Abstract
We evaluated the FDA-approved Roche Cobas AmpliPrep/Cobas TaqMan (CAP/CTM) HIV-1 viral load assay for sensitivity, reproducibility, linearity, HIV-1 subtype detection, and correlation to the Roche Amplicor HIV-1 monitor test, version 1.5 (Amplicor). The limit of detection calculated by probit analysis was 23.8 copies/ml using the 2nd International WHO Standard and 30.8 copies/ml using Viral Quality Assurance (VQA) standard material. Serial dilutions of six patient samples were used to determine inter- and intra-assay reproducibility and linearity, which were very good (<8% coefficient of variation [CV]; between ∼1.7 and 7.0 log10 copies/ml). Subtype detection was evaluated in the CAP/CTM, Amplicor, and Bayer Versant HIV-1 bDNA 3.0 (Versant) assays using a commercially available panel. Versant averaged 0.829 log10 copies/ml lower than CAP/CTM and Amplicor averaged 0.427 log10 copies/ml lower than CAP/CTM for the subtype panel. Correlation with samples previously tested by Amplicor was excellent (R2 = 0.884; average difference [Amplicor value minus CAP/CTM value], 0.008 log10 copies/ml). Of the 305 HIV samples tested, 7 samples generated CAP/CTM titers between 1.0 and 2.75 log10 copies/ml lower than those for Amplicor. Three of these samples revealed primer and probe mismatches that could account for the discrepancies. Otherwise, the CAP/CTM assay exhibits excellent sensitivity, dynamic range, reproducibility, and correlation with Amplicor in an automated format.
Measurement of HIV-1 RNA in the plasma of infected patients is critical for guiding treatment. Over the past decade, several commercial quantitative HIV-1 RNA assays have become available that utilize endpoint PCR, isothermal amplification, or signal amplification techniques. Most recently, Abbott Molecular (Des Plaines, IL) and Roche Molecular Systems (Branchburg, NJ) received FDA approval for their real-time PCR-based systems, the RealTime HIV-1 assay and Cobas AmpliPrep/Cobas TaqMan HIV-1 Test (CAP/CTM), respectively. Each assay has its own advantages and disadvantages in terms of sensitivity, equipment requirements, throughput, dynamic range, subtype detection, and cost (1a, 4, 6, 7, 8, 11, 13, 15, 16).
The CAP/CTM test includes a “docked” version that permits automated “sample in—results out” analysis of specimens without user intervention. We evaluated this configuration and compared its performance to those of the Roche Amplicor HIV-1 monitor test, version 1.5 (Amplicor), and the Bayer Versant HIV-1 bDNA 3.0 assay (Versant). Seven samples which gave discrepant Amplicor versus CAP/CTM results were further evaluated by sequencing analysis.
MATERIALS AND METHODS
Limit of detection.
To determine the limit of detection in the CAP/CTM assay, nine dilutions of the WHO reference material (HIV-1 RNA, 2nd International Standard, 97/650; NIBSC, Potters Bar, United Kingdom) or Viral Quality Assurance (VQA) standard (Rush University Medical Center, Chicago, IL) were prepared in Basematrix diluent (SeraCare Life Sciences, Milford, MA). Fourteen replicates at each concentration (25, 20, 15, 10, 7.5, 5, 2.5, 1, 0.5 copies/ml for WHO; 40, 35, 30, 25, 20, 10, 5, 2, 0.5 copies/ml for VQA) were tested. The conversion of IU to copies for the WHO standard used the conversion listed in the CAP/CTM product insert (1.7 IU/copy). Probit analysis was used to determine the limit of detection (95% detection rate) and 95% confidence interval.
Reproducibility and linearity.
Six patient samples with high viral titers, serially diluted 7-fold in Basematrix, were used to evaluate intra- and interassay reproducibility and linearity of the CAP/CTM assay. Sufficient volume for five aliquots of each dilution was prepared and stored at 4°C until use. For each sample, three of five replicates were tested on the same input rack to evaluate intra-assay reproducibility and linearity. Interassay reproducibility was evaluated by testing two additional aliquots, each on a separate day. The average values for the intra-assay replicates were combined with the values for the two additional aliquots to determine interassay reproducibility and linearity. Reproducibility was evaluated by determining the mean and standard error of the mean for each dilution. Linearity was evaluated by linear regression analysis, excluding the aliquots with the highest measured concentration, which were set at identity to establish the expected values.
Subtype panels.
To assess the ability of the CAP/CTM assay to measure viral loads in samples containing different group M subtype viruses, the members of the HIV RNA genotype performance panel PRD202 (SeraCare Life Sciences, Milford, MA) were diluted 3-fold in Basematrix. Each subtype was tested in triplicate by the Amplicor, CAP/CTM, and Versant assays using standard procedures.
Correlation samples.
Archived frozen plasma samples previously tested by Amplicor were retrieved from storage, deidentified, and stored at −70°C before testing. Prior to being tested in the CAP/CTM assay, the samples were thawed at room temperature, vortexed briefly, and transferred to the S-input tubes. A total of 218 patient samples were selected to evaluate assay correlation. An additional 63 samples with viral loads below the limit of detection of the UltraSensitive Amplicor assay (<50 copies/ml) were selected for retesting in the CAP/CTM assay.
Eighty-seven archived specimens that had previously been tested by Amplicor were also tested by CAP/CTM after the instrument was relocated to a different location in the laboratory.
Linearity panel.
Ten aliquots for each of the members of the AcroMetrix (Benicia, CA) ValiQuant HIV-1 RNA quantification panel were pooled to obtain a homogenous sample for testing. Each member was tested in triplicate on each of three days. The data were analyzed by Deming regression.
Roche Amplicor assay.
The Roche Amplicor HIV-1 monitor test, version 1.5, was performed as directed by the manufacturer using manual extraction (using either the UltraSensitive or Standard procedure, as specified by the ordering physician) and PCR setup. The amplifications were performed on Applied Biosystems GeneAmp PCR system 9600 or 9700 thermal cyclers. The microwell plates were washed using a Bio-Tek Instruments ELP-40 and read using a Bio-Tek Instruments ELx800 microtiter plate reader.
Roche CAP/CTM assay.
Nucleic acid extraction, PCR preparation, and amplification for the CAP/CTM assay were performed using the “docked” configuration of the Cobas AmpliPrep/Cobas TaqMan HIV-1 test system using protocols, reagents, and software provided by the manufacturer. The assay extracts nucleic acid from 0.85 ml of plasma using magnetic particles, previously described by Schumacher et al. (14).
Versant HIV-1 RNA 3.0 assay (bDNA).
The Versant assay was performed as directed by the manufacturer, using the System 340 bDNA analyzer (Siemens Healthcare Diagnostics, Deerfield, IL).
Sequence analysis.
Nucleic acids from 200 μl of plasma were extracted from the samples using reagents from the Amplicor HIV-1 monitor kit. Briefly, 600 μl lysis reagent was mixed with 200 μl sample and the mixture was incubated for 10 min at room temperature. Then, 800 μl isopropanol was added to each sample. Nucleic acids were pelleted by centrifugation at room temperature for 15 min in a microcentrifuge. The pellets were washed with 1 ml 70% ethanol and centrifuged for 5 min. The alcohol was removed, the tubes were pulse spun for 30 s, and residual ethanol was removed. The pellets were resuspended in 100 μl HIV-1 sample diluent and stored at −80°C.
An HIV-1 sequence flanking the CAP/CTM HIV-1 target region in gag was amplified by reverse transcriptase PCR (RT-PCR). MuLV reverse transcriptase was used for the RT reaction, and PCR amplification was carried out using Taq DNA polymerase. Hemi-nested amplifications were then performed to increase amplification sensitivity. The amplicons were treated with ExoSAP-IT (GE Healthcare, United Kingdom) and then were sequenced. The cycle sequencing reaction was set up using ABI BigDye Terminator v3.1 Chemistry (Applied Biosystems Inc., Foster City, CA) on the GeneAmp PCR System 9700 or 9800 fast thermal cycler (Applied Biosystems Inc., Foster City, CA).
To determine the subtype of the virus in each sample, the gag sequence from each sample was aligned to sequences from HIV isolates of known genotypes from the GenBank sequence database using the ClustalW sequence alignment program, which also generates a phylogenetic tree, via the Lasergene software package (MegAlign version 5.07; DNASTAR Inc.).
Sequence analysis was performed by Roche's Global Surveillance Program (RGSP) in Pleasanton, CA, using residual patient samples. The interpretation of sequencing results was done by Roche personnel in the context of Roche's proprietary HIV database and experience with the CAP/CTM assay. ARUP personnel were not granted access to the sequence data due to the proprietary nature of the data.
RESULTS
The limit of detection (LOD) of the CAP/CTM assay was determined using 14 replicates of nine concentrations of 2nd International WHO Standard material ranging between 0.5 and 25 copies/ml and VQA standard material ranging between 0.5 and 40 copies/ml (Table 1). Probit analysis predicts a 95% limit of detection of 23.8 copies/ml (95% confidence interval, 20.1 to 27.5) for WHO material and 30.8 copies/ml (95% confidence interval, 26.4 to 35.3) for VQA material.
TABLE 1.
Limit of detection and probit analysisa
| 2nd International WHO Standardb |
Viral Quality Assurance standardc |
||||||
|---|---|---|---|---|---|---|---|
| Copies/ml | No. tested | No. detected | % detected | Copies/ml | No. tested | No. detected | % detected |
| 25 | 14 | 14 | 100.0 | 40 | 14 | 14 | 100.0 |
| 20 | 14 | 12 | 85.7 | 35 | 14 | 13 | 92.9 |
| 15 | 14 | 10 | 71.4 | 30 | 14 | 12 | 85.7 |
| 10 | 14 | 8 | 57.1 | 25 | 14 | 13 | 92.9 |
| 7.5 | 14 | 5 | 35.7 | 20 | 14 | 13 | 92.9 |
| 5 | 14 | 5 | 35.7 | 10 | 14 | 8 | 57.1 |
| 2.5 | 14 | 4 | 28.6 | 5 | 14 | 6 | 42.9 |
| 1 | 10d | 1 | 10.0 | 2 | 14 | 4 | 28.6 |
| 0.5 | 14 | 3 | 21.4 | 0.5 | 14 | 1 | 7.1 |
Nine dilutions of the 2nd International Standard or Viral Quality Assurance standard were prepared in Basematrix diluent. Fourteen replicates at each concentration were tested. Probit analysis was used to determine the limit of detection (95% detection rate) and 95% confidence interval.
Probit results: 23.8 copies/ml (95% CI: 20.1 to 27.5).
Probit results: 30.8 copies/ml (95% CI: 26.4 to 35.3).
Four samples were not tested due to an instrument error.
The reproducibility and linearity of the CAP/CTM assay were determined by replicate measurements of dilution series derived from six high-titer patient samples (Tables 2 and 3). Intra-assay slopes ranged from 0.888 ± 0.049 to 1.020 ± 0.023, and interassay slopes ranged from 0.916 ± 0.033 to 1.037 ± 0.049. The intercept ranges were −0.056 ± 0.089 to 0.440 ± 0.067 and −0.005 ± 0.151 to 0.412 ± 0.143 for the intra- and interassay regressions, respectively.
TABLE 2.
Intra- and interassay precisiona
| Dilution no. and comparison type | Mean ± SEM log10 HIV RNA copies/ml in the indicated sample (n)b |
|||||
|---|---|---|---|---|---|---|
| A | B | C | D | E | F | |
| Intra-assay | ||||||
| 1 | 4.792 ± 0.028 | 5.774 ± 0.053 | 5.522 ± 0.018 | >ULQ | 6.177 ± 0.092 | 6.714 ± 0.034 |
| 2 | 3.897 ± 0.015 | 4.950 ± 0.117 | 4.932 ± 0.071 | 6.073 ± 0.023 | 5.307 ± 0.033 | 5.898 ± 0.138 |
| 3 | 3.033 ± 0.053 | 4.205 ± 0.061 | 4.045 ± 0.068 | 5.196 ± 0.012 | 4.626 ± 0.027 | 4.967 ± 0.046 |
| 4 | 2.396 ± 0.069 | 3.146 ± 0.125 | 3.125 ± 0.005 | 4.440 ± 0.039 | 3.624 ± 0.064 | 4.163 ± 0.166 |
| 5 | <LLQ | 2.559 ± 0.079 | 2.214 ± 0.052 | 3.583 ± 0.016 | 2.850 ± 0.009 | 3.375 ± 0.041 |
| 6 | <LLQ | 1.732 (1) | 1.998 ± 0.044 (2) | 2.982 ± 0.033 | 1.861 ± 0.048 (2) | 2.555 ± 0.033 |
| 7 | <LLQ | <LLQ | <LLQ | 2.083 ± 0.064 | <LLQ | <LLQ |
| Interassay | ||||||
| 1 | 4.704 ± 0.049 | 5.774 (1) | 5.589 ± 0.067 (2) | >ULQ | 6.177 (1) | 6.729 ± 0.009 |
| 2 | 4.014 ± 0.062 | 4.883 ± 0.034 | 4.897 ± 0.032 | 6.172 ± 0.061 | 5.209 ± 0.051 | 5.852 ± 0.057 |
| 3 | 3.082 ± 0.030 | 4.141 ± 0.032 | 4.079 ± 0.024 | 5.204 ± 0.006 | 4.538 ± 0.066 | 4.986 ± 0.010 |
| 4 | 2.262 ± 0.078 | 3.192 ± 0.059 | 3.255 ± 0.096 | 4.402 ± 0.023 | 3.581 ± 0.023 | 4.297 ± 0.069 |
| 5 | <LLQ | 2.544 ± 0.021 | 2.359 ± 0.102 | 3.636 ± 0.028 | 2.936 ± 0.048 | 3.278 ± 0.070 |
| 6 | <LLQ | 1.789 ± 0.056 (2) | 1.998 (1) | 2.830 ± 0.076 | 1.969 ± 0.065 | 2.572 ± 0.103 |
| 7 | <LLQ | <LLQ | <LLQ | 1.986 ± 0.049 | <LLQ | <LLQ |
The intra- and interassay precision were evaluated by 7-fold serial dilutions of six high-titer patient samples (A to F). Five replicates were prepared and tested in the CAP/CTM assay. Three replicates were tested on a single rack to evaluate intra-assay reproducibility, and the remaining two replicates were tested on a second and third day (and compared to the mean of the intra-assay data) to evaluate interassay reproducibility. The expected values were calculated by setting the highest measured concentration at identity and using the dilution factor to calculate the remaining expected concentrations. The data assigned to identity in this manner were excluded from the regression analysis.
>ULQ, greater than the upper limit of quantification (>7 log10 copies/ml). <LLQ, less than the lower limit of quantification (<48 copies/ml) or not detected. n = 3 unless noted otherwise in parentheses (the number of replicates that produced quantitative results was less than the number of samples tested).
TABLE 3.
Intra- and interassay linear regression analysisa
| Sample | Intra-assay regression analysis |
Interassay regression analysis |
||||||
|---|---|---|---|---|---|---|---|---|
| Slope | Slope standard error | Intercept | Intercept standard error | Slope | Slope standard error | Intercept | Intercept standard error | |
| A | 0.888 | 0.049 | 0.354 | 0.156 | 1.037 | 0.049 | −0.005 | 0.151 |
| B | 0.964 | 0.046 | 0.190 | 0.170 | 0.916 | 0.033 | 0.376 | 0.100 |
| C | 0.938 | 0.050 | 0.438 | 0.166 | 0.938 | 0.041 | 0.412 | 0.143 |
| D | 0.909 | 0.018 | 0.440 | 0.067 | 0.948 | 0.015 | 0.165 | 0.057 |
| E | 1.020 | 0.023 | −0.056 | 0.089 | 0.988 | 0.035 | 0.195 | 0.113 |
| F | 0.980 | 0.034 | 0.098 | 0.149 | 0.978 | 0.027 | 0.094 | 0.118 |
See Table 2 for an explanation of the method and calculations.
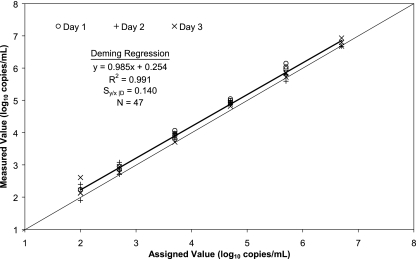
In addition to the patient sample dilutions to evaluate linearity, the six members of the ValiQuant panel, ranging from 100 to 5,000,000 copies/ml, were tested in triplicate on each of three days (Fig. 1). Some replicates of the 5,000,000-copies/ml panel member produced results above the upper limit of quantification for the assay (10,000,000 copies/ml). Deming regression analysis of the quantitative results produced the following results: measured log10 copies/ml = 0.985 (assigned log10 copies/ml) + 0.254, Sy/x |D = 0.140 (R2 = 0.991, n = 47). All negative panel members were “not detected” by the CAP/CTM assay.
FIG. 1.
ValiQuant panel. The members of the AcroMetrix ValiQuant HIV-1 RNA quantification panel were tested in triplicate on each of 3 days. All replicates of the negative panel member were “not detected.” The data were analyzed by Deming regression, shown by the heavy line. The light line represents the line of identity.
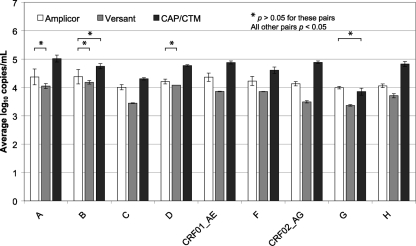
A group M subtype panel was evaluated in the CAP/CTM, Amplicor, and Versant assays. This panel represents group M subtypes, including some common circulating recombinant forms (CRFs). As shown in Fig. 2, there were statistically significant differences (P < 0.05) between assays for nearly every subtype. The following assay pairs were not significantly different (P > 0.05): subtype A, Amplicor-Versant; subtype B, Amplicor-Versant and Amplicor-CAP/CTM; subtype D, Amplicor-Versant; subtype G, Amplicor-CAP/CTM. Versant averaged 0.829 log10 copies/ml lower than CAP/CTM, and Amplicor averaged 0.427 log10 copies/ml lower than CAP/CTM.
FIG. 2.
Subtype panel. The members of the HIV RNA genotype performance panel PRD202 (SeraCare Life Sciences, Milford, MA) were diluted 3-fold in Basematrix and tested in triplicate by the Amplicor, Versant, and CAP/CTM assays. The subtype designations and average log10 copies/ml are shown. The error bars represent 1 standard deviation. No HIV-1 RNA was detected by any assay for the negative-control panel member. For the pairs marked with an asterisk, P > 0.05; all other pairs, P < 0.05.
Sixty-three samples with UltraSensitive Amplicor results below the limit of detection (<50 copies/ml) were retested with the CAP/CTM assay. One sample had an invalid QS (internal standard). Forty-eight samples had no detectable HIV-1 RNA. Ten samples were detected but were below the quantification limit of the assay (48 copies/ml). Four samples were quantitated, measuring 60, 64, 94, and 116 copies/ml. These samples had insufficient volume for repeat testing.
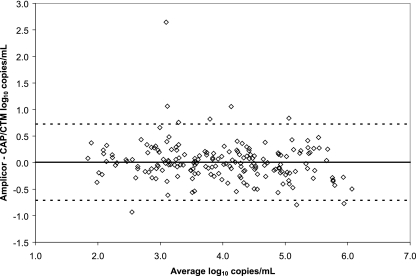
The results of the correlation of the CAP/CTM assay to the Amplicor assay using archived samples are shown in Fig. 3. A total of 218 samples were tested in the CAP/CTM assay; 1 sample had an invalid QS, 3 had clots detected, 17 were detected but not quantified, and 10 were not detected. A total of 187 samples produced valid quantitative results, with a Deming regression (measured as log10 copies/ml) of CAP/CTM = 1.056 (Amplicor) − 0.230 (R2 = 0.884, Sy/x |D = 0.366). On average, the Amplicor assay results were 0.008 log10 copies/ml higher than those of the CAP/CTM assay. Ninety-six percent of the samples differed by fewer than 0.5 log10 copies/ml; 98% of the samples differed by fewer than 1.0 log10 copies/ml.
FIG. 3.
Correlation samples. A total of 218 samples previously tested in the Amplicor assay were tested in the CAP/CTM assay. The data were analyzed by the method of Bland and Altman (1). The bold line indicates the average difference, and the dashed lines indicate the average ± 2 standard deviations.
One correlation sample (RIU025) had highly discrepant results in the correlation study. The Amplicor result was 26,000 copies/ml (4.4 log10 copies/ml), while the CAP/CTM result was 59 copies/ml (1.8 log10 copies/ml). The sample was retested with Amplicor, with a result of 46,000 copies/ml (4.7 log10 copies/ml). There was insufficient sample for retesting in the CAP/CTM assay. Roche was notified of the discrepancy, and the remaining sample was sent to RGSP for sequence analysis (Table 4).
TABLE 4.
Summary of samples with discrepanciesa
| Sample | Amplicor log (copies/ml) | CAP/CTM log (copies/ml) | Difference log (copies/ml) | No. of mismatches present |
Mismatches likely to affect quantification |
Comments | ||
|---|---|---|---|---|---|---|---|---|
| Primers | Probe | Primers | Probe | |||||
| RIU025 | 4.4/4.7e | 1.8 | 2.75e | 1 | 3 | Yes | No | Same sequence as sample 3 |
| 1 | 4.9 | 3.8 | 1.1 | 1 | 0 | No | N/A | |
| 2 | 4.1 | 3.1 | 1.0 | 2 | 1 | No | No | |
| 3 | 3.9 | 1.7 | 2.2 | 1 | 3 | Yes | No | Same sequence as RIU025 |
| 4 | 6.4 | 5.2 | 1.2 | 0 | 2 | N/A | No | |
| 5Ab | 4.1 | 1.9 | 2.2 | N/A | N/A | QNS; not sequenced | ||
| 5Bb | 2.6/2.7c,d,e | 2.0c | 0.65e | Unknown | Unknown | Sequencing was not successful | ||
| 6 | 5.2 | 4.1 | 1.1 | 1 | 4 | No | Yes | Mismatches not seen before by Roche |
The seven samples that had large discrepancies between Amplicor and CAP/CTM results were further analyzed by sequencing analysis where possible. N/A, not applicable.
Sample 5A was of a quantity not sufficient (QNS) for retesting. However, a sample from the same patient drawn 2 weeks later (5B) was available.
These results were produced by Roche.
In addition to these results, four replicates had “not detected” results.
Sample was tested twice. The difference is based on the average of the results for the replicates.
Sequence information, including the gag target region of the CAP/CTM test, was obtained from the sample. Phylogenetic analysis determined the virus to be subtype B. Alignment of this sequence against the proprietary sequences of the CAP/CTM assay primers and probe revealed mismatches to one of the primers and the probe. The three mismatches to the probe alone were determined to be unlikely to affect quantification of this specimen based on the location of the mismatches and known mismatch tolerance of the assay probe (data not shown). In addition, quantification data from other HIV-1 samples with at least the same three probe mismatches showed a nominal titer difference (0.2 to 0.5 log10) between the CAP/CTM and Amplicor tests. The primer mismatch found in sample RIU025 is likely to affect quantification by the CAP/CTM test, since the mismatch is located near the 3′ end of the primer. Other HIV-1 samples that contain the same mismatch have been shown to give titers that are 1.2 to 2.8 log10 lower by the CAP/CTM test than by the Amplicor test.
After completion of the evaluation described, the CAP/CTM instrument was moved to a different location in the laboratory. To verify proper performance, additional experiments were performed. This included testing 87 additional archived specimens that had previously been tested using Amplicor (data not shown) (Deming regression [measured as log10 copies/ml], CAP/CTM value = 1.087 [Amplicor value] − 0.534; R2 = 0.829; Sy/x |D = 0.496; n = 86; average difference [Amplicor value minus CAP/CTM value], 0.186 log10 copies/ml). Although these were new samples, since they are deidentified, the possibility that one or more are from the same patient as the prior batch of samples cannot be excluded. Of the 87 samples, 6 had viral load measurements that were at least 1 log10 copies/ml lower in the CAP/CTM assay than by Amplicor (Table 4, samples 1 to 6). To determine if these specimens also contained polymorphisms that may have interfered with quantification, aliquots were sent to RGSP for further testing. One sample (5A) was no longer available, but a plasma sample drawn from the same patient 2 weeks later was available. This sample (5B) was sent to RGSP for sequencing and testing in both Amplicor and CAP/CTM assays.
Sequence surrounding the CAP/CTM target region was obtained from five of the six samples. No sequence information was obtained from sample 5B. This is likely due to the low titer of this sample. (Sample 5A was unable to be sequenced due to insufficient sample.) The sequences for the remaining five discrepant samples were determined to be subtype B based on phylogenetic analysis of their gag sequences. Each of the sequences from these samples contains at least one nucleotide mismatch to a primer or probe.
For sample 1, there is one mismatch located near the 5′ end of one of the primers. The location of this mismatch makes it unlikely to interfere with amplification. In addition, the identical mismatch has been found in five other samples previously analyzed by the RGSP that generated similar titers (titer differences, −0.6 to 0.3 log10) between the CAP/CTM test and the Amplicor test.
The sequence from sample 2 was found to have mismatches to one of the primers and the probe. The primer mismatches are located in the 5′ half of the primer. A previous sample analyzed by the RGSP that contains the same primer mismatches plus one additional mismatch located closer to the 3′ end of the primer generated titers that are nearly identical (difference, 0.1 log10) between the CAP/CTM test and the Amplicor test, indicating that the primer mismatches seen in sample 2 are unlikely to be the cause of the titer discrepancy. The probe mismatch found in the sequence for sample 2 is common. There have been 40 samples sequenced by the RGSP that contain only this same probe mismatch, but all of these samples generated titers by the CAP/CTM test within one log10 of the titers from the Amplicor test (titer differences, −0.44 to 0.63 log10). Therefore, the probe mismatch found in the sequence of sample 2 is not likely to be the cause of the titer discrepancy. However, the possibility that the combination of the primer and probe mismatches may have contributed to the observed titer difference cannot be ruled out.
The HIV-1 sequence from sample 4 contains two mismatches to the probe of the CAP/CTM test. Four additional samples (not connected with the current study) that have been sequenced by the RGSP have shown the same probe mismatches, with and without additional mismatches. These four samples generated similar titers (titer differences, −0.20 to 0.58 log10) between the CAP/CTM test and the Amplicor test. Therefore, it is not likely that the probe mismatches found in sample 4 account for the titer discrepancy observed between the CAP/CTM and Amplicor tests.
Based on the location of the mismatches and additional samples tested with the same mismatches, the mismatches found in samples 1, 2, and 4 are unlikely to be the cause of the underquantification of these samples. For samples 1, 2, and 4, the difference between Amplicor and CAP/CTM results for these samples was between 1.0 and 1.2 log10 copies/ml, which could be accounted for by inherent variability in the two assays and might be resolved by replicate testing if sufficient material is available.
The HIV-1 sequence obtained from sample 3 contains the same primer and probe mismatches as already described for sample RIU025. The gag sequences generated from these two samples were identical when aligned to each other. It is very unlikely for two different HIV-infected patients to contain identical gag sequences, suggesting that samples RIU025 and 3 are from the same source. Contamination is not likely the cause of the identical sequences, as the samples were analyzed at different locations and nearly 1 year apart. Since the samples were deidentified, it cannot be determined unequivocally if these are independent samples from the same patient.
The HIV-1 sequence obtained from sample 6 contains four mismatches to the CAP/CTM probe and one mismatch to the downstream primer. The downstream primer mismatch is located near the 5′ end of the primer. Based on its location, this mismatch is not likely to affect amplification efficiency. In addition, two samples analyzed previously by the RGSP contain at least the same downstream primer mismatch and were found to generate very similar titers (titer differences, −0.1 to −0.3 log10) between the CAP/CTM test and the Amplicor test. However, based on their number and location, the four probe mismatches found in sample 6 may account for the titer discrepancy observed between the two assays. Although this exact mismatch pattern has not been observed in other samples collected by the RGSP, samples with sequences that contain similar numbers of mismatches in similar locations have been found to be underquantified by the CAP/CTM test by approximately one log10 compared to those quantified by the Amplicor test (data not shown).
DISCUSSION
The standard of care for HIV-infected patients has long included viral load testing. Accurate measurements are critical for determining disease prognosis and guiding the course of treatment. Failure to accurately quantify HIV viral load can lead to inappropriate clinical management of patients and unnecessary virus transmission (3, 5).
Viral load assays have evolved dramatically over the past decade. A trend for all quantitative viral testing during this period has been the adoption of real-time chemistries and instrumentation. Two assays (Abbott Molecular and Roche Molecular Systems) have received FDA approval for HIV-1 viral load testing in the real-time format. Compared to previous PCR testing which used endpoint detection, these real-time assays offer closed-tube automation with a wider analytical dynamic range.
We evaluated several key performance characteristics of the FDA-approved CAP/CTM assay. This study documents the overall good performance of the Roche assay for clinical isolates and defined HIV subtype isolates. The LOD was determined by probit analysis to be slightly lower than the 40 copies/ml previously reported (14). Differences between the 1st and 2nd WHO standards could account for this difference. The CAP/CTM assay exhibited excellent reproducibility and linearity over a 5-log10 copies/ml range with variability of 10% or less. There were statistically significant differences between assays for most subtype panel members, with CAP/CTM measurements generally highest while Versant measurements were generally lowest. However, genetic variability between individual samples, as we have shown in this study, may have a greater consequence than variability of results between subtype members. Therefore, the results of subtype panel testing should be interpreted cautiously.
Prior comparisons of the Amplicor and CAP/CTM assays have shown average differences (between Amplicor and CAP/CTM) of −0.215 (1a), −0.12 (8), −0.05 (11), 0.098 (16), −0.01 (14), 0.48 (2), and 0.28 (6). Although the subtypes of the correlation samples in this study are unknown, they probably consist mainly of subtype B viruses, since they were derived from a pool of samples submitted from across the United States (reference 12 and unpublished observations at ARUP).
This study also reveals the challenge of sequence diversity in the real-time format. We identified seven samples (from a total of 305 that were tested) with between 1.0 and 2.7 log10 copies/ml underquantification by the CAP/CTM assay. Sequence analysis of these samples was successful in six of seven cases; unfortunately, no sequence was obtained from sample 5. The sequences from samples 1, 2, and 4 contain polymorphisms in the CAP/CTM primer and/or probe regions which are deemed by RGSP to be unlikely to contribute to the observed titer differences based on the location of the mismatches and data generated from other HIV-1 samples evaluated by RGSP.
The sequences from samples RIU025, 3, and 6 contain polymorphisms that are likely to be contributing to the underquantification by the CAP/CTM test. The sequences generated from samples RIU025 and 3 are identical to each other, strongly suggesting that they were obtained from the same patient. This analysis suggests that 2 of the 304 (0.66%) unique HIV samples tested by the CAP/CTM test were confirmed to contain mismatches that are likely to interfere with quantification.
The majority of the discrepant samples in this study had insufficient volume for repeat testing, with the exception of samples RIU025 and 5B (which were measured twice and six times in Amplicor, respectively). For samples containing similar sequence mismatches which have been previously analyzed by RGSP, the details of sample handling such as freeze/thaw cycles, age, and storage conditions of the samples are not available. These problems are compounded by the imperfect science of predicting effects of polymorphisms on PCR performance and the proprietary nature of the assay sequences, which require investigators to rely on manufacturer analysis (such as in this study) or to make assumptions about the primer and probe locations to interpret sequencing results (10). These factors pose a significant obstacle to achieving a rigorous understanding of effects of sequence variation on commercial HIV assays. The process of producing RNA transcripts from cloned PCR products as a surrogate for patient sample is one pathway for reducing the uncertainty of assay comparisons when sample is limiting. The approach is impractical for most laboratories and still leaves in question the basis for test failure for proprietary assays. However, the challenge posed by the continued diversification of the highly variable HIV genome in clinical isolates worldwide and its impact on testing is critically important and has increasingly become the focus of study. While most reports have emphasized underquantification of non-B subtypes (2, 3, 5, 6, 7, 9), the discrepant samples in this study and others (2, 10) showed that underquantification can also occur in subtype B and is not restricted to specific subtypes.
Versions of the CAP/CTM product insert subsequent to the investigation of the discrepant samples described in this study have included a statement that “though rare, mutations within the highly conserved region of the viral genome covered by the Cobas AmpliPrep/Cobas TaqMan HIV-1 Test primers and/or probe may result in the underquantitation of or failure to detect the virus.” Current practice dictates determining new baseline viral loads for patients when switching to a different assay when there is a significant average bias between the assays. Even when there is no average bias between the assays (such as in this study), it may be necessary to determine relative quantification between the methods for some patients, especially when test results do not agree with clinical symptoms. The frequency, severity, and subtype distribution of underquantified samples requires additional study.
Acknowledgments
This study was performed in compliance with regulations concerning human subject research and was approved by the University of Utah Institutional Review Board.
All components for Roche CAP/CTM testing, including the use of the instruments, disposables, reagents, genotype panels, reference standards, and additional Amplicor and Versant testing, were provided or funded by Roche. The discrepant sample sequencing analysis and interpretation were performed by Roche.
We extend our thanks to Denise Jones for preparing samples, the staff of the Molecular Hepatitis/Retrovirus Laboratory at ARUP for performing the Amplicor and Versant testing, Andrew Wilson for assistance with statistics, and Jody Harris and Monica Lin at Roche Molecular Systems for sequencing the discrepant samples.
Footnotes
Published ahead of print on 23 June 2010.
REFERENCES
- 1.Bland, J. M., and D. G. Altman. 1986. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet i:307-310. [PubMed] [Google Scholar]
- 1a.Braun, P., R. Ehret, F. Wiesmann, F. Zabbai, M. Knickmann, R. Kühn, S. Thamm, G. Warnat, and H. Knechten. 2007. Comparison of four commercial quantitative HIV-1 assays for viral load monitoring in clinical daily routine. Clin. Chem. Lab. Med. 45:93-99. [DOI] [PubMed] [Google Scholar]
- 2.Damond, F., B. Roquebert, A. Bénard, G. Collin, M. Miceli, P. Yéni, F. Brun-Vezinet, and D. Descamps. 2007. Human immunodeficiency virus type 1 (HIV-1) plasma load discrepancies between the Roche COBAS AMPLICOR HIV-1 MONITOR version 1.5 and the Roche COBAS AmpliPrep/COBAS TaqMan HIV-1 assays. J. Clin. Microbiol. 45:3436-3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Delaugerre, C., B. Denis, G. Peytavin, P. Palmer, T. Mourez, J. Le Goff, J. Molina, and F. Simon. 2009. Clinical and resistance consequences of misquantification of plasma and cerebrospinal fluid human immunodeficiency virus type 1 (HIV-1) RNA in an HIV-1 subtype G-infected patient. J. Clin. Microbiol. 47:3763-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foulongne, V., B. Montes, M.-N. Didelot-Rousseau, and M. Segondy. 2006. Comparison of the LCx human immunodeficiency virus (HIV) RNA Quantitative, RealTime HIV, and COBAS AmpliPrep-COBAS TaqMan assays for quantitation of HIV type 1 RNA in plasma. J. Clin. Microbiol. 44:2963-2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geelen, S., J. Lange, J. Borleffs, T. Wolfs, A. Weersink, and R. Schuurman. 2003. Failure to detect a non-B HIV-1 subtype by the HIV-1 Amplicor Monitor test, version 1.5: a case of unexpected vertical transmission. AIDS 17:781-782. [DOI] [PubMed] [Google Scholar]
- 6.Gueudin, M., J. C. Plantier, V. Lemée, M. P. Schmitt, L. Chartier, T. Bourlet, A. Ruffault, F. Damond, M. Vray, and F. Simon. 2007. Evaluation of the Roche Cobas TaqMan and Abbott RealTime extraction-quantification systems for HIV-1 subtypes. J. Acquir. Immune Defic. Syndr. 44:500-505. [DOI] [PubMed] [Google Scholar]
- 7.Holguín, A., M. López, M. Molinero, and V. Soriano. 2008. Performance of three commercial viral load assays, Versant human immunodeficiency virus type 1 (HIV-1) RNA bDNA v3.0, Cobas AmpliPrep/Cobas TaqMan HIV-1, and NucliSens HIV-1 EasyQ v1.2, testing HIV-1 non-B subtypes and recombinant variants. J. Clin. Microbiol. 46:2918-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katsoulidou, A., M. Petrodaskalaki, V. Sypsa, E. Papachristou, C. G. Anastassopoulou, P. Gargalianos, A. Karafoulidou, M. Lazanas, T. Kordossis, A. Andoniadou, and A. Hatzakis. 2006. Evaluation of the clinical sensitivity for the quantification of human immunodeficiency virus type 1 RNA in plasma: comparison of the new COBAS TaqMan HIV-1 with three current HIV-RNA assays—LCx HIV RNA quantitative, VERSANT HIV-1 RNA 3.0 (bDNA) and COBAS Amplicor HIV-1 Monitor v1.5. J. Virol. Methods 131:168-174. [DOI] [PubMed] [Google Scholar]
- 9.Kim, J. E., B. Beckthold, Z. Chen, J. Mihowich, L. Malloch, and M. J. Gill. 2007. Identification of a novel HIV type 1 subtype H/J recombinant in Canada with discordant HIV viral load (RNA) values in three different commercial assays. AIDS Res. Hum. Retroviruses 23:1309-1313. [DOI] [PubMed] [Google Scholar]
- 10.Korn, K., B. Weissbrich, C. Henke-Gendo, A. Heim, C. M. Jauer, N. Taylor, and J. Eberle. 2009. Single-point mutations causing more than 100-fold underestimation of human immunodeficiency virus type 1 (HIV-1) load with the Cobas TaqMan HIV-1 real-time PCR assay. J. Clin. Microbiol. 47:1238-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oliver, A. R., S. F. Pereira, and D. A. Clark. 2007. Comparative evaluation of the automated Roche TaqMan real-time quantitative human immunodeficiency virus type 1 RNA PCR assay and the Roche AMPLICOR version 1.5 conventional PCR assay. J. Clin. Microbiol. 45:3616-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osmanov, S., C. Pattou, N. Waler, B. Schwardländer, J. Esparza, and the WHO-UNAIDS Network for HIV Isolation and Characterization. 2002. Estimated global distribution and regional spread of HIV-1 genetic subtypes in the year 2000. J. Acquir. Immune Defic. Syndr. 29:184-190. [DOI] [PubMed] [Google Scholar]
- 13.Pyne, M. T., E. Q. Konnick, A. Phansalkar, and D. R. Hillyard. 2009. Evaluation of the Abbott investigational use only RealTime HIV-1 assay and comparison to the Roche Amplicor HIV-1 monitor test, version 1.5. J. Mol. Diagn. 11:347-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schumacher, W., E. Frick, M. Kauselmann, V. Maier-Hoyle, R. van der Vliet, and R. Babiel. 2007. Fully automated quantification of human immunodeficiency virus (HIV) type 1 RNA in human plasma by the COBAS AmpliPrep/COBAS Taqman system. J. Clin. Virol. 38:304-312. [DOI] [PubMed] [Google Scholar]
- 15.Schutten, M. 2008. Comparison of the Abbott Realtime HIV-1 and HCV viral load assays with commercial competitor assays. Expert Rev. Mol. Diagn. 8:369-377. [DOI] [PubMed] [Google Scholar]
- 16.Wolff, D., and A. Gerritzen. 2007. Comparison of the Roche COBAS Amplicor Monitor, Roche COBAS Ampliprep/COBAS Taqman and Abbott RealTime test assays for quantification of hepatitis C virus and HIV RNA. Clin. Chem. Lab. Med. 45:917-922. [DOI] [PubMed] [Google Scholar]