In a screening study for methicillin-resistant Staphylococcus aureus (MRSA), nasal, pharyngeal, and rectal swabs were collected from healthy dogs at the University of Saskatchewan. Multiple solid media, including CHROMagar Staph aureus (CHROMagar, Paris, France), were inoculated; S. aureus grows as distinctive mauve colonies on CHROMagar Staph aureus (8). The inclusion of 4 μg/ml oxacillin (Sigma-Aldrich, St. Louis, MO) in the medium allows for the selective culture of MRSA.
Colonies typical of S. aureus, grown from a nasal swab on CHROMagar plus oxacillin, were presumptively identified as MRSA. While this isolate was oxidase negative, other biochemical characteristics were inconsistent with S. aureus including the following: no hemolysis on 5% sheep blood agar (Becton, Dickinson and Company, Sparks, MD) and lack of DNase, hyaluronidase, coagulase, and acetoin production.
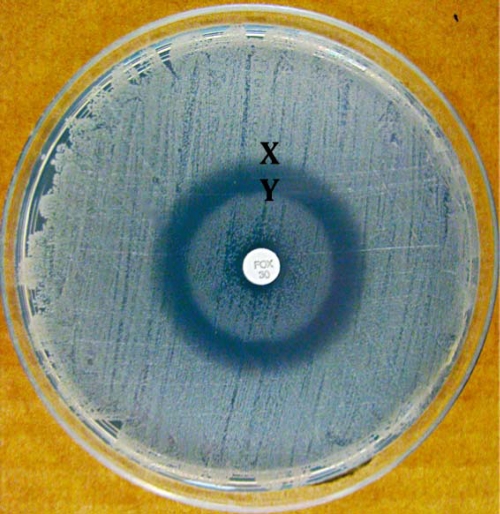
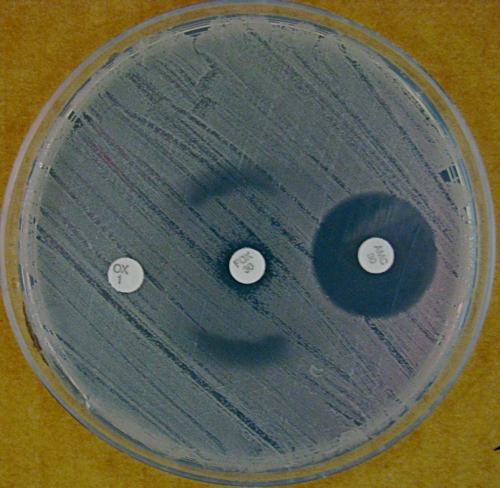
Analysis of the 16S rRNA gene (GenBank accession no. GU994136) using previously published primers revealed 99% similarity to a previously deposited Macrococcus caseolyticus sequence (1, 6). However, analysis of the cpn60 gene (GenBank accession no. GU994137), which has superior discrimination of staphylococci and macrococci, found only 89% similarity with the same Macrococcus entry (1, 10, 12). Phenotypic methicillin resistance was confirmed with oxacillin disk testing; when cefoxitin disks (30 μg) were used, a target zone of inhibition was seen (Fig. 1) and postulated to indicate inducible resistance. Target phenotypes resulting from inducible resistance are reported for MRSA with nafcillin and coagulase-negative staphylococci with imipenem (2, 3). Growth inside the inhibitory zone occurred when oxacillin or amoxicillin-clavulanate disks were placed adjacent to cefoxitin disks, indicating more potent resistance induction by these drugs (Fig. 2). Using a commercially prepared nitrocefin test (Hardy Diagnostics, Santa Maria, CA), the isolate was found to produce β-lactamase which may be responsible for the resistance seen. The contrast and brightness of both images were enhanced for clarity.
FIG. 1.
Target zone of inhibition associated with cefoxitin disk (FOX) (30 μg) on antibiotic-free Mueller-Hinton agar. The cefoxitin concentration at X is the MIC resulting in a zone of clearing, while induction of resistance at supra-MIC concentrations Y accounts for the inner zone of growth.
FIG. 2.
Blunting of the cefoxitin zone of inhibition, suggesting resistance induction by oxacillin (OX) (1 μg) and amoxicillin-clavulanate (AMC) (30 μg).
Methicillin resistance has been found in M. casolyticus, the most closely related organism identified, where a gene with 72% homology to the S. aureus mecA gene was identified (1). Perhaps due to a similarly low homology, mecA was not amplified from our isolate with previously published primers (5). It is not known whether the inducible cefoxitin resistance in this isolate was due to extended-spectrum β-lactamase production or a mecA-like mechanism. It is possible that like S. aureus ATCC 43300, this isolate produces both altered penicillin binding proteins and β-lactamase (4, 9). While the mechanism leading to this putative inducible resistance phenotype is unknown, this may be an example of “Eagle-type” resistance. Eagle-type resistance describes bacterial growth at high, but paradoxically not low, drug concentrations; in the case of MRSA, “derepression” of the mecA gene is purportedly causal (7, 11).
This report highlights the need for confirmatory testing when using selective/differential media for the identification of S. aureus (MRSA), as false-positive results may have therapeutic or infection control implications. Additional tests, including analysis of the cpn60 sequence may be indicated, particularly in bacterial species in veterinary animals, where colonizing bacterial populations are ill defined compared to those in humans (12). Finally, as with inducible clindamycin among S. aureus, further testing is essential to accurately characterize isolates with unusual resistance phenotypes (4).
Footnotes
Published ahead of print on 16 June 2010.
REFERENCES
- 1.Baba, T., K. Kuwahara-Arai, I. Uchiyama, F. Takeuchi, T. Ito, and K. Hiramatsu. 2009. Complete genome sequence of Macrococcus caseolyticus strain JSCS5402, reflecting the ancestral genome of the human-pathogenic staphylococci. J. Bacteriol. 191:1180-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blumenthal, R. M., R. Raeder, C. D. Takemoto, and E. H. Freimer. 1983. Occurrence and expression of imipemide (N-formimidoyl thienamycin) resistance in clinical isolates of coagulase-negative staphylococci. Antimicrob. Agents Chemother. 24:61-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyce, J. M., A. A. Medeiros, E. F. Papa, and C. J. O'Gara. 1990. Induction of beta-lactamase and methicillin resistance in unusual strains of methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 25:73-81. [DOI] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. 2008. Performance standards for antimicrobial susceptibility testing. M100-S18. Clinical and Laboratory Standards Institute, Wayne, PA.
- 5.de Neeling, A. J., W. J. van Leeuwen, L. M. Schouls, C. S. Schot, A. van Veen-Rutgers, A. J. Beunders, A. G. Buiting, C. Hol, E. E. Ligtvoet, P. L. Petit, L. J. Sabbe, A. J. van Griethuysen, and J. D. van Embden. 1998. Resistance of staphylococci in The Netherlands: surveillance by an electronic network during 1989-1995. J. Antimicrob. Chemother. 41:93-101. [DOI] [PubMed] [Google Scholar]
- 6.Dorsch, M., and D. Stackebrandt. 1992. Some modifications in the procedure of direct sequencing of PCR amplified 16S rDNA. J. Microbiol. Methods 16:271-279. [Google Scholar]
- 7.Eagle, H., and A. D. Musselman. 1948. The rate of bactericidal action of penicillin in vitro as a function of its concentration, and its paradoxically reduced activity at high concentrations against certain organisms. J. Exp. Med. 88:99-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaillot, O., M. Wetsch, N. Fortineau, and P. Berche. 2000. Evaluation of CHROMagar Staph. aureus, a new chromogenic medium, for isolation and presumptive identification of Staphylococcus aureus from human clinical specimens. J. Clin. Microbiol. 38:1587-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hardy Diagnostics. Nitrocef MatchBook. Product insert. Hardy Diagnostics, Santa Maria, CA.
- 10.Hill, J. E., A. Paccagnella, K. Law, P. L. Melito, D. L. Woodward, L. Price, A. H. Leung, L. K. Ng, S. M. Hemmingsen, and S. H. Goh. 2006. Identification of Campylobacter spp. and discrimination from Helicobacter and Arcobacter spp. by direct sequencing of PCR-amplified cpn60 sequences and comparison to cpnDB, a chaperonin reference sequence database. J. Med. Microbiol. 55:393-399. [DOI] [PubMed] [Google Scholar]
- 11.Kondo, N., K. Kuwahara-Arai, H. Kuroda-Murakami, E. Tateda-Suzuki, and K. Hiramatsu. 2001. Eagle-type methicillin resistance: new phenotype of high methicillin resistance under mec regulator gene control. Antimicrob. Agents Chemother. 45:815-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwok, A. Y., and A. W. Chow. 2003. Phylogenetic study of Staphylococcus and Macrococcus species based on partial hsp60 gene sequences. Int. J. Syst. Evol. Microbiol. 53:87-92. [DOI] [PubMed] [Google Scholar]