT 960 system for second-line drug susceptibility testing of Mycobacterium tuberculosis complex. J. Clin. Microbiol. 47:3630-3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Migliori, G. B., A. Matteelli, D. Cirillo, and M. Pai. 2008. Diagnosis of multidrug-resistant tuberculosis and extensively drug-resistant tuberculosis: current standards and challenges. Can. J. Infect. Dis. Med. Microbiol. 19:169-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mokrousov, I., T. Otten, O. Manicheva, Y. Potapova, B. Vishnevsky, O. Narvskaya, and N. Rastogi. 2008. Molecular characterization of ofloxacin-resistant Mycobacterium tuberculosis strains from Russia. Antimicrob. Agents Chemother. 52:2937-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morgan, M., S. Kalantri, L. Flores, and M. Pai. 2005. A commercial line probe assay for the rapid detection of rifampicin resistance in Mycobacterium tuberculosis: a systematic review and meta-analysis. BMC Infect. Dis. 5:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sugawara, I., J. Zhang, and C. Li. 2009. Cross-resistance of Mycobacterium tuberculosis isolates among streptomycin, kanamycin and amikacin. Indian J. Exp. Biol. 47:520-522. [PubMed] [Google Scholar]
- 12.van Doorn, H. R., D. D. An, M. D. de Jong, N. T. Lan, D. V. Hoa, H. T. Quy, N. V. Chau, P. M. Duy, D. Q. Tho, N. T. Chinh, J. J. Farrar, and M. Caws. 2008. Fluoroquinolone resistance detection in Mycobacterium tuberculosis with locked nucleic acid probe real-time PCR. Int. J. Tuber. Lung Dis. 12:736-742. [PubMed] [Google Scholar]
- 13.Via, L. E., S. Cho, S. Hwang, H. Bang, S. K. Park, H. S. Kang, D. Jeon, S. Y. Min, T. Oh, Y. Kim, Y. M. Kim, V. Rajan, S. Y. Wong, I. C. Shamputa, M. Carroll, L. Goldfeder, S. A. Lee, S. M. Holland, S. Eum, H. Lee, and C. E. Barry III. 2010. Polymorphisms associated with resistance and cross-resistance to aminoglycosides and capreomycin in Mycobacterium tuberculosis isolates from South Korean patients with drug-resistant tuberculosis. J. Clin. Microbiol. 48:402-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.World Health Organization. 2009. Global Tuberculosis Control-epidemiology, strategy, and financing. WHO report 2009. WHO/HTM/TB/2009.411. World Health Organization, Geneva, Switzerland. http://www.who.int/tb/publications/global_report/2009/en/.
- 15.World Health Organization. 2008. Anti-tuberculosis drug resistance in the world. Fourth Global Report. WHO/HTM/TB/2008.394. The World Health Organization/International Union Against Tuberculosis and Lung Disease (WHO/UNION) Global Project on Anti-Tuberculosis Drug Resistance Surveillance 2002-2007. World Health Organization, Geneva, Switzerland. http://whqlibdoc.who.int/hq/2008/WHO_HTM_TB_2008.394_eng.pdf.
- 16.World Health Organization. 2008. Molecular line probe assays for rapid screening of patients at risk of multi-drug resistant tuberculosis (MDR TB). Expert Group Report. World Health Organization and the UNICEF/UNDP/World Bank/WHO Special Programme for Research and Training in Tropical Diseases (TDR). World Health Organization, Geneva, Switzerland. http://www.who.int/tb/features_archive/expert_group_report_june08.pdf.
- 17.Zaunbrecher, M. A., R. D. Sikes, Jr., B. Metchock, T. M. Shinnick, and J. E. Posey. 2009. Overexpression of the chromosomally encoded aminoglycoside acetyltransferase eis confers kanamycin resistance in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 106:20004-20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang, Y., and W. W. Yew. 2009. Mechanisms of drug resistance in Mycobacterium tuberculosis. Int. J. Tuber. Lung Dis. 13:1320-1330. [PubMed] [Google Scholar]