Abstract
In the Global Polio Eradication Initiative, laboratory diagnosis plays a critical role by isolating and identifying poliovirus (PV) from the stool samples of patients with acute flaccid paralysis (AFP). In this study, we developed a particle agglutination (PA) method with a soluble human PV receptor (hPVR) in the form of an immunoadhesin (PVR-IgG2a) for the simple and rapid identification of PV. Sensitized gelatin particles with PVR-IgG2a showed specific agglutination with the culture fluid of PV-infected cells within 2 h of reaction in a one-step procedure. Detection limits for type 1, 2, and 3 PV(Sabin) strains were 1.5 × 106 50% cell culture infectious doses (CCID50), 5.3 × 105 CCID50, and 9.1 × 105 CCID50, respectively. Wild-type PVs and PV isolates from acute flaccid paralysis cases examined were identified correctly with this PA method, except for some samples with a mixture of different serotypes of PVs, where a minor population of PV failed to be detected. These results suggest that this PA method is useful for the simple and rapid identification of PV, although the sensitivity was not high enough to detect a minor population of PV (<1/10 of the major population) among mixed PVs.
In the Global Polio Eradication Initiative, laboratory diagnosis plays a critical role by isolating and identifying poliovirus (PV) from stool samples of patients with acute flaccid paralysis (AFP). In the World Health Organization (WHO) Global Polio Laboratory Network, PV isolation and identification are performed at WHO national polio laboratories by use of a cell culture system with a human rhabdomyosarcoma cell line (RD cells) and a mouse L cell line expressing the human PV receptor (hPVR) (L20B cells) (17, 18), followed by differentiation of the isolated strains into oral PV vaccine (OPV)-related PVs, vaccine-derived PVs, and wild-type PVs at WHO regional reference laboratories by several methods, including enzyme-linked immunosorbent assay (ELISA), probe hybridization, and reverse transcription-PCR (RT-PCR) (7, 18).
For the identification of PV, a neutralization test with anti-PV antibodies has been performed with the cell culture system and takes 3 to 5 days for results (18). The latest procedure for the laboratory diagnosis of PV (called New Algorithm) gives priority to minimizing the time to report. Accordingly, PV identification by neutralization testing at the national laboratories is no longer necessary, in principle, before the differentiation, except for samples with a mixture of different serotypes of PVs. In this procedure, RT-PCR systems have a critical role in identification and differentiation to minimize the overall time for analysis (on the order of hours) (7, 16). However, identification by RT-PCR, based on the mobility of PCR products, has 4 steps (i.e., viral RNA preparation, RT reaction, PCR, and gel electrophoresis) and might be laborious for laboratories with high workloads. Real-time RT-PCR requires additional equipment for the measurement and quality control system. In the posteradication era of PV, the availability of simple and rapid identification procedures at the national laboratories would be helpful for rapid confirmation of polio cases.
In the present study, we developed a novel particle agglutination (PA) method with a soluble hPVR for the simple and rapid identification of PV. With this PA method, PV strains, including Sabin vaccine strains, wild-type PVs, and PV isolates from AFP cases, were identified within 2 h of reaction in a one-step procedure.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
RD cells (human rhabdomyosarcoma cell line), HEp-2c cells (human larynx epidermoid carcinoma cell line), and GP2-293 cells (Clontech) were cultured as monolayers in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS). HEK293 cells were cultured as monolayers in Opti-Pro SFM medium (Gibco) supplemented with 2% FCS. RD cells were used for the titration of PV, coxsackievirus B (CVB), echoviruses, and enterovirus 71 (EV71). HEp-2c cells were used for the titration of coxsackievirus A (CVA). GP2-293 cells were used for the production of a recombinant retrovirus for the expression of PVR-IgG2a. HEK293 cells were used for the stable expression of PVR-IgG2a. RD cells, HEp-2c cells, and L20B cells infected with viruses were collected after freezing and thawing and then used as virus samples. For characterization of the PA method for the identification of PV, we examined vaccine strains [PV1(Sabin), PV2(Sabin), and PV3(Sabin)], wild-type strains [PV1(Mahoney), PV1(Brunhilde), PV2(MEF-1), PV3(Leon), PV3(Saukett), and PV3(Suwa-3)], and isolates from AFP cases. Nonpoliovirus enteroviruses examined to show the specificity of the PA method were as follows: echovirus 11 (strain Gregory; 6.8 × 105 50% cell culture infectious doses [CCID50]/μl), echovirus 25 (strain JV-4; 1.0 × 106 CCID50/μl), CVB3 (strain Nancy; 5.6 × 104 CCID50/μl), CVB4 (strain JVB; 3.2 × 104 CCID50/μl), EV71 (strains BrCr-TR, Nagoya, C7-Osaka, 1095, and 75-Yamagata-2003 [1.8 × 104 CCID50/μl, 6.3 × 104 CCID50/μl, 3.3 × 105 CCID50/μl, 1.2 × 105 CCID50/μl, and 6.3 × 105 CCID50/μl, respectively]), CVA17 (CAM2163 isolate; 1.8 × 104 CCID50/μl), and CVA20 (CAM1976 isolate; 3.2 × 104 CCID50/μl). Virus titers were determined by measuring the CCID50 at 35°C by a microtitration assay (13). Type-specific anti-PV antibody (RIVM PV typing antiserum) was reconstituted with 0.5 ml of distilled water and then diluted by adding 4.5 ml of 2% FCS-DMEM. Anti-PV1+PV2+PV3 (anti-P1+P2+P3), -P1+P2, -P2+P3, and -P3+P1 antibody pools were prepared by mixing equal volumes of each type-specific antibody and by adding a 1/5 volume of FCS (final FCS concentration, 18.2%).
Purification of PVR-IgG2a.
PVR-IgG2a, which is an immunoadhesin molecule consisting of the extracellular domains of hPVR and the Fc domains of mouse IgG2a (1), was expressed in HEK293 cells by use of a retrovirus expression system. A DNA fragment of the coding region of PVR-IgG2a was obtained by a PCR using primers 5′CCATAGATCTACCATGGCCCGAGCCATGGC3′ and 5′ATGAATCGATTCATTTACCCGGAGTCCGGG3′ (BglII and ClaI sites in the primers are underlined), with a baculovirus expression vector for PVR-IgG2a as the template (1). The DNA fragment was digested with BglII and ClaI and then was cloned into the corresponding sites of plasmid pLEGFP-N1 (BD Biosciences Clontech). The resultant plasmid was designated pPVR-IgG2a. GP2-293 cells were cotransfected with pPVR-IgG2a and pVSV-G (Clontech). The cell culture supernatant of the transfected cells was collected at 96 h posttransfection. HEK293 cells were inoculated with the collected supernatant and then used for the expression of PVR-IgG2a. For the purification of PVR-IgG2a, the cell culture supernatant of PVR-IgG2a-expressing HEK293 cells was collected on day 5 after passage. PVR-IgG2a was purified from 100 ml of the supernatant by use of protein A Sepharose Fast Flow (GE Healthcare) in 4 ml of elution buffer at a concentration of 0.03 mg/ml, as described previously (1).
Quantification of PVR-IgG2a.
The amount of PVR-IgG2a was quantified by a dot blot analysis, with purified mouse IgG2a (BD Pharmingen) as a standard sample. Purified PVR-IgG2a and purified mouse IgG2a (0.05, 0.025, and 0.0125 mg/ml) in a 5-μl volume were adsorbed to an Immobilon-P transfer membrane (Millipore) by use of an SRC 96 D Minifold I dot blotter (Schleicher & Schuell). The filter was blocked in phosphate-buffered saline (PBS) (10 mM phosphate buffer [pH 7.0], 135 mM NaCl, and 2.6 mM KCl) containing 5% nonfat dry milk and then incubated at room temperature for 20 min. The filter was washed with PBS containing 0.1% Tween 20 three times for 5 min each and then subjected to detection by use of a SuperSignal West Femto maximum sensitivity substrate kit (Pierce). The filter was incubated with goat anti-mouse IgG antibodies conjugated with horseradish peroxidase (1:1,000 dilution in PBS containing 0.1% Tween 20 and 0.5% nonfat dry milk) at room temperature for 1 h. The filter was washed with PBS containing 0.1% Tween 20 three times for 5 min each and then treated with substrate solution for detection of the signal by use of an LAS-3000 imaging system (Fujifilm). The concentration of PVR-IgG2a was estimated from that of mouse IgG2a and presented as the corresponding concentration of mouse IgG2a.
Sensitization of gelatin particles with PVR-IgG2a.
Gelatin particles which had been activated with tannic acid were mixed with 5.8 to 12.6 μg/ml of PVR-IgG2a in phosphate buffer solution (pH 6.0) and incubated at 37°C for 1 h. After being washed, the gelatin particles were suspended in phosphate buffer solution (pH 6.0) supplemented with 2% normal rabbit serum and lyophilized.
PA procedure.
For the PA procedure, 12 μl of anti-PV antibody solution or 10% FCS-DMEM without anti-PV antibodies (positive control for PA) was added to the reaction plates (Fastec U-bottomed microplate; Fujirebio Inc.), and then 12 μl of PV solution (cell culture fluid obtained after freezing and thawing of the infected cells) was added to the anti-PV antibody solutions. Finally, 25 μl of reconstituted sensitized gelatin particle solution was added to the plate. The plates were mixed in a plate mixer (Micro Mixer P; Taitec) for 30 s and then incubated at room temperature for 2 h. Agglutination of the sensitized gelatin particles was judged by visual observation.
RESULTS
Development of a PA method for the identification of PV.
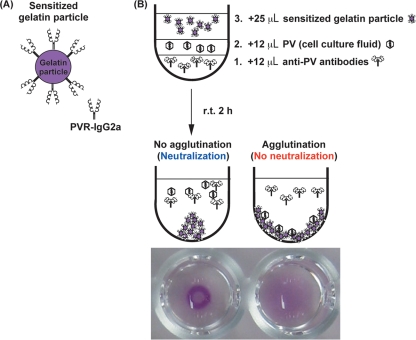
To detect all the serotypes of PV by a PA method, we used soluble hPVR in the form of an immunoadhesin (PVR-IgG2a), consisting of extracellular domains of hPVR and the hinge and Fc portions of mouse IgG2a (1, 3), for the sensitization of gelatin particles (Fig. 1A). In the PA method, anti-PV antibody solutions or 10% FCS-DMEM without anti-PV antibodies (a positive control for PA) was added to the reaction plates, and then PV solutions (cell culture fluid obtained after freezing and thawing of infected cells) were added to the anti-PV antibody solutions. Finally, the reconstituted sensitized gelatin particle suspension was added to the plates, and then the plates were mixed and incubated at room temperature for 2 h to observe the agglutination and inhibition of agglutination by anti-PV antibodies (Fig. 1B).
FIG. 1.
PA method for the identification of PV. (A) Schematic view of a sensitized gelatin particle with a soluble PVR (PVR-IgG2a). (B) Procedure of the PA method for the identification of PV and the appearance of agglutination by PV. The order of sample addition to the reaction plate is as follows: 1, anti-PV antibodies; 2, PV solution; and 3, sensitized gelatin particle solution. r.t., room temperature.
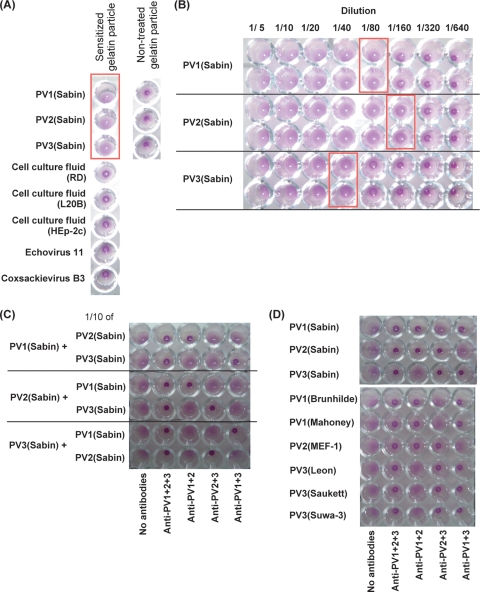
We examined the specificity of this PA method and the detection limit for PV(Sabin) strains. Agglutination was observed for PV(Sabin) strains but not for cell culture fluids of mock-infected RD cells, L20B cells, HEp-2c cells, and RD cells infected with other enteroviruses, including echoviruses 11 and 25, CVB3 and -4, EV71, and CVA17 and -20) (Fig. 2A; data not shown). Nontreated gelatin particles did not show any apparent agglutination. Detection limits of this PA method for type 1, 2, and 3 PV(Sabin) strains were 1.5 × 106 CCID50, 5.3 × 105 CCID50, and 9.1 × 105 CCID50, respectively (Fig. 2B). These results suggested that the observed agglutination was caused by the specific interaction of PV with its soluble receptor.
FIG. 2.
Characterization of PA method and identification of PV strains. (A) Specificity of sensitized gelatin particles for PV. The agglutination activity of gelatin particles was examined with nonpoliovirus enteroviruses (echovirus 11 and CVB3) and nontreated gelatin particles. Wells that showed agglutination are shown in a box. (B) Sensitivity of the PA method for PV(Sabin) strains. Virus solutions of PV1(Sabin), PV2(Sabin), and PV3(Sabin) (virus titers of 9.5 × 106 CCID50/μl [1.1 × 108 CCID50 in 12 μl], 6.9 × 106 CCID50/μl [8.2 × 107 CCID50 in 12 μl], and 2.9 × 106 CCID50/μl [3.5 × 107 CCID50 in 12 μl], respectively) were diluted 1/5 to 1/640 and then examined by the PA method. The detection limit of the PA method for each sample observed on the reaction plate is shown with a box. (C) Identification of mixed PV(Sabin) strains by the PA method. Virus solutions of PV1(Sabin), PV2(Sabin), and PV3(Sabin) (virus titers of 9.5 × 106 CCID50/μl [1.1 × 108 CCID50 in 12 μl], 6.9 × 106 CCID50/μl [8.2 × 107 CCID50 in 12 μl], and 2.9 × 106 CCID50/μl [3.5 × 107 CCID50 in 12 μl], respectively) were mixed with a 1/10 volume (1.2 μl) of each PV(Sabin) strain and then subjected to the PA method for identification. (D) Identification of wild-type PV strains by the PA method. Virus titers of wild-type strains examined [PV1(Brunhilde), PV1(Mahoney), PV2(MEF-1), PV3(Leon), PV3(Saukett), and PV3(Suwa-3)] were 3.2 × 106 CCID50/μl, 1.8 × 106 CCID50/μl, 1.0 × 107 CCID50/μl, 1.0 × 106 CCID50/μl, 1.0 × 106 CCID50/μl, and 5.6 × 105 CCID50/μl, respectively.
Identification of PV strains by the PA method.
Next, we examined PV samples consisting of mixed Sabin strains. A minor population of different serotypes of Sabin strains (1/10 of the major strains) was detected and correctly identified (Fig. 2C). We examined wild-type PVs and also PV isolates from AFP cases. All of the wild-type PVs examined were identified correctly (Fig. 2D). PV isolates with single PV strains showed consistent results with those obtained by neutralization test in cell culture, irrespective of the cell line (RD cells or L20B cells) used for the isolation (Table 1). However, for some isolates with a mixture of different serotypes of PV (CAM2554-R1 and CAM2970-L2), minor PV strains were not detected by the PA method. These results suggest that the PA method is useful for the identification of PV, although minor strains might not be detected in samples with a mixture of different serotypes of PV.
TABLE 1.
PV identification by cell culture and PA method
| Sample no. | PV isolatea | Virus titerb (log10 CCID50/50 μl) | Virus identification by cell culturec | Virus identification by PA method |
|---|---|---|---|---|
| 1 | CAM1967-R1L1 | 6.25 | P3 | P3 |
| 2 | CAM1967-R2 | 7.25 | P3 | P3 |
| 3 | CAM2057-L2 | 6.5 | P3 | P3 |
| 4 | CAM2057-R2 | 6.75 | P3 | P3 |
| 5 | CAM2057-R1L1 | NA | P3 | P3 |
| 6 | CAM2058-L2 | 6.25 | P3 + NPV | P3 |
| 7 | CAM2058-R2 | 7.75 | P3 + NPV | P3 |
| 8 | CAM2058-R1L1 | NA | P3 + NPV | P3 |
| 9 | CAM2294-L1 | NA | P3 | P3 |
| 10 | CAM2294-R2 | 7.5 | P3 | P3 |
| 11 | CAM2553-L1 | 6.5 | P2 | P2 |
| 12 | CAM2554-L1 | 7.0 | P1 + P2 | P1 + P2 |
| 13 | CAM2554-R1 | 7.5 | P1 + P2 | P2 |
| 14 | CAM2840-L2 | 7.25 | P2 | P2 |
| 15 | CAM2906-L3 | 7.25 | P3 | P3 |
| 16 | CAM2907-L3 | 7.0 | P3 | P3 |
| 17 | CAM2936-L2 | 7.0 | P3 + NPV | P3 |
| 18 | CAM2936-R2 | 7.25 | P3 + NPV | P3 |
| 19 | CAM2937-L2 | 6.25 | P3 + NPV | P3 |
| 20 | CAM2937-R2 | 7.25 | P3 + NPV | P3 |
| 21 | CAM2970-L2 | 6.25 | P1 + P3 | P1 |
| 22 | CAM2995-L2 | 7.0 | P3 + NPV | P3 |
| 23 | CAM2995-R2 | NA | P3 + NPV | P3 |
| 24 | CAM3017-L2 | 8.0 | P2 | P2 |
| 25 | CAM3017-R2 | NA | P2 | P2 |
| 26 | CAM3017-R2L1 | NA | P2 | P2 |
Passage histories of the isolates are shown in the sample names. R, recovered from RD cells; L, recovered from L20B cells. Numbers after R and L represent the numbers of passage of the virus in the cells (e.g., R1L1 means one passage in RD cells followed by one passage in L20B cells).
NA, not available.
NPV, non-PV.
DISCUSSION
In this study, we developed a novel PA method for the identification of PV, using a soluble hPVR and gelatin particles. We used a soluble hPVR in the form of an immunoadhesin (PVR-IgG2a) (1) rather than anti-PV antibodies to sensitize gelatin particles, because hPVR binds all serotypes of PV with its specific interaction and uniform affinity (4, 8, 10, 11). For PVR-IgG2a, some avidity to PV would be expected that could not be attained by monomeric forms of soluble hPVR, which have relatively weak affinities for a single binding site on the virion (Kd [dissociation constant] of 6.7 × 10−7 to 4.5 × 10−8 M) (2, 9). Gelatin particles have little or no nonspecific reaction caused by their own epitopes, and their utility has been well established for the diagnosis of a number of infectious diseases, including strongyloidiasis (infection with Strongyloides stercoralis) (15), leprosy (infection with Mycobacterium leprae) (5), human immunodeficiency virus infection (19), human T-cell leukemia virus infection (6), infection with Mycoplasma pneumoniae (Fujirebio Inc.), and exposure to diphtheria, pertussis, and tetanus toxins (12), by detection of antibodies against infectious agents in the sera of suspected patients. In accordance with the given properties of hPVR and gelatin particles, sensitized gelatin particles formed specific agglutination with all serotypes of PV, with comparable sensitivities (Fig. 2).
The advantages of the PA method developed in this study over currently available PV identification procedures are as follows: (i) it is simple (one-step procedure, mixing PV isolates, anti-PV antibodies, and sensitized particles), (ii) it is rapid (2 h of incubation of the sample at room temperature, which is comparable to the time for analysis by RT-PCR systems and faster than a cell culture-based neutralization test [3 to 5 days]), and (iii) it allows easy evaluation of the results (visual observation without equipment). Disadvantages of the PA method compared to neutralization testing with a cell culture system are as follows: (i) it has a lower sensitivity (105 CCID50 versus about 1.5 CCID50) and (ii) it cannot isolate single serotypes of PV in a mixture of different serotypes of PVs. Actually, PV isolates examined in this study showed virus titers of 106.25 to 108.0 CCID50/50 μl, and all samples were correctly identified by the PA method, except for some mixtures (Table 1). For the identification of major PV strains in the samples, isolates from both L20B cells and RD cells are detectable with the PA method. For the isolation of minor strains, isolates from RD cells might be more suitable than those from L20B cells because of their higher titers (14). However, one isolate from L20B cells (CAM2554-L1), but not other isolates, from L20B cells and from RD cells (CAM2970-L2 and CAM2554-R1), was correctly identified by the PA method. In the current procedure for laboratory diagnosis of PV, identification in a cell culture system is indispensable for isolation of each PV strain for differentiation (18). Utilization of type-specific monoclonal antibodies in the PA method might allow differentiation of PVs along with genetic differentiation by RT-PCR systems (7, 16). The PA method is not an alternative to identification by the cell culture system or RT-PCR systems in the current procedure but could provide supportive information as a rapid and simple confirmation test for PV at national polio laboratories without facilities for molecular diagnosis. The PA method would be useful in cases with a limited time for reporting (within a day) and where reliable specificity is required, e.g., for identification at the national laboratories in the posteradication era of PV.
In summary, we have developed a PA method for the identification of PV. This PA method is useful for the simple and rapid identification of PV.
Acknowledgments
We are grateful to Junko Wada for her excellent technical assistance. We are grateful to Akio Nomoto for kindly providing a baculovirus expression vector for PVR-IgG2a.
This study was supported in part by Grants-in-Aid for the Promotion of Polio Eradication and Research on Emerging and Re-Emerging Infectious Diseases from the Ministry of Health, Labor and Welfare, Japan.
Footnotes
Published ahead of print on 2 June 2010.
REFERENCES
- 1.Arita, M., H. Horie, M. Arita, and A. Nomoto. 1999. Interaction of poliovirus with its receptor affords a high level of infectivity to the virion in poliovirus infections mediated by the Fc receptor. J. Virol. 73:1066-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arita, M., S. Koike, J. Aoki, H. Horie, and A. Nomoto. 1998. Interaction of poliovirus with its purified receptor and conformational alteration in the virion. J. Virol. 72:3578-3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arita, M., S. Ohka, Y. Sasaki, and A. Nomoto. 1999. Multiple pathways for establishment of poliovirus infection. Virus Res. 62:97-105. [DOI] [PubMed] [Google Scholar]
- 4.Bibb, J. A., G. Witherell, G. Bernhardt, and E. Wimmer. 1994. Interaction of poliovirus with its cell surface binding site. Virology 201:107-115. [DOI] [PubMed] [Google Scholar]
- 5.Escobar-Gutierrez, A., M. E. Amezcua, S. Pasten, F. Pallares, J. V. Cazares, R. M. Pulido, O. Flores, E. Castro, and O. Rodriguez. 1993. Comparative assessment of the leprosy antibody absorption test, Mycobacterium leprae extract enzyme-linked immunosorbent assay, and gelatin particle agglutination test for serodiagnosis of lepromatous leprosy. J. Clin. Microbiol. 31:1329-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujino, R., K. Kawato, M. Ikeda, H. Miyakoshi, M. Mizukoshi, and J. Imai. 1991. Improvement of gelatin particle agglutination test for detection of anti-HTLV-I antibody. Jpn. J. Cancer Res. 82:367-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kilpatrick, D. R., C. F. Yang, K. Ching, A. Vincent, J. Iber, R. Campagnoli, M. Mandelbaum, L. De, S. J. Yang, A. Nix, and O. M. Kew. 2009. Rapid group-, serotype-, and vaccine strain-specific identification of poliovirus isolates by real-time reverse transcription-PCR using degenerate primers and probes containing deoxyinosine residues. J. Clin. Microbiol. 47:1939-1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koike, S., H. Horie, I. Ise, A. Okitsu, M. Yoshida, N. Iizuka, K. Takeuchi, T. Takegami, and A. Nomoto. 1990. The poliovirus receptor protein is produced both as membrane-bound and secreted forms. EMBO J. 9:3217-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McDermott, B. M., Jr., A. H. Rux, R. J. Eisenberg, G. H. Cohen, and V. R. Racaniello. 2000. Two distinct binding affinities of poliovirus for its cellular receptor. J. Biol. Chem. 275:23089-23096. [DOI] [PubMed] [Google Scholar]
- 10.Mendelsohn, C., B. Johnson, K. A. Lionetti, P. Nobis, E. Wimmer, and V. R. Racaniello. 1986. Transformation of a human poliovirus receptor gene into mouse cells. Proc. Natl. Acad. Sci. U. S. A. 83:7845-7849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mendelsohn, C. L., E. Wimmer, and V. R. Racaniello. 1989. Cellular receptor for poliovirus: molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell 56:855-865. [DOI] [PubMed] [Google Scholar]
- 12.Miyamura, K., S. Sadahiro, T. Konda, M. Takahashi, R. Fujino, Y. Nishimura, H. Miyakoshi, K. Horiuchi, Y. Furuya, T. Kubota, H. Watanabe, S. Inoue, and S. Yamazaki. 1995. Development and usefulness of the gelatin-particle-agglutination test for titration of antibodies against diphtheria, pertussis and tetanus toxins. Jpn. J. Med. Sci. Biol. 48:49-59. [DOI] [PubMed] [Google Scholar]
- 13.Nagata, N., H. Shimizu, Y. Ami, Y. Tano, A. Harashima, Y. Suzaki, Y. Sato, T. Miyamura, T. Sata, and T. Iwasaki. 2002. Pyramidal and extrapyramidal involvement in experimental infection of cynomolgus monkeys with enterovirus 71. J. Med. Virol. 67:207-216. [DOI] [PubMed] [Google Scholar]
- 14.Pipkin, P. A., D. J. Wood, V. R. Racaniello, and P. D. Minor. 1993. Characterisation of L cells expressing the human poliovirus receptor for the specific detection of polioviruses in vitro. J. Virol. Methods 41:333-340. [DOI] [PubMed] [Google Scholar]
- 15.Sato, Y., H. Toma, S. Kiyuna, and Y. Shiroma. 1991. Gelatin particle indirect agglutination test for mass examination for strongyloidiasis. Trans. R. Soc. Trop. Med. Hyg. 85:515-518. [DOI] [PubMed] [Google Scholar]
- 16.van der Avoort, H. G., B. P. Hull, T. Hovi, M. A. Pallansch, O. M. Kew, R. Crainic, D. J. Wood, M. N. Mulders, and A. M. van Loon. 1995. Comparative study of five methods for intratypic differentiation of polioviruses. J. Clin. Microbiol. 33:2562-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wood, D. J., and B. Hull. 1999. L20B cells simplify culture of polioviruses from clinical samples. J. Med. Virol. 58:188-192. [PubMed] [Google Scholar]
- 18.World Health Organization. 2004. Polio laboratory manual, 4th ed., WHO/IVB/04.10, and supplement to the WHO polio laboratory manual. World Health Organization, Geneva, Switzerland.
- 19.Yoshida, T., T. Matsui, S. Kobayashi, and N. Yamamoto. 1987. Evaluation of passive particle agglutination test for antibody to human immunodeficiency virus. J. Clin. Microbiol. 25:1433-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]