Abstract
To develop an immunofluorescence assay for identification of the 2009 H1N1 influenza A virus, we generated a number of monoclonal antibodies (MAbs) by using inactivated H1N1 2009 virus (A/California/07/2009) as the immunogen. Two MAbs that target two different epitopes of the 2009 H1N1 hemagglutinin (HA) were selected to make the D3 Ultra 2009 H1N1 Influenza A ID kit (2009 H1N1 ID kit; Diagnostic Hybrids, Inc., Athens, OH), which is intended for the identification of the 2009 H1N1 virus by indirect immunofluorescence assay (IFA). The kit does not detect any of 14 seasonal H1N1 or H3N2 prototype influenza virus strains and is also not reactive with seven other major respiratory viruses. Clinical respiratory specimens were evaluated using both the 2009 H1N1 ID kit and the CDC human influenza virus real-time reverse transcription-PCR swine flu panel (CDC rRT-PCR) and showed 100% agreement between the two assays. Four of these clinical specimens, however, were positive by the 2009 H1N1 ID kit but were identified as presumptively positive by the CDC rRT-PCR by virtue of showing threshold cycle (CT) values only with universal InfA and swInfA primers, not with swH1 primers. Sequence analysis of the HA genes of these four specimens revealed point mutations in both the primer and probe regions. In addition, unlike the CDC rRT-PCR, the 2009 H1N1 ID kit can differentiate the 2009 H1N1 virus from a swine-derived H1 influenza A virus (A/New Jersey/8/76). The 2009 H1N1 ID kit offers clinical laboratories an alternative to RT-PCR for the identification of the 2009 H1N1 influenza A virus.
The novel H1N1 influenza A virus spread rapidly across the United States and around the world in 2009 (1, 3). The ensuing pandemic created a need for laboratories to subtype influenza A virus, because among other reasons, there has been concern that the 2009 H1N1 virus has a unique pattern of transmission and pathogenesis (6, 13). This new, emerging virus can be detected by direct fluorescent-antibody assay (DFA) and virus culture assays designed for the detection of influenza A viruses; however, these tests cannot differentiate this new virus from other seasonal influenza viruses. The most commonly used methods for subtyping the 2009 H1N1 virus are molecular tests, such as the Centers for Disease Control and Prevention human influenza virus real-time reverse transcription-PCR swine flu panel (CDC rRT-PCR), in which subtype specificity is achieved through amplification of a region of the hemagglutinin (HA) gene or M gene, using primers specific to the 2009 H1N1 subtype (5, 7, 8, 16).
Monoclonal antibody (MAb)-based technology has been approved for use in in vitro diagnostic tests for almost 3 decades. Only a few reports, however, have been published on the use of MAbs for influenza virus subtyping in clinical laboratories (15, 19). Studies on antigenic and genetic characteristics of swine-origin 2009 H1N1 viruses revealed that the novel virus contains a unique combination of gene segments from human, swine, and avian influenza viruses (6, 11). The H1 HA gene of the 2009 H1N1 virus is highly divergent from the H1 genes of seasonal H1N1 viruses, while homology among the 2009 H1N1 virus isolates is high (6, 11). This suggested to us that MAbs could be generated that are specific for the 2009 H1N1 virus.
In order to provide clinical laboratories with a rapid and inexpensive test to identify the 2009 H1N1 influenza A virus, we developed MAbs that specifically recognize this virus. These MAbs were used to develop an indirect immunofluorescence assay (IFA) kit (D3 Ultra 2009 H1N1 Influenza A ID kit) that can accurately identify 2009 H1N1 influenza A viral antigens in influenza A virus-infected cells from nasopharyngeal swabs or aspirates or from cell culture.
MATERIALS AND METHODS
Cells and viruses.
Virus cultures were done in R-Mix and R-Mix Too cells in 96-well or shell vial format (Diagnostic Hybrids, Inc. [DHI], Athens, OH). The cells were cultured in a 35 to 37°C CO2 incubator. Sf9 cells (Invitrogen, Carlsbad, CA) were grown following the manufacturer's instructions. Influenza A viruses and other prototype respiratory virus strains used in experiments were from the DHI virus repository, and most of them were originally from the American Type Culture Collection (ATCC). The 2009 H1N1 A/California/07/2009 and A/Mexico/4108/2009 viruses were obtained from the CDC. Virus stocks were propagated in MDCK cells (ATCC, Manassas, VA).
Clinical isolates and specimens.
Clinical isolates were obtained from various laboratories, and viruses were propagated in MDCK cells for 1 to 3 passages. Forty-three clinical isolates positive for 2009 H1N1 virus by RT-PCR were obtained from the Charleston Area Medical Center Health System, Inc. (Charleston, WV). Forty-five clinical isolates acquired before the 2009 H1N1 outbreak were from the DHI virus repository and were confirmed to contain influenza A virus by use of a DFA Respiratory Virus Screening & ID kit (Diagnostic Hybrids, Inc., Athens, OH). One hundred twenty-five influenza A virus-positive frozen clinical specimens collected during the 2009 H1N1 outbreak were provided by Kaiser Permanente Hospital (North Hollywood, CA). Fifty nasopharyngeal swab specimens were collected in November 2009 during the 2009 H1N1 outbreak from local hospitals in southeast Ohio. Two-well slides were prepared for immunofluorescence staining within 24 h of specimen collection. A portion of the original specimens from clinical isolates and of clinical specimens was used for viral RNA isolation and rRT-PCR testing.
Generation of 2009 H1N1-specific MAbs.
BALB/c mice were immunized with β-propiolactone-inactivated 2009 H1N1 A/California/07/2009 virus. Hybridoma production was carried out by previously described methods (9). Hybridoma supernatants were screened by IFA on both R-Mix monolayers infected with a mixture of H1N1 A/California/07/2009 and H1N1 A/Mexico/4108/2009 and R-Mix monolayers infected with a mixture of H3N2 strains. Positive hybridoma cells were expanded and then screened on R-Mix cell monolayers infected with 2009 H1N1 strains, four seasonal H1N1 strains, and four seasonal H3N2 strains. The hybridoma cells that were positive for both H1N1 A/California/07/2009 and H1N1 A/Mexico/4108/2009, without cross-reactivity to seasonal H1N1 strains and H3N2 strains, were selected for cloning by limiting dilution. The same screening strategy was applied to hybridoma clone selection. The candidate clones were expanded for MAb production. The MAbs were purified from hybridoma supernatants by protein G affinity chromatography, using fast-performance liquid chromatography (FPLC). The isotypes of the MAbs were determined using an isotyping enzyme-linked immunosorbent assay (ELISA) kit (Southern Biotech, Birmingham, AL).
Recombinant HA proteins.
Primers were designed to target the HA gene sequence of A/Mexico/4108/2009. The full-length HA gene was amplified by RT-PCR from RNA extracted from the A/Mexico/4108/2009 virus. SpHI and EcoRI restriction enzyme sites were inserted into the primer sequences for directional cloning into the baculovirus transfer vector pFastBac HT-A (Invitrogen, Carlsbad, CA). The 2009 H1N1 HA recombinant baculovirus was constructed using the Bac-to-Bac baculovirus expression system (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. To express the 2009 H1N1 full-length HA protein, Sf9 cells were infected with the recombinant baculovirus stock. The protein was purified under denaturing conditions, using a nickel affinity chromatography column (Qiagen, Valencia, CA).
A truncated 2009 H1N1 gene (HA1 subunit) was also synthesized with codons optimized for expression in Escherichia coli. Multiple overlapping complementary oligonucleotides which encompass the HA1 coding sequences were synthesized, and the gene was assembled by PCR and then amplified using outermost primers in a second PCR. The PCR product was cloned into the TA vector (Invitrogen, Carlsbad, CA), followed by DNA sequencing. The HA1 gene was then subcloned into the pQE expression vector (Qiagen, Valencia, CA), and the HA1 protein was purified using nickel affinity chromatography (Qiagen, Valencia, CA).
Western blotting.
The purified full-length HA (A/Mexico/4108/2009) recombinant protein expressed in Sf9 cells and the 2009 H1N1 HA1 subunit recombinant protein expressed in E. coli were separated by SDS-PAGE. The separated proteins were transferred electrophoretically to polyvinylidene difluoride (PVDF) membranes. The blotted membranes were blocked with phosphate-buffered saline (PBS) containing 0.05% Tween 20 and 5% skim milk and then incubated with 5 μg/ml of each 2009 H1N1 candidate MAb. The MAbs that bound to the membrane were detected by anti-mouse IgG-horseradish peroxidase (HRP) conjugate, followed by development with a WesternC chemiluminescence detection kit (Bio-Rad, Hercules, CA).
ELISA.
Multiwell plates (EIA/RIA; Costar, Lowell, MA) were coated with purified recombinant 2009 H1N1 HA1 subunit protein at 10 μg/ml in coating buffer (0.1 M NaHCO3, pH 9.5). Plates were blocked with PBS containing 1% bovine serum albumin (BSA). After washing of the plates, the 2009 H1N1 candidate MAbs were added in duplicate at 5 μg/ml in PBS-0.1% Tween 20. After 1 h of incubation at room temperature, the binding of the MAbs to the immobilized HA1 protein was detected by anti-mouse IgG-HRP conjugate, followed by color development with BM Blue POD substrate (Roche, Mannheim, Germany). Optical density was read at 450 nm with a multiwell spectrophotometer (Molecular Devices, Sunnyvale, CA).
IFA.
Clinical isolates and frozen specimens were inoculated into 96-well R-Mix plates or R-Mix Too shell vials (Diagnostic Hybrids, Inc., Athens, OH). The cells were centrifuged at 700 × g for 60 min and incubated overnight in a 35 to 37°C humidified 5.0% CO2 incubator. The cell monolayers were fixed with acetone for 10 min and then incubated with the 2009 H1N1-specific MAbs at 35°C to 37°C for 30 min. After washing of the cells with PBS, anti-mouse IgG-fluorescein isothiocyanate (FITC) was added, and the cells were incubated for another 15 to 30 min. Stained cells were washed with PBS and preserved with mounting fluid. Cells were examined at a magnification of ×200, using a Nikon TS100 microscope with an FITC wide-pass filter set and a 50-W mercury lamp to detect the presence of fluorescently stained cells. For direct specimen testing, cell suspensions were spotted onto each well of a 2-well slide, dried on a 37°C warming plate, and fixed with acetone. Duplicate slides were prepared for each clinical specimen, and two qualified lab technicians viewed the slides independently and without knowledge of the CDC rRT-PCR results.
DFA.
Cells inoculated with virus isolates or frozen specimen and direct specimen slides were stained with D3 Ultra influenza A virus identification reagent (Diagnostic Hybrids, Inc., Athens, OH) according to the instructions in the product insert.
RT-PCR.
RNAs were isolated from clinical specimens by use of a QIAamp Viral RNA Mini kit (Qiagen, Valencia, CA) according to the instructions in the product insert. Briefly, 140 μl of supernatant from clinical specimens or viral isolates was used for RNA isolation. RNA was eluted in a final volume of 60 μl. Five microliters was used for each rRT-PCR, following the CDC protocol for the detection of the 2009 H1N1 subtype (16). As recommended by the CDC, all reactions used an Invitrogen Superscript III Platinum one-step quantitative kit and a Stratagene QPCR instrument. All RT-PCR runs included no-template controls (water) and positive-template controls (2009 H1N1 Mexico viral RNA). According to the CDC protocol, a sample was considered presumptively positive for 2009 H1N1 influenza A virus if both the influenza A virus (InfA) primers that target the matrix gene and the respective subtype-specific primers recognizing either the swine influenza A virus (swInfA) nucleocapsid gene or the H1N1 swine influenza A virus (swH1) hemagglutinin gene had amplification curves which crossed the threshold (CT values) within 40 cycles.
Sequence analysis.
The four presumptively positive specimens that failed to produce CT values with swH1 primers in the CDC rRT-PCR were amplified using either the CDC swH1 primers or the WHO sequencing primers. PCR amplicons derived from clinical specimens J769, J773, H464, and H465 were cloned using a TOPO T/A cloning kit (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Sequencing was performed at the Ohio University Genomics Facility (Athens, OH). Sequence analysis was performed using Clone Manager Suite 7 (SciEd Central, Durham, NC).
RESULTS
Generation of 2009 H1N1-specific MAbs.
MAbs generated from mice immunized with β-propiolactone-inactivated A/California/07/2009 virus were selected after several rounds of IFA screening with numerous seasonal H3N2 and H1N1 strains and 2009 H1N1 isolates. This diligent screening process enabled us to identify MAbs that were specific to the 2009 H1N1 virus. Sixteen MAbs exhibited 2009 H1N1 specificity, based on IFA testing of infected cell monolayers by use of 11 2009 H1N1 isolates, 8 seasonal H1N1 strains, and 6 seasonal H3N2 strains.
To determine whether the MAbs targeted the influenza virus HA protein, 16 MAbs were tested by IFA on Sf9 cells infected with a recombinant baculovirus that expresses full-length 2009 H1N1 HA (rBAC-HA). All of the MAbs showed positive staining of rBAC-HA-infected Sf9 cells but not uninfected Sf9 cells, demonstrating that the MAbs indeed recognized the 2009 H1N1 HA protein (data not shown). The MAbs were also analyzed by an indirect ELISA using an E. coli-expressed HA1 protein subunit. Only one MAb, 9F8C7, showed strong binding to the HA1 subunit, indicating that this MAb recognized a different epitope from that for the other 15 MAbs tested (data not shown). For further studies, we selected MAb 9F8C7 and one other MAb, 16E11B3, based on the intensity of immunofluorescence staining of virus-infected cells. These two MAbs were analyzed by Western blotting, using full-length HA expressed in Sf9 cells and the HA1 subunit protein expressed in E. coli as targets. Results from Western blotting confirmed that MAb 9F8C7 bound to the HA1 fragment, while MAb 16E11B3 bound only to the full-length HA protein and therefore must recognize a linear epitope in HA2 (Fig. 1). These two MAbs were blended to make the 2009 H1N1 ID kit.
FIG. 1.
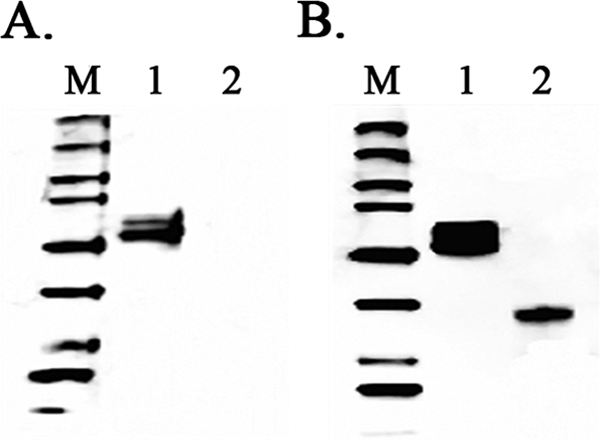
Western blot analysis of 2009 H1N1 MAbs with recombinant HA proteins. (A) Western blot probed with MAb 16E11B3. (B) Western blot probed with MAb 9F8C7. Lanes M, Precision Plus WesternC standards (Bio-Rad); lanes 1, purified 2009 H1N1 full-length HA recombinant protein (∼66 kDa); lanes 2, purified 2009 H1N1 HA subunit 1 recombinant protein (∼37 kDa).
Performance of the 2009 H1N1 ID kit.
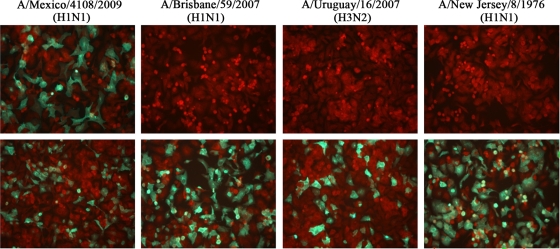
Specificity of the 2009 H1N1 ID kit was evaluated on R-Mix cell monolayers infected with 11 2009 H1N1 clinical isolates, 8 seasonal H1N1 influenza A virus strains, and 6 seasonal H3N2 influenza A virus strains individually. The kit demonstrated 100% specificity for the 2009 H1N1 subtype (Table 1; Fig. 2). In addition to influenza A virus, the D3 Ultra Respiratory Virus Screening & ID kit targets influenza B virus, adenovirus, respiratory syncytial virus (RSV), parainfluenza viruses, and human metapneumovirus (hMPV), so we tested a panel of 29 isolates of these viruses by use of the 2009 H1N1 ID kit on infected cell monolayers, using specific MAbs to each individual virus as positive controls. The kit showed no cross-reactivity to any of these viruses (Table 2). Interestingly, the kit did not cross-react with the swine-derived H1N1 A/NewJersey/8/76 strain that was associated with a human outbreak in New Jersey in 1976. The A/New Jersey/8/76 strain did test positive, however, in the CDC rRT-PCR assay (Table 1).
TABLE 1.
Subtype specificity of D3 Ultra 2009 H1N1 Influenza A Virus ID kit
| Influenza A virus subtype | Strain | Test result |
||
|---|---|---|---|---|
| Influenza A virus DFA | 2009 H1N1 IFA | CDC rRT-PCRa | ||
| 2009 H1N1 | A/California/07/2009 | + | + | + for swH1 |
| A/Mexico/4108/2009 | + | + | + for swH1 | |
| 2009 Clinical isolates | J0790 | + | + | + for swH1 |
| J0819 | + | + | + for swH1 | |
| J0859 | + | + | + for swH1 | |
| J0869 | + | + | + for swH1 | |
| CAMC4 | + | + | + for swH1 | |
| CAMC9 | + | + | + for swH1 | |
| CAMC10 | + | + | + for swH1 | |
| CAMC17 | + | + | + for swH1 | |
| CAMC19 | + | + | + for swH1 | |
| Seasonal H1N1 | A/Brisbane/59/2007 | + | − | NA |
| A/Solomon Islands/03/2006 | + | − | NA | |
| A/Puerto Rico/8/1934 | + | − | NA | |
| A/New Jersey/8/1976 | + | − | + for swH1 | |
| A/Denver/1/1957 | + | − | NA | |
| A/Malaya/302/1954 | + | − | NA | |
| A/NWS/1933 | + | − | NA | |
| A/Wilson-Smith/1933 | + | − | NA | |
| Seasonal H3N2 | A/Uruguay/716/2007 | + | − | NA |
| A/Wisconsin/67/2005 | + | − | NA | |
| A/Victoria/3/75 | + | − | NA | |
| A/Port Chalmers/1/73 | + | − | NA | |
| A/Hong Kong/8/68 | + | − | NA | |
| A/Aichi/2/68 | + | − | NA | |
NA, not available.
FIG. 2.
Images of influenza A virus-infected R-Mix cell monolayers detected with the 2009 H1N1 ID kit (top) and the DFA Respiratory Virus Screening & ID kit (bottom). R-Mix cells were infected with different strains of influenza A virus for 20 h. The cell monolayers were stained with both reagents, side by side, after acetone fixation. The images were taken at a magnification of ×200, using a fluorescence microscope.
TABLE 2.
Cross-reactivity testing of D3 Ultra 2009 H1N1 Influenza A Virus ID kit with other respiratory virusesa
| Virus | Strain or type |
|---|---|
| Adenovirus | 1 |
| 3 | |
| 5 | |
| 6 | |
| 7 | |
| 8 | |
| 10 | |
| 13 | |
| 14 | |
| 18 | |
| 31 | |
| Influenza B virus | Hong Kong |
| Maryland | |
| Mass | |
| GL | |
| Taiwan | |
| JH-001 isolate | |
| Russia | |
| RSV | Long |
| Wash | |
| 9320 | |
| Parainfluenza virus type 1 | C-35 |
| Parainfluenza virus type 2 | Greer |
| Parainfluenza virus type 3 | C-243 |
| Parainfluenza virus type 4 | 4a M-25 |
| 4b CH-19503 | |
| Metapneumovirus | A1 |
| B1 | |
| B2 |
All viruses tested negative in the 2009 H1N1 IFA.
Evaluation of the 2009 H1N1 ID kit by use of clinical specimens.
Eighty-eight confirmed influenza A virus-positive clinical isolates in cell culture (45 isolates collected from 2005 to 2008 and 43 isolates collected from October to November during the 2009 H1N1 outbreak) were tested with the 2009 H1N1 ID kit. An aliquot of each specimen was saved for rRT-PCR analysis before the specimen was used to inoculate two wells each of 96-well R-Mix plates. One well was stained with the D3 Ultra Respiratory Virus Screening & ID kit to confirm the presence of influenza A virus in each sample, and a second well was stained with the 2009 H1N1 ID kit. There was 100% concordance between the H1N1 kit and the CDC rRT-PCR assay (Table 3). An additional study was also performed with 125 frozen clinical specimens that had previously tested positive for influenza A virus by the clinical virology laboratory at Kaiser Permanente Hospital. After 24 h in culture on R-Mix Too cells, all specimens tested positive for 2009 H1N1 by both CDC rRT-PCR and IFA (Table 3).
TABLE 3.
Results of testing of cultures of clinical isolates and frozen specimens with D3 Ultra 2009 H1N1 Influenza A Virus ID kit and CDC rRT-PCR assay
| D3 Ultra 2009 H1N1 Influenza A Virus ID kit result | No. of specimens with CDC rRT-PCR assay result for 2009 H1N1 virus |
Total | |
|---|---|---|---|
| Positive | Negative | ||
| Clinical isolates collected before and during the 2009 H1N1 outbreak | |||
| Positive | 43 | 0 | 43 |
| Negative | 0 | 45 | 45 |
| Total | 43 | 45 | 88 |
| Frozen clinical specimens collected during the 2009 H1N1 outbreak | |||
| Positive | 125 | 0 | 125 |
| Negative | 0 | 0 | 0 |
| Total | 125 | 0 | 125 |
Clinical respiratory specimens were also collected from a local clinic in southeast Ohio during the 2009 H1N1 outbreak. The unfrozen specimens were directly screened by the D3 Ultra Respiratory Virus Screening & ID kit, and then 50 influenza A virus-positive specimens were selected for testing with the 2009 H1N1 ID kit. Slides were prepared with cells from each specimen, and the supernatants were used for CDC rRT-PCR testing. Results of each assay were 100% in agreement. One of the 50 specimens was identified as a non-2009 H1N1 strain of influenza A virus by both methods. The remaining 49 clinical specimens were positive by both the 2009 H1N1 ID kit and the CDC rRT-PCR assay (Table 4). Interestingly, four specimens (J769, J773, H464, and H465) tested in the CDC rRT-PCR assay were positive with the two sets of primers targeting either the matrix (InfA) or nucleocapsid (swInfA) gene but failed to produce CT values with the swH1 primers, which are specific for the 2009 H1N1 HA gene (Table 5). The results of these four specimens were considered presumptively positive for 2009 H1N1 virus according to CDC guidance for interpretation of rRT-PCR results.
TABLE 4.
Results of direct testing of unfrozen clinical specimens with D3 Ultra 2009 H1N1 Influenza A Virus ID kit and CDC rRT-PCR assay
| D3 Ultra 2009 H1N1 Influenza A Virus ID kit result | No. of specimens with CDC rRT-PCR assay result for 2009 H1N1 virus |
Total | |
|---|---|---|---|
| Positive | Negative | ||
| Positive | 49a | 0 | 49 |
| Negative | 0 | 1b | 1 |
| Total | 49 | 1 | 50 |
Four specimens identified as presumptively positive by the CDC rRT-PCR assay showed CT values only with universal InfA and swInfA primers, not with swH1 primers.
One sample was positive for seasonal influenza A virus by the DFA Respiratory Virus Screening & ID kit and the CDC rRT-PCR assay.
TABLE 5.
CDC rRT-PCR results for four presumptive 2009 H1N1 variants that tested negative with the swine H1 primers
| Sample |
CT value with CDC primer set |
|||
|---|---|---|---|---|
| Universal InfA | Swine InfA | Swine H1 | Human Rnase P | |
| J769 | 14.82 | 14.8 | 30.98 | |
| J773 | 16.31 | 16.06 | 30.07 | |
| H464 | 32.28 | 33.89 | 23.9 | |
| H465 | 31.51 | 32.27 | 25.42 | |
| A/Mexico/4108/2009 | 22.06 | 23.3 | 25.8 | 29.16 |
| No template | ||||
Sequence analysis of the four variants.
The four clinical specimens (J769, J773, H464, and H465) that presumptively contained variant 2009 H1N1 viruses were analyzed further. To confirm the presence of the 2009 H1N1 virus in these specimens, conventional RT-PCR was performed on the specimens, using the CDC swH1 primers. Appropriately sized bands were visualized after gel electrophoresis only for specimens J769 and J773. PCR products of these two specimens were cloned and sequenced. Alignment of the PCR amplicon sequences confirmed that these two specimens contained the 2009 H1N1 HA gene and that both contained two nucleotide polymorphisms in the probe-binding region (Fig. 3 A). One of these mutations causes a T316I amino acid change that is not found in other reported 2009 H1N1 sequences.
FIG. 3.
Alignment of HA gene sequences of two 2009 H1N1 variants against the CDC swH1 primer and probe sequences. (A) Variant identified in clinical samples J769 and J773. (B) Variant identified in clinical samples H464 and H465. Nucleotide variations in clinical specimens compared to the CDC swH1 primer or probe region are highlighted.
RT-PCR of the other two specimens, H464 and H465, by use of the CDC swH1 primers did not result in a visible amplicon. We therefore performed RT-PCR by using the WHO-recommended sequencing primers for the 2009 H1N1 HA gene with the same nucleic acid extraction sample that was negative with the CDC swH1 primers. An appropriately sized PCR product (825 bp) was observed, and it was cloned and subjected to DNA sequencing. The WHO PCR product spans the CDC swH1 primer and probe regions. DNA sequence alignment of specimens H464 and H465 showed that they both contained two mutations in the 3′ swH1 primer region (Fig. 3B), which most likely explains our failure to obtain a PCR product for these two specimens by using the CDC swH1 primers. H464 and H465 also contained two nucleotide mutations in the probe-binding region, one of which (sw1 probe nucleotide [nt] 16) was shared by all four variants and the other of which (sw1 probe nt 13) was unique to the H464 and H465 variants (Fig. 3B) and results in a P314S amino acid change that has been reported for other 2009 H1N1 viruses.
DISCUSSION
In response to the 2009 influenza pandemic, we have developed MAbs that are specific for the 2009 H1N1 influenza A virus. Based on the high antigenic stability of the 2009 H1N1 virus and the high degree of divergence of the H1 HA gene of the 2009 H1N1 virus compared with those for recent seasonal H1 influenza A viruses, we hypothesized that MAbs that target the HA protein might be capable of subtyping the 2009 H1N1 virus (6, 12, 13). After several rounds of careful screening on cell monolayers infected with 2009 H1N1 viruses and seasonal influenza A viruses, two MAbs (16E11B3 and 9F8C7) that react with different epitopes of the 2009 H1N1 HA protein (Fig. 1) were selected for use in the subtyping kit. A mixture of the two MAbs was used to enhance the likelihood that this kit would identify virtually all of the circulating 2009 H1N1 virus isolates.
The 2009 H1N1 ID kit did not react with 14 H3 or H1 seasonal strains of influenza A virus or with 29 strains or serotypes of eight other respiratory viruses (Tables 1 and 2). The 2009 H1N1 ID kit did not react with the A/New Jersey/8/1976 swine-derived H1N1 strain which produces a positive result in the CDC rRT-PCR assay. These results suggest that the 2009 H1N1 ID kit is highly specific for the 2009 H1 HA antigen and that any new swine-derived virus that might circulate in the human population in the future probably will not react with this kit unless the virus contains an H1 HA gene from the same lineage. Interestingly, a recent study on the antigenic structure of the 2009 H1N1 HA revealed that the antigenic sites within HA share conserved antigenic epitopes with human and swine H1 viruses from the early 20th century (17). These structural characteristics suggest the possibility that the 2009 H1N1 ID kit would recognize the 1918 H1N1 virus.
Mutations in the primer- and/or probe-annealing region for RT-PCR tests that target the M gene have been reported during the 2009 H1N1 pandemic (5, 18). The HA gene is an appropriate target for identification of newly emerged virus subtypes, but its high mutation rate can make HA-directed tests, whether molecular or MAb-based, problematic because the frequent occurrence of new variants increases the probability of obtaining false-negative results. Here we report four 2009 H1 HA variants that were not able to be subtyped definitively by the CDC rRT-PCR test but were identified correctly by the 2009 H1N1 ID kit. Aligning the sequences of the four PCR products with the CDC 2009 swH1 probe and primer sequences revealed two 2009 H1 HA variants (Fig. 3). These variants had amino acid substitutions in the HA protein, at positions 316 and 314. The P314S variant has been reported previously, but to our knowledge the T316I variant has not been documented in the NCBI database and is a newly emerged variant.
Prior to the expansion of the availability of molecular testing, influenza A virus subtyping was relatively cumbersome to perform, was not done routinely in clinical laboratories, and was rarely carried out on direct specimens. There are a few reports of the utilization of MAbs for influenza A virus subtyping, but the method had not been developed fully for use in clinical laboratories (14, 19). Subtyping of influenza A virus first became clinically relevant with the emergence of oseltamivir-resistant seasonal H1N1 virus in 2007 (2, 4). The 2009 pandemic further emphasized the need for clinical laboratories to be able to subtype viruses. Recently, a report was published on two MAbs that were generated using a recombinant nucleoprotein of H5N1 virus and serendipitously were found to react with the 2009 H1N1 virus, without apparent cross-reactivity with other human seasonal influenza A viruses (10). These MAbs were used in an immunochromatographic assay for specifically detecting and identifying the 2009 H1N1 virus. The 2009 H1N1 ID kit described in the present report represents the only nonmolecular kit that has received emergency use authorization by the FDA for the identification of the 2009 pandemic virus in specimens or culture samples that have been shown to contain influenza A virus by DFA. This kit will provide clinical laboratories a very useful tool for identifying the 2009 H1N1 virus, especially if molecular tests are not a practical option.
The excellent performance of the 2009 H1N1 ID kit on all clinical specimens tested suggests that the MAbs in the kit probably recognize conserved epitopes within the 2009 H1 HA gene. This feature may make the kit less susceptible to the genetic drift of 2009 H1N1 viruses, particularly since one of the MAbs used appears to recognize the HA2 subunit, which is not usually associated with antigenic drift changes. However, the 2009 H1N1 ID kit will require constant monitoring to ensure its performance.
Acknowledgments
We thank Yung Huang for help in preparing the manuscript and Andrew Pekosz for a critical review of the manuscript.
All authors are employees of Diagnostic Hybrids, Inc., which is a wholly owned subsidiary of Quidel Corporation, San Diego, CA.
Footnotes
Published ahead of print on 2 June 2010.
REFERENCES
- 1.CDC. 2009. 2009 H1N1 flu: international situation update. Centers for Disease Control and Prevention, Atlanta, GA.
- 2.CDC. 2009. FluView: 2008-2009 influenza season week 24 ending June 20, 2009. CDC, Atlanta, GA.
- 3.Dawood, F. S., S. Jain, L. Finelli, M. W. Shaw, S. Lindstrom, R. J. Garten, L. V. Gubareva, X. Xu, C. B. Bridges, and T. M. Uyeki. 2009. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N. Engl. J. Med. 360:2605-2615. [DOI] [PubMed] [Google Scholar]
- 4.Dharan, N. J., L. V. Gubareva, J. J. Meyer, M. Okomo-Adhiambo, R. C. McClinton, S. A. Marshall, K. St. George, S. Epperson, L. Brammer, A. I. Klimov, J. S. Bresee, and A. M. Fry. 2009. Infections with oseltamivir-resistant influenza A(H1N1) virus in the United States. JAMA 301:1034-1041. [DOI] [PubMed] [Google Scholar]
- 5.Dhiman, N., M. J. Espy, C. Irish, P. Wright, T. F. Smith, and B. S. Pritt. 2009. Mutability in the matrix gene of novel influenza A H1N1 virus detected using a FRET probe-based real-time reverse transcriptase PCR assay. J. Clin. Microbiol. 48:677-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garten, R. J., C. T. Davis, C. A. Russell, B. Shu, S. Lindstrom, A. Balish, W. M. Sessions, X. Xu, E. Skepner, V. Deyde, M. Okomo-Adhiambo, L. Gubareva, J. Barnes, C. B. Smith, S. L. Emery, M. J. Hillman, P. Rivailler, J. Smagala, M. de Graaf, D. F. Burke, R. A. Fouchier, C. Pappas, C. M. Alpuche-Aranda, H. Lopez-Gatell, H. Olivera, I. Lopez, C. A. Myers, D. Faix, P. J. Blair, C. Yu, K. M. Keene, P. D. Dotson, Jr., D. Boxrud, A. R. Sambol, S. H. Abid, K. St. George, T. Bannerman, A. L. Moore, D. J. Stringer, P. Blevins, G. J. Demmler-Harrison, M. Ginsberg, P. Kriner, S. Waterman, S. Smole, H. F. Guevara, E. A. Belongia, P. A. Clark, S. T. Beatrice, R. Donis, J. Katz, L. Finelli, C. B. Bridges, M. Shaw, D. B. Jernigan, T. M. Uyeki, D. J. Smith, A. I. Klimov, and N. J. Cox. 2009. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325:197-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall, R. J., M. Peacey, Q. S. Huang, and P. E. Carter. 2009. Rapid method to support diagnosis of swine origin influenza virus infection by sequencing of real-time PCR amplicons from diagnostic assays. J. Clin. Microbiol. 47:3053-3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kubo, T., M. Agoh, Q. Mai Le, K. Fukushima, H. Nishimura, A. Yamaguchi, M. Hirano, A. Yoshikawa, F. Hasebe, S. Kohno, and K. Morita. 2010. Development of a reverse transcription-loop-mediated isothermal amplification assay for detection of pandemic (H1N1) 2009 virus as a novel molecular method for diagnosis of pandemic influenza in resource-limited settings. J. Clin. Microbiol. 48:728-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miao, L. Y., C. Pierce, J. Gray-Johnson, J. DeLotell, C. Shaw, N. Chapman, E. Yeh, D. Schnurr, and Y. T. Huang. 2009. Monoclonal antibodies to VP1 recognize a broad range of enteroviruses. J. Clin. Microbiol. 47:3108-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miyoshi-Akiyama, T., K. Narahara, S. Mori, H. Kitajima, T. Kase, S. Morikawa, and T. Kirikae. 2010. Development of an immunochromatographic assay specifically detecting pandemic H1N1 (2009) influenza virus. J. Clin. Microbiol. 48:703-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peiris, J. S., L. L. Poon, and Y. Guan. 2009. Emergence of a novel swine-origin influenza A virus (S-OIV) H1N1 virus in humans. J. Clin. Virol. 45:169-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Russell, C. A., T. C. Jones, I. G. Barr, N. J. Cox, R. J. Garten, V. Gregory, I. D. Gust, A. W. Hampson, A. J. Hay, A. C. Hurt, J. C. de Jong, A. Kelso, A. I. Klimov, T. Kageyama, N. Komadina, A. S. Lapedes, Y. P. Lin, A. Mosterin, M. Obuchi, T. Odagiri, A. D. Osterhaus, G. F. Rimmelzwaan, M. W. Shaw, E. Skepner, K. Stohr, M. Tashiro, R. A. Fouchier, and D. J. Smith. 2008. The global circulation of seasonal influenza A (H3N2) viruses. Science 320:340-346. [DOI] [PubMed] [Google Scholar]
- 13.Smith, G. J., D. Vijaykrishna, J. Bahl, S. J. Lycett, M. Worobey, O. G. Pybus, S. K. Ma, C. L. Cheung, J. Raghwani, S. Bhatt, J. S. Peiris, Y. Guan, and A. Rambaut. 2009. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459:1122-1125. [DOI] [PubMed] [Google Scholar]
- 14.Tkacova, M., E. Vareckova, I. C. Baker, J. M. Love, and T. Ziegler. 1997. Evaluation of monoclonal antibodies for subtyping of currently circulating human type A influenza viruses. J. Clin. Microbiol. 35:1196-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ueda, M., A. Maeda, N. Nakagawa, T. Kase, R. Kubota, H. Takakura, A. Ohshima, and Y. Okuno. 1998. Application of subtype-specific monoclonal antibodies for rapid detection and identification of influenza A and B viruses. J. Clin. Microbiol. 36:340-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.WHO. 2009. CDC protocol of real-time RT-PCR for influenza A (H1N1). World Health Organization, Geneva, Switzerland.
- 17.Xu, R., D. C. Ekiert, J. C. Krause, R. Hai, J. E. Crowe, Jr., and I. A. Wilson. 2010. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science 328:357-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng, X., K. M. Todd, B. Yen-Lieberman, K. Kaul, K. Mangold, and S. T. Shulman. 2009. Unique finding of a 2009 H1N1 influenza virus-positive clinical sample suggests matrix gene sequence variation. J. Clin. Microbiol. 48:665-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ziegler, T., H. Hall, A. Sanchez-Fauquier, W. C. Gamble, and N. J. Cox. 1995. Type- and subtype-specific detection of influenza viruses in clinical specimens by rapid culture assay. J. Clin. Microbiol. 33:318-321. [DOI] [PMC free article] [PubMed] [Google Scholar]