Abstract
Loop-mediated isothermal amplification (LAMP) of DNA offers the ability to detect very small quantities of pathogen DNA following minimal tissue sample processing and is thus an attractive methodology for point-of-care diagnostics. Previous attempts to diagnose malaria by the use of blood samples and LAMP have targeted the parasite small-subunit rRNA gene, with a resultant sensitivity for Plasmodium falciparum of around 100 parasites per μl. Here we describe the use of mitochondrial targets for LAMP-based detection of any Plasmodium genus parasite and of P. falciparum specifically. These new targets allow routine amplification from samples containing as few as five parasites per μl of blood. Amplification is complete within 30 to 40 min and is assessed by real-time turbidimetry, thereby offering rapid diagnosis with greater sensitivity than is achieved by the most skilled microscopist or antigen detection using lateral flow immunoassays.
Examination of thick and thin blood smears by microscopy remains the most common tool for diagnosis of malarial infections due to its historical utility, low startup and running costs, and the lack of simple but more sensitive alternatives. This technique is, however, completely dependent upon the availability of trained microscopists and can be time consuming (requiring up to half an hour for a single slide) when very low levels of parasitemia are involved (19). Accurate diagnosis by microscopy is also dependent upon the implementation of effective quality control (QC) and quality assurance (QA) schemes (19). It is no surprise, therefore, that the levels of quality of microscopy-based diagnosis have been shown to differ widely among different diagnostic facilities (5, 6, 12, 15, 29). At its best, microscopy can routinely detect as few as 10 to 50 parasites per μl of blood (unpublished data); however, detection limits of 50 to 100 parasites per μl, even for well-trained microscopists, are currently reported (7, 14, 18).
The labor-intensive nature of microscopy, and the requirement for high levels of operator training, has led to the development of antigen-based rapid diagnostic tests (RDTs). Such tests make fast and accurate point-of-care diagnostic testing possible for medical staff with no formal training in microscopy and in areas without well-equipped diagnostic facilities. The majority of such tests are formatted as lateral-flow immunoassays. The drawbacks of this format include its susceptibility to degradation in suboptimal storage conditions, limited shelf life, an apparent failure to detect some parasites due to mutations within the target antigen (11), and the possibility of false-positive results due to persistence of the antigen (2). RDTs offer moderate sensitivity of around 100 to 200 parasites per μl (1, 3, 9, 10, 13, 16, 23-25, 30, 31).
PCR amplification of parasite DNA allows detection with higher sensitivity than either RDTs or microscopy (26). The inhibition of PCR by heme means that the purification of DNA from blood samples is an absolute requirement. This, and the cost and complexity of conventional DNA amplification methods, limits its application outside reference facilities. Loop-mediated isothermal amplification (LAMP) is an alternative method for rapid and sensitive DNA amplification that can be performed with simplified and inexpensive specimen processing (17). In addition, the reaction is performed under isothermal conditions, requires only simple hardware, and can be formatted for visual detection without the need for instrumentation (for an in-depth description of LAMP and a schematic representing loop-mediated amplification methods, see reference 13). Commercial LAMP reaction kits have recently been developed for numerous viral, bacterial, and protozoan pathogens. Previously reported LAMP assays for malaria species employed primer sets targeting the small-subunit (SSU) rRNA (18S) locus (8, 22). While the reported assay was highly specific, the sensitivity for Plasmodium falciparum was limited to around 100 parasites per μl. Here we report the development of primer sets for two mitochondrion-based LAMP targets, one specific for P. falciparum and one capable of amplifying all species of the genus Plasmodium. These primer sets routinely allow the positive identification of densities of 5 parasites per μl. Amplification was detected by real-time turbidimetry in 30 min, which is faster than has been reported for 18S ribosomal DNA (rDNA) targets, while the primer sets offer high (100%) specificity. Thus, LAMP detection of malarial mitochondrial DNA sequences offers sensitivity greater than microscopy and combines speed with utility approaching that of a rapid diagnostic test.
MATERIALS AND METHODS
Sample selection, preparation, and ethical approval.
Lyophilized blood constituting the WHO international standard for P. falciparum (ISPf) was obtained from the National Institute for Biological Standards and Control (NIBSC) (Potters Bar, United Kingdom) and reconstituted in water per the instructions of the manufacturer (21). P. knowlesi DNA was a kind gift from Graham Mitchell.
The LAMP assay was developed using archived parasite DNA extracted from venous blood collected for routine malaria diagnosis by microscopy from parasite-positive patients at the Hospital for Tropical Diseases (HTD). The performance of the LAMP assay was characterized using 143 sequential venous blood samples collected in EDTA tubes between 10 October 2008 and 13 November 2008 from patients presenting with fever at the HTD for routine malaria diagnosis by microscopy (27). These included samples from patients later found to have no malaria parasites after examination of blood films.
This study was granted ethical approval by the Research Ethics Committee (REC) of University College London Hospitals (REC reference 07/Q0505/60).
Processing of samples for LAMP, PCR, and microscopy.
Aliquots (200 μl) of blood were used both for rapid processing prior to LAMP and for DNA extraction prior to PCR. For LAMP, samples were mixed with equal volumes of 40 mM Tris (pH 6.5)-0.4% sodium dodecyl sulfate (SDS) and vortexed for 10 s before being heated at 95°C for 5 min. After heating, the samples were sedimented by centrifugation at 14,000 × g for 5 min and the top clear supernatant layer was removed for LAMP (for immediate use or after short-term storage at −80°C). DNA from 200-μl aliquots of the ISPf and human blood samples was extracted for SSU ribosomal DNA nested PCR (26) using a Qiamp DNA blood mini-kit and a Qiacube automated extraction system (Qiagen, Germany). This robotic platform carried out the exact procedures listed in the manual extraction protocol, with the exception that the protocol used two 50-μl volumes of Qiagen's EB buffer, each with a 2.5-min incubation period, to elute the purified DNA from the columns. We recently described two distinct species of P. ovale; one of the two species was not detected by the SSU rRNA gene nested PCR (28). Any samples which were microscopy positive for P. ovale but PCR negative were retested with an adaptation of the Snounou assay (26), utilizing the P. ovale reverse primer of Padley et al. (20). All blood samples were examined by both thick- and thin-film microscopy using rapid Field's and Giemsa staining to determine the level of parasitemia and species of infection. Each blood film was read by 2 separate readers for 15 min per slide. There were no discrepant microscopy readings in our sample set.
A dilution series of the ISPf was produced using blood from a noninfected volunteer before DNA extraction. Similar dilutions were made for selected infected blood samples from humans to determine the limits of sensitivity of the assays with different malaria species. The parasite samples chosen for these dilutions contained initial densities ranging between 0.1 and 1 parasitized cells per 100 red blood cells (referred to as 0.1% and 1% parasitemia, respectively), as determined by microscopic examination of stained thin blood films treated by rapid Field's staining. The initial numbers of parasites per microliter were calculated using an estimated 5 × 106 red blood cells/μl, since exact red cell or white cell counts were not available for many of the samples, and dilutions were performed such that samples contained 5,000, 500, 50, 5, and 2 parasites per μl.
DNA from samples archived in the form of blood spots on glass fiber filter papers were extracted in 100 μl of 40 mM Tris (pH 6.5)-0.4% SDS. A 3-mm-diameter punch sample of the blood spot was heated in extraction buffer at 95°C for 5 min before centrifugal sedimentation was performed at 14,000 × g for 5 min. The clear supernatant was removed and used directly for LAMP testing. A dilution series of the 3D7 strain of cultured P. falciparum was created by using cultured ring stages of known parasite densities diluted in uninfected volunteer blood. Specimens from the dilution series were aliquoted onto the filter papers before drying, and processing was performed in order to determine the sensitivity of this method. Filter papers (printed Filtermat A; Wallac) were prepared by aliquoting 10 μl of blood directly onto the center of the square. The blood was allowed to dry at room temperature overnight in a class I safety cabinet before storage at room temperature within cardboard sleeves together with desiccant pouches in zip-lock bags for 2 months before usage.
Locus selection, primer design, and LAMP reaction conditions.
The entire mitochondrial genome sequence of P. falciparum was aligned against the P. vivax sequence, and primers were designed using Clustal V alignment (produced using MegAlign [DNAStar]). Primer sequences for LAMP were generated using Primer Explorer version 4.0 software (freely available via http://primerexplorer.jp/elamp4.0.0/index.html) with the “specific” and “common” settings to produce P. falciparum-specific and pan-genus primer sets capable of supporting loop primers. Similarly, 18 selected genomic targets comprising both multiple- and single-copy loci were examined using Primer Explorer version 4.0 (Table 1) to find either species-specific or pan-genus primer sets. All targets which produced primer sets capable of supporting the loop primers were logged and reanalyzed to see whether additional primer sets with lower self-compatibility (3′ ΔG) and improved annealing temperature (Tm) could be identified. The primer set with the highest Tm and lowest self-compatibility with respect to each target logged was synthesized and empirically tested against the dilution series of DNA purified from all six species of human malaria parasites to determine the sensitivity and selectivity of amplification. When multiple primer sets identified in silico had very similar predicted kinetics, up to three separate primer sets were synthesized and tested.
TABLE 1.
Loci analyzed by PrimerSelect V4.0 and activity of generated primer sets
| Genea | Primer setb | Amplification result (no. of parasites/μl)c |
Notes | |||
|---|---|---|---|---|---|---|
| P. falciparum | P. vivax | P. ovale | P. malariae | |||
| Pf 18S | 50 | None | None | None | Published (22) | |
| Pf Mt | 869 | 5 | None | None | None | |
| Pf G3P | 171 | 5 | None | None | XR | Isolates faild |
| Pf G3P | 114 | 5 | None | None | None | |
| Pf G3P | 137 | 50 | ND | ND | ND | |
| Pf G3P | 81 | 50 | ND | ND | ND | |
| Pf 28S | 1 | 50 | None | None | None | |
| Pf Stevor | 556 | 50 | ND | ND | ND | Isolates fail |
| Pf Ogene | ND | ND | ND | ND | ||
| Pf Ngene | 49 | 5 | None | None | None | |
| Pf 2TM | ND | ND | ND | ND | ||
| Pf plasmepsin | ND | ND | ND | ND | ||
| Pf rif | ND | ND | ND | ND | ||
| Pf Vir | ND | ND | ND | ND | ||
| Pm 18S | None | None | None | 500 | Published (8) | |
| Pm G3P | 72 | None | None | None | 5 | |
| Pm CSP | ND | ND | ND | ND | ||
| Po 18S | None | None | 50 | None | Published (8) | |
| Po Ook25 | 1 | None | 50 | None | None | |
| Po Ook25 | 6 | None | 50 | None | None | |
| Pv 18S | None | 50 | None | None | Published (8) | |
| Pv Ook25 | 59 | None | 50 | None | None | |
| Pv SERA | 1 | None | 5 | None | None | Isolates fail |
| Pv SERA | 20 | None | 50+ | None | None | |
| Pv 28s | 95 | None | 5 | None | XR | |
| Pv 28s | 72 | None | 50 | None | None | |
| Pv 28s | 50 | None | 50 | None | None | |
| Pv G3P | 7 | ND | 50 | ND | ND | |
| Pv G3P | 12 | ND | 50 | ND | ND | |
| Pv G3P | 997 | ND | 50 | ND | ND | |
| Pv Mt | 287 | ND | 50 | ND | ND | |
| Pg 18S | 5 | 5 | 50 | 5 | Published (8) | |
| Pg Mt | 41 | 50 | 50 | 50 | 50 | |
| Pg Mt | 19 | 5 | 5 | 5 | 5 | |
Pf, P. falciparum; Mt, mitochondrial sequence; Pm, P. malariae; Po, P. ovale; Pv, P. vivax; Pg, pan-genus.
For loci with no primer set listed, in silico analysis failed to identify any suitable primer set(s) (except for published primer sets).
50, successful amplification from a sample containing 50 parasites per μl; 5, successful amplification from a sample containing 5 parasites per μl; None, no amplification; ND, not done; XR, inappropriate cross-reaction.
Isolates fail, the primer set failed to amplify all the clinical isolates against which it was tested.
LAMP reactions were performed at a range of temperatures (60, 63, and 65°C) with a range of primer concentrations to determine the optimal conditions for sensitivity and selectivity. LAMP reaction mixtures (25 μl) contained 2 μl of crude boiled blood supernatant, purified DNA, or water and were set up per the instructions of the manufacturer (Mast Diagnostics, United Kingdom) supplied with the LAMP reaction kit, with the exception that a reaction mix lacking betaine was produced and used. Amplification and detection were performed using a real-time turbidimeter (LA360Ce; Eiken Chemical Ltd., Tokyo, Japan). A positive result was recorded at the first time point at which the change in turbidity increased by 0.1 optical density (OD) units/s.
Statistical analysis.
The sensitivity and specificity of LAMP diagnosis using these novel primer sets were compared to those of conventional nested PCR (26) as the gold standard, and 95% confidence intervals (CI) were calculated using STATA software.
RESULTS
Sensitivity and specificity of LAMP diagnosis of malaria.
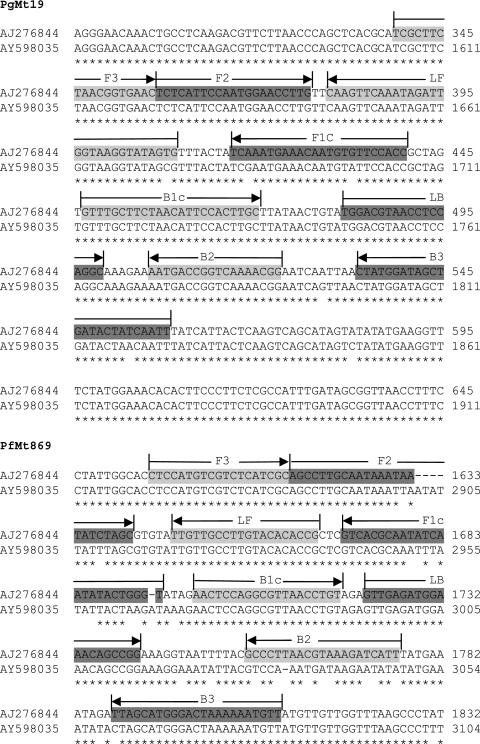
When the entire 8 kb of the mitochondrial genome was examined, only a single 250-bp region to which primer sets containing both core and loop primers could bind in order to selectively amplify P. falciparum DNA was identified. A single primer set for this region (labeled PfMt869) was designed and synthesized (Fig. 1). Similarly, only a single 250-bp region to which pan-genus primer sets containing both core and loop primers could bind in order to amplify both P. vivax and P. falciparum DNA was identified. A single pan-genus primer set was designed, synthesized, and labeled PgMt19 (Fig. 1). From the chromosomal loci analyzed in silico, 23 different primer sets that could provide either species-specific or pan-genus amplification were identified and subsequently synthesized for testing (Table 1).
FIG. 1.
Primer sets for PgMt19 and PfMt869 superimposed on alignments of P. vivax (EMBL AY598035) and P. falciparum (EMBL AJ276844) mitochondrial sequences. The core F3/B3, F2/B2, and F1c/B1c primer sequences, together with those of the LF/LB loop primers, are shown superimposed on the P. falciparum sequence.
Synthesized primer sets were tested against DNA purified from samples containing (per microliter) 5, 50, and 500 parasites. P. falciparum parasites (both the international standard and three patient samples), P. vivax parasites (three patient samples), P. ovale parasites (three patient samples that included both species), P. malariae parasites (three patient samples) and P. knowlesi parasites (rhesus monkey sample) were tested with each primer set. Only four species-specific primer sets routinely and specifically amplified samples containing 5 parasite genome equivalents of DNA (Table 1). Three of these primer sets (PfMt869, PfG3P114, and PfNgn49) were P. falciparum specific. When tested against samples from a serial dilution of the WHO international standard, only the N gene locus primers (PfNgn49) and the mitochondrial sequence primers (PfMt869) routinely amplified DNA from 5 parasite genome equivalents of DNA in under 60 and 30 min, respectively. For pan-genus primers, both the PgMt19 pan-genus primer set and a published 18S pan-genus primer set (8) amplified samples containing 5 parasites per μl from all five species (with the exception of one P. ovale sample for the published 18S primer set; with this primer set, amplification was seen only with the sample with 50 parasites per μl). Against the ISPf, both primer sets amplified 2 parasite genome equivalents of DNA in less than either 30 min (PgMt19 primer; Fig. 2) or 60 min (18S primer). A single primer set designed to amplify GAPDH (glyceraldehyde 3-phosphate dehydrogenase) of P. malariae gave specific amplification of P. malariae parasite samples containing 5 parasites per μl. All other primer sets, including published primer sets, failed to amplify 5 parasites per μl, failed to amplify all the clinical isolates against which they were tested, or were nonspecific for a given malaria species (Table 1).
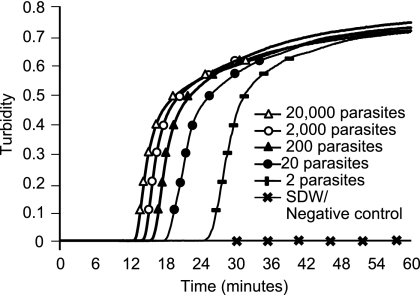
FIG. 2.
Amplification of known parasite equivalents of the Padley international standard with the PfMt19 primer. The figure shows the increase in turbidity with time in reaction wells containing 20,000, 2,000, 200, 20, or 2 parasite genome equivalents of purified DNA. The ISPf was diluted in blood to a given concentration (assuming an initial parasite density of 5 × 105 parasites per μl in the ISPf starting material) and then extracted using QiaAMP blood mini-kits. Reactions were performed using an LA-320CE turbidimeter. SDW, sterile distilled water.
Purified DNA from the WHO international P. falciparum standard, diluted to fixed parasite densities, was tested 25 times against the PgMt19 and PfMt869 primer sets. The PgMt19 primer set amplified all 25 replicates containing 2 parasite genome equivalents of DNA in each reaction. The PfMt869 amplified 23 out of 25 replicates containing 5 parasite genome equivalents of DNA. For dried blood spot samples on filter paper, routine amplification could be obtained with blood containing as few as 10 parasites per μl.
The PgMt19 and PfMt869 primer sets were tested against 27 rapid-boil preparations of archived blood samples representing 8 P. ovale spp. (including both subspecies of P. ovale) and 15 P. vivax and 6 P. malariae infections. Amplification with the PgMt19 primer set was seen with 26 out of the 27 samples when analyzed by real-time turbidimetry. The sample that failed to amplify comprised a P. vivax infection with a parasite burden of 4 parasites per μl, according to the results of analysis by an in-house real-time PCR. No amplification was seen with the PfMt869 primer set.
Sensitivity and specificity of LAMP diagnosis of malaria compared to nested PCR.
Between 10 October 2008 and 13 November 2008, 143 blood samples received by the HTD Department of Clinical Parasitology staff for malaria diagnosis were tested with the LAMP assays described above. All samples were also analyzed by microscopy according to standard diagnostic practice and by nested PCR. Using nested PCR as the gold standard, 30 samples were positive for P. falciparum, 1 was positive for P. vivax, none were positive for P. malariae, and 2 were positive for P. ovale spp. For the detection of P. falciparum, both the PfMt869 LAMP assay and microscopy failed to identify 2 of the 30 PCR-positive samples, giving sensitivity for both tests of 93.3% (95% CI, 89.3% to 97.4%) (Table 2). Both techniques were 100% specific. For the detection of Plasmodium species, both microscopy and the pan-genus PgMt19 primer set yielded a sensitivity of 93.9% (95% CI, 90.0% to 97.9%) compared to nested PCR (Table 3). Both techniques were 100% specific against the appropriate nested PCR.
TABLE 2.
Results of malaria diagnosis by PCR, P. falciparum LAMP, and microscopya
| Comparison | No. of samples: |
|
|---|---|---|
| PCR positive | PCR negative | |
| PCR vs LAMP (n = 143) | ||
| LAMP positive | 28 | 0 |
| LAMP negative | 2b | 113 |
| Microscopy vs PCR (n = 143) | ||
| Microscopy positive | 28 | 0 |
| Microscopy negative | 2 | 113 |
A total of 143 clinical blood samples were analyzed by rapid boiling and LAMP using the PfMt869 primer set and by microscopy using rapid thick films treated by Field's staining.
One sample initially negative by LAMP tested positive when reanalyzed using the PfMt869 primers. Specificity for both microscopy and LAMP with the 113 samples that were PCR negative for P. falciparum was 100%.
TABLE 3.
Results of malaria diagnosis by PCR, Plasmodium (genus) LAMP, and microscopya
| Comparison | No. of samples: |
|
|---|---|---|
| PCR positive | PCR negative | |
| PCR vs LAMP (n = 143) | ||
| LAMP positive | 31 | 0 |
| LAMP negative | 2b | 110 |
| Microscopy vs LAMP (n = 143) | ||
| Microscopy positive | 31 | 0 |
| Microscopy negative | 2 | 110 |
Analysis was performed as described for Table 2 except that LAMP was performed using the PgMt19 primer set.
One sample tested positive when reanalyzed using the PgMt19 primers. Two isolates of P. ovale and one of P. vivax were among the 33 PCR-positive samples tested.
DISCUSSION
LAMP has a number of advantages over current methods for the molecular diagnosis of disease. The Bst polymerase catalyzing the LAMP reaction is more robust with respect to inhibition than Taq polymerase. It is therefore possible to use a simple, rapid, and inexpensive regimen for sample preparation, in contrast to that required for standard PCR. This ease and speed of sample preparation is coupled with rapid reaction kinetics and the ability to monitor positivity either by real-time turbidimetry or with a simple visible-endpoint assay using the fluorescent compound calcein and a UV light source. In addition, the isothermal nature of the reaction removes the requirements for expensive thermal cyclers, which require stable power supplies. While such requirements of equipment and power may not be a major consideration for many diagnostic facilities in the developed world, the availability of a simple and rapid diagnosis system is an obvious advantage for resource- or infrastructure-poor locations in areas where malaria is endemic.
Previous published LAMP primer sets for malaria species take up to 1 h to produce positive amplification in samples with low parasite densities. Both the P. falciparum and pan-genus mitochondrial primer sets described here gave a positive result in under 30 min when tested with samples with parasite densities of 5 parasites per μl. That reaction time, coupled with a rapid-boil preparation method, means that it would be technically feasible to both process and analyze a sample within 1 h. Such a time span, while significantly longer than the time required for RDTs, is equivalent to that achieved with microscopy and far shorter than that required for current nested PCR protocols (typically 9 to 16 h). The sensitivity of these novel mitochondrial primers rivals that of microscopy performed by even the most highly trained microscopist, while requiring only minimal training for the operator to process the blood sample and set up the LAMP reaction. While nested PCR would appear to be the most sensitive method currently available, the requirement for lengthy extraction, amplification, and analysis protocols limits its use to reference facilities and eliminates the possibility of on-the-spot testing for very sick patients in common hospital settings. In addition, the closed-tube nature of the LAMP detection systems means that the prospect of cross-contamination leading to false-positive results is greatly reduced compared to nested PCR. In an internal audit of nested PCR versus microscopy over a 2-month period at the HTD, only a single patient presenting for malaria diagnosis was found to be positive by PCR but not by microscopy. The increase in sensitivity seen with nested PCR compared to LAMP would, therefore, most likely have a limited impact in such a routine diagnostic setting. Furthermore, the use of nested PCR as a first-line method for malaria diagnosis would be prohibitively expensive in terms of staff time.
The mitochondrial primer sets used here recognized all microscopy-positive clinical samples tested, and the high-density sequence data available for numerous malarial mitochondrial sequences isolated from parasites from diverse geographical locations showed no mutations within the region containing these primer sequences that would prevent amplification (noncoding intergenic regions between cox1 and cox 3 and between cytb and cox1 for PfMt869 and PgMt19, respectively). It is likely, therefore, that this assay would be transferable to any location where malaria is endemic and would detect the full repertoire of wild-type parasites. This would represent an advantage compared to some current HRP2 antigen-based rapid diagnostics that fail to detect certain wild-type isolates due to mutations within the HRP2 locus (11). Even if rare mutations do exist that have not been identified, the LAMP reaction is robust against single nucleotide polymorphisms, provided that these do not occur in the extreme 5′ ends of the F1c and B1c sequences or the extreme 3′ ends of the F3, B3, F2, and B2 sequences.
The combination of improved sample preparation and endpoint assays (currently under development) with highly sensitive, rapid primer sets offers a tangible route to the development of a Plasmodium LAMP diagnostic for routine field use. The development of such a reaction into a lyophilized format would minimize the preparation time for the reaction components, limit the training required by the operator, and remove the necessity for storage of the reaction components in a frozen form. The combination of these technologies into a “lab on a chip” format, where sample collection, processing, and analysis are all performed using a single disposable device, would offer an equally sensitive alternative to microscopic examination, while overcoming many of the limitations seen with current RDTs. Furthermore, the recent commercial production of battery-powered turbidimeters partially alleviates concerns about the availability of reliable power supplies, resulting in a technology with the potential to be truly transportable. In addition, the adaptation of this technology to high-throughput format may facilitate large-scale screening programs and thus facilitate the current drive for malaria elimination.
In some United Kingdom laboratories, after-hours diagnosis of malaria infection is currently performed using RDTs (4). The greater sensitivity and ability to read LAMP objectively when a turbidimeter is employed would offer significant advantages in these situations. It would also be of great use to many nonexpert microscopists, who form the bulk of on-call personnel outside major tropical and infectious disease units and do not regularly receive malaria films to analyze. Such a technology may well remove the need for time-consuming training and the requirement for regular examinations to ensure continued quality of microscopy-based diagnosis, as well as addressing the differences in diagnostic accuracy which exist between diagnostic facilities.
Acknowledgments
This work was supported by the Foundation for Innovative New Diagnostics (FIND). S.D.P. is supported by FIND. C.J.S. is supported by the United Kingdom Health Protection Agency. P.L.C. is supported by UCL Hospitals Comprehensive Biomedical Research Centre Infection Theme.
Footnotes
Published ahead of print on 16 June 2010.
REFERENCES
- 1.Abeku, T. A., M. Kristan, C. Jones, J. Beard, D. H. Mueller, M. Okia, B. Rapuoda, B. Greenwood, and J. Cox. 2008. Determinants of the accuracy of rapid diagnostic tests in malaria case management: evidence from low and moderate transmission settings in the East African highlands. Malar. J. 7:202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell, D. R., D. W. Wilson, and L. B. Martin. 2005. False-positive results of a Plasmodium falciparum histidine-rich protein 2-detecting malaria rapid diagnostic test due to high sensitivity in a community with fluctuating low parasite density. Am. J. Trop. Med. Hyg. 73:199-203. [PubMed] [Google Scholar]
- 3.Bharti, P. K., N. Silawat, P. P. Singh, M. P. Singh, M. Shukla, G. Chand, A. P. Dash, and N. Singh. 2008. The usefulness of a new rapid diagnostic test, the First Response Malaria Combo (pLDH/HRP2) card test, for malaria diagnosis in the forested belt of central India. Malar. J. 7:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chilton, D., A. N. Malik, M. Armstrong, M. Kettelhut, J. Parker-Williams, and P. L. Chiodini. 2006. Use of rapid diagnostic tests for diagnosis of malaria in the UK. J. Clin. Pathol. 59:862-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dini, L., and J. Frean. 2003. Quality assessment of malaria laboratory diagnosis in South Africa. Trans. R. Soc. Trop. Med. Hyg. 97:675-677. [DOI] [PubMed] [Google Scholar]
- 6.Durrheim, D. N., P. J. Becker, and K. Billinghurst. 1997. Diagnostic disagreement—the lessons learnt from malaria diagnosis in Mpumalanga. S. Afr. Med. J. 87:1016. [PubMed] [Google Scholar]
- 7.Endeshaw, T., T. Gebre, J. Ngondi, P. M. Graves, E. B. Shargie, Y. Ejigsemahu, B. Ayele, G. Yohannes, T. Teferi, A. Messele, M. Zerihun, A. Genet, A. W. Mosher, P. M. Emerson, and F. O. Richards. 2008. Evaluation of light microscopy and rapid diagnostic test for the detection of malaria under operational field conditions: a household survey in Ethiopia. Malar. J. 7:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han, E. T., R. Watanabe, J. Sattabongkot, B. Khuntirat, J. Sirichaisinthop, H. Iriko, L. Jin, S. Takeo, and T. Tsuboi. 2007. Detection of four Plasmodium species by genus- and species-specific loop-mediated isothermal amplification for clinical diagnosis. J. Clin. Microbiol. 45:2521-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hopkins, H., L. Bebell, W. Kambale, C. Dokomajilar, P. J. Rosenthal, and G. Dorsey. 2008. Rapid diagnostic tests for malaria at sites of varying transmission intensity in Uganda. J. Infect. Dis. 197:510-518. [DOI] [PubMed] [Google Scholar]
- 10.Kamugisha, M. L., H. Msangeni, E. Beale, E. K. Malecela, J. Akida, D. R. Ishengoma, and M. M. Lemnge. 2008. Paracheck Pf compared with microscopy for diagnosis of Plasmodium falciparum malaria among children in Tanga City, north-eastern Tanzania. Tanzan. J. Health Res. 10:14-19. [DOI] [PubMed] [Google Scholar]
- 11.Lee, N., J. Baker, K. T. Andrews, M. L. Gatton, D. Bell, Q. Cheng, and J. McCarthy. 2006. Effect of sequence variation in Plasmodium falciparum histidine-rich protein 2 on binding of specific monoclonal antibodies: implications for rapid diagnostic tests for malaria. J. Clin. Microbiol. 44:2773-2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKenzie, F. E., J. Sirichaisinthop, R. S. Miller, R. A. Gasser, Jr., and C. Wongsrichanalai. 2003. Dependence of malaria detection and species diagnosis by microscopy on parasite density. Am. J. Trop. Med. Hyg. 69:372-376. [PMC free article] [PubMed] [Google Scholar]
- 13.Mendoza, N. M., M. Garcia, L. J. Cortes, C. Vela, R. Erazo, P. Perez, O. L. Ospina, and J. D. Burgos. 2007. Evaluation of two rapid diagnostic tests, NOW ICT Malaria Pf/Pv and OptiMAL, for diagnosis of malaria. Biomedica 27:571-580. (In Spanish.) [PubMed] [Google Scholar]
- 14.Metzger, W. G., S. Vivas-Martinez, I. Rodriguez, J. Goncalves, E. Bongard, C. I. Fanello, L. Vivas, and M. Magris. 2008. Malaria diagnosis under field conditions in the Venezuelan Amazon. Trans. R. Soc. Trop. Med. Hyg. 102:20-24. [DOI] [PubMed] [Google Scholar]
- 15.Milne, L. M., M. S. Kyi, P. L. Chiodini, and D. C. Warhurst. 1994. Accuracy of routine laboratory diagnosis of malaria in the United Kingdom. J. Clin. Pathol. 47:740-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moody, A. 2002. Rapid diagnostic tests for malaria parasites. Clin. Microbiol. Rev. 15:66-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Notomi, T., H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino, and T. Hase. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohrt, C., W. P. O'Meara, S. Remich, P. McEvoy, B. Ogutu, R. Mtalib, and J. S. Odera. 2008. Pilot assessment of the sensitivity of the malaria thin film. Malar. J. 7:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohrt, C., P. Obare, A. Nanakorn, C. Adhiambo, K. Awuondo, W. P. O'Meara, S. Remich, K. Martin, E. Cook, J. P. Chretien, C. Lucas, J. Osoga, P. McEvoy, M. L. Owaga, J. S. Odera, and B. Ogutu. 2007. Establishing a malaria diagnostics centre of excellence in Kisumu, Kenya. Malar. J. 6:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Padley, D., A. H. Moody, P. L. Chiodini, and J. Saldanha. 2003. Use of a rapid, single-round, multiplex PCR to detect malarial parasites and identify the species present. Ann. Trop. Med. Parasitol. 97:131-137. [DOI] [PubMed] [Google Scholar]
- 21.Padley, D. J., A. B. Heath, C. Sutherland, P. L. Chiodini, and S. A. Baylis. 2008. Establishment of the 1st World Health Organization International Standard for Plasmodium falciparum DNA for nucleic acid amplification technique (NAT)-based assays. Malar. J. 7:139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poon, L. L., B. W. Wong, E. H. Ma, K. H. Chan, L. M. Chow, W. Abeyewickreme, N. Tangpukdee, K. Y. Yuen, Y. Guan, S. Looareesuwan, and J. S. Peiris. 2006. Sensitive and inexpensive molecular test for falciparum malaria: detecting Plasmodium falciparum DNA directly from heat-treated blood by loop-mediated isothermal amplification. Clin. Chem. 52:303-306. [DOI] [PubMed] [Google Scholar]
- 23.Rakotonirina, H., C. Barnadas, R. Raherijafy, H. Andrianantenaina, A. Ratsimbasoa, L. Randrianasolo, M. Jahevitra, V. Andriantsoanirina, and D. Menard. 2008. Accuracy and reliability of malaria diagnostic techniques for guiding febrile outpatient treatment in malaria-endemic countries. Am. J. Trop. Med. Hyg. 78:217-221. [PubMed] [Google Scholar]
- 24.Randrianasolo, L., P. B. Tafangy, L. A. Raharimalala, A. C. Ratsimbasoa, A. Randriamanantena, and M. Randrianarivelojosia. 2007. Rapid diagnostic test for malaria: preliminary study in Madagascar in 2003. Sante 17:69-73. (In French.) [PubMed] [Google Scholar]
- 25.Ratnawati, M. Hatta, and H. L. Smits. 2008. Point-of-care testing for malaria outbreak management. Trans. R. Soc. Trop. Med. Hyg. 102:699-704. [DOI] [PubMed] [Google Scholar]
- 26.Snounou, G. 1996. Detection and identification of the four malaria parasite species infecting humans by PCR amplification. Methods Mol. Biol. 50:263-291. [DOI] [PubMed] [Google Scholar]
- 27.Sutherland, C. J., H. Fifer, R. J. Pearce, F. bin Reza, M. Nicholas, T. Haustein, N. E. Njimgye-Tekumafor, J. F. Doherty, P. Gothard, S. D. Polley, and P. L. Chiodini. 2009. Novel pfdhps haplotypes among imported cases of Plasmodium falciparum malaria in the United Kingdom. Antimicrob. Agents Chemother. 53:3405-3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sutherland, C. J., N. Tanomsing, D. Nolder, M. Oguike, C. Jennison, S. Pukrittayakamee, D. Dolecek, T. T. Hein, V. E. do Rosario, A. P. Arez, J. Pinto, P. Michon, A. A. Escalante, F. Nosten, M. Burke, R. Lee, M. Blaze, T. D. Otto, J. Barnwell, A. Pain, J. E. Williams, N. J. White, N. P. Day, G. Snounou, P. L. Lockhart, P. L. Chiodini, M. Imwong, and S. D. Polley. 2010. Two nonrecombining sympatric forms of the human malaria parasite Plasmodium ovale occur globally. J. Infect. Dis. 201:1544-1550. [DOI] [PubMed] [Google Scholar]
- 29.Thomson, S., R. C. Lohmann, L. Crawford, R. Dubash, and H. Richardson. 2000. External quality assessment in the examination of blood films for malarial parasites within Ontario, Canada. Arch. Pathol. Lab. Med. 124:57-60. [DOI] [PubMed] [Google Scholar]
- 30.Wanji, S., H. K. Kimbi, J. E. Eyong, N. Tendongfor, and J. L. Ndamukong. 2008. Performance and usefulness of the Hexagon rapid diagnostic test in children with asymptomatic malaria living in the Mount Cameroon region. Malar. J. 7:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zerpa, N., R. Pabon, A. Wide, M. Gavidia, M. Medina, J. L. Caceres, J. Capaldo, M. Baker, and O. Noya. 2008. Evaluation of the OptiMAL test for diagnosis of malaria in Venezuela. Invest. Clin. 49:93-101. [PubMed] [Google Scholar]