Abstract
We described the colonization dynamics of Staphylococcus aureus in a group of 266 healthy carriers over a period of approximately 1 year. We used precise genotyping methods, i.e., amplified fragment length polymorphism (AFLP), spa typing, and double-locus sequence typing (DLST), to detect changes in strain identity. Strain change took place rather rarely: out of 89 carriers who had initially been colonized, only 7 acquired a strain different from the original one. Approximately one-third of the carriers eliminated the colonization, and a similar number became newly colonized. Some of these events probably represent detection failure rather than genuine colonization loss or acquisition. Lower bacterial counts were associated with increased probability of eliminating the colonization. We have confirmed a high mutation rate in the spa locus: 6 out of 53 strains underwent mutation in the spa locus. There was no overall change in S. aureus genotype composition.
Staphylococcus aureus is an important human pathogen responsible for both nosocomial and community-acquired infections, but it also asymptomatically colonizes a large fraction of humans (20 to 60%). Although most carriers never experience health problems related to colonization, it is estimated that 80% of infections with S. aureus are endogenous, i.e., caused by the colonizing strain (19, 20). Understanding of the interplay between nasal carriage and infection with S. aureus is far from complete. For example, it is not known whether prior nasal carriage of S. aureus (methicillin-susceptible S. aureus [MSSA]) might prevent hospital-acquired colonization and subsequent infection with methicillin-resistant S. aureus (MRSA).
Particularly little is known about persistence and transmission. Many studies have described temporal patterns of carriage, but only a handful explicitly monitored the temporal changes in strain identity (17, 18). Persistence and transmissibility should be the main determinants of long-term fitness of S. aureus. Low transmission of S. aureus in comparison with other staphylococcal human colonizers such as S. epidermidis has been suggested to play a major role in its evolutionary increase in virulence (9). Moreover, the differences in transmissibility can have implications for human health care. For example, MRSA with superb transmission abilities might be much more problematic than virulent MRSA with low transmission.
Explicit studies of transmission of S. aureus are difficult for logistic reasons, but those which have been carried out have brought interesting insights. For example, Peacock et al. (11) have found that for healthy newborn babies the primary source of S. aureus is the mother. Documenting the persistence of a bacterial strain in a human host requires longitudinal sampling and precise genotyping of bacterial strains.
Here we have documented the colonization dynamics of healthy adult carriers with S. aureus strains. We have also explored host and bacterial characteristics responsible for differences in persistence. In addition, we have monitored microevolutionary changes in the spa and clfB genes, which are used for genotyping.
MATERIALS AND METHODS
Sample collection.
The first sampling was described in detail previously (12). Ethical clearance was sought and obtained from the Ethical Committee of the University of Lausanne. Briefly, nasal swabs were collected from 405 newly employed hospital personnel during their first medical checkup at a tertiary care hospital in Lausanne, Switzerland, over a period of 9 months. The second sampling was conducted after 6 to 15 months. All available participants were swabbed again using the same methods. From each participant, one nasal swab was obtained (Amies agar transport swabs; Copan). Each swab was vigorously rubbed in 1 ml Tris-EDTA (TE) buffer, and then the dilutions were quantitatively plated on a SAID (bioMérieux) specific chromogenic plate. After 24 to 48 h of incubation at 37°C, green colonies were counted and isolated for further use. To detect colonization with even a very low number of bacteria, the swab was transferred to 5 ml of Bacto m staphylococcus broth (Difco) and incubated overnight at 37°C. The enrichment broth was than plated on a SAID plate and incubated overnight at 37°C. Eight isolates per participant were collected from directly inoculated plates, as well as another isolate from the plate inoculated with enrichment broth. To screen for MRSA, the enrichment broth was plated on MRSASelect (Bio-Rad) selective chromogenic agar. A PCR screen for an internal fragment of the mecA gene was performed as described previously (12).
Genotyping.
DNA was extracted from 600-μl to 1-ml overnight cultures grown in brain heart infusion broth as described previously (2). Amplified fragment length polymorphism (AFLP) was performed as described previously (12). AFLP electropherograms were analyzed with the help of GeneMapper software (Applied Biosystems). At the first sampling, the enrichment isolate and eight direct isolates were genotyped. At the second sampling, the enrichment isolate and one direct isolate were also genotyped. If the genotypes of the isolates from the second sampling were not identical to those of the isolates from the first sampling, all available direct isolates of a participant were genotyped by AFLP to maximize the chance of finding multiple genotypes. Neighbor-joining trees based on Nei and Li distances were constructed using Phylip (4). Multilocus sequence typing (MLST) analysis was performed as previously described (3) on selected isolates. Sequence types (STs) were identified by consulting the S. aureus MLST database (http://www.mlst.net/). Double-locus sequence typing (DLST) was performed as described previously (8). Briefly, highly polymorphic fragments of the spa and clfB genes were amplified and sequenced. Similarly to the MLST scheme, each unique sequence is given a consecutive number, and a combination of the two numbers uniquely identifies the DLST genotype. The repeat region of the spa gene was amplified as previously described (6, 14). The spa types were assigned with the online spa database (http://www.spaserver.ridom.de/). Initially, spa and clfB sequencing was performed on all isolates which appeared to be distinct after AFLP genotyping. If a different allele was detected in the isolates from the first and second samplings from the same carrier, the sequencing was also done on an additional isolate from the same carrier. All statistical analyses were done with the R package (http://www.R-project.org; Foundation for Statistical Computing).
RESULTS
Colonization dynamics.
Carriage patterns in the first sampling round were described in detail previously (12). The median time between the two samplings was 281 days (range, 141 to 463 days). In the second sampling round, 267 of the original 405 volunteers (66%) could be resampled, among whom 89 were carriers at the first sampling. The carriage rates were very similar at the two sampling occasions, i.e., 32% at the first sampling and 34% at the second sampling. The participants who were not resampled did not differ in any characteristics that we recorded from those who were resampled (data not shown; for the list of characteristics, see reference 12). In 27 (30%) of the volunteers who were previously carriers, S. aureus was not detected in the second sampling round. Thirty previous noncarriers became colonized (Table 1).
TABLE 1.
Colonization status of 267 participants at the two sampling occasions
| Status at sampling I | n | Status at sampling II | n |
|---|---|---|---|
| Carrier | 89 | Carrier | 62 |
| Noncarrier | 27 | ||
| Noncarrier | 177 | Carrier | 30 |
| Noncarrier | 148 |
The time between the two samplings did not affect the change in colonization status (Kruskal-Wallis chi-square test = 1.96; df = 2 [not significant]). In addition, for several participants we tested characteristics which might affect retention, loss, or new acquisition of S. aureus: none proved to be significant (Kruskal-Wallis rank sum test) (Table 2).
TABLE 2.
Number of participants with various characteristics by colonization status
| Characteristic | No. of participants with characteristic/no. without characteristica for the following colonization status: |
||
|---|---|---|---|
| Retained | Lost | Acquired | |
| Age, yr (mean) | 28.0 | 26.5 | 31.5 |
| Gender (no. male/no. female) | 21/35 | 14/20 | 18/20 |
| Contact with patients | 22/55 | 11/32 | 13/37 |
| Hospitalization between the two samplings | 4/55 | 0/32 | 2/36 |
| Antibiotic treatment between the two samplings | 10/54 | 3/32 | 7/37 |
Except as indicated for age and gender.
Strain replacement (i.e., the strain found on the second sampling occasion was entirely different from the strain found on the first sampling occasion) took place in only 5 out of 62 participants who were colonized at both sampling occasions (Table 3).
TABLE 3.
Fate of bacterial strains from participants who were colonized at the first sampling occasion
| Status at sampling I | n | Status at sampling II | n |
|---|---|---|---|
| Colonized with two strains | 2 | One strain lost | 2 |
| Colonized with one strain | 87 | Colonized with same strain | 53 |
| Colonized with different strain | 5 | ||
| Colonized with additional strain | 2 | ||
| Uncolonized | 27 |
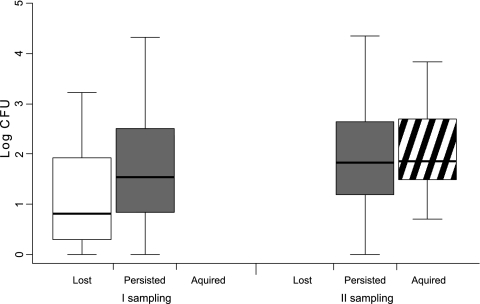
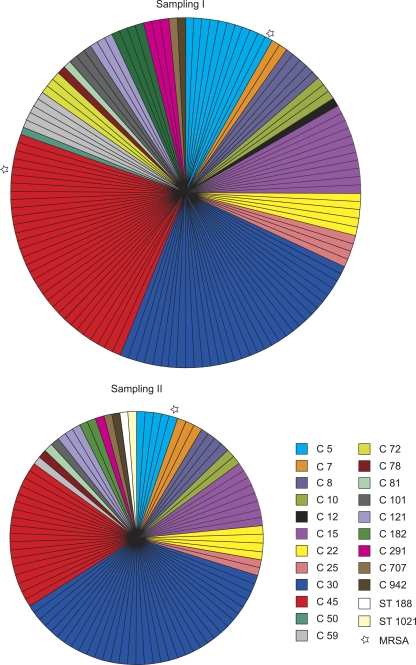
The number of CFU was significantly higher in the second sampling (Kruskal-Wallis chi-square = 7.1; df = 1 [P < 0.01]). The strains which persisted in colonization had higher CFU than those which were no longer detectable at the second sampling (Kruskal-Wallis chi-square = 7.6; df = 1 [P < 0.01]), but there was no difference between the strains which persisted and those which were newly found on the second sampling occasion (Kruskal-Wallis chi-square = 0.30; df = 1 [not significant]) (Fig. 1). The genotype composition of S. aureus remained essentially unchanged. AFLP analysis unambiguously assigned all isolates except two to the clusters which were shown earlier to strictly correspond to MLST clonal complexes (12). The two isolates which did not cluster to previously identified groups were genotyped by MLST. They were STs 188 and 1021 (Fig. 2).
FIG. 1.
In vivo abundance of strains according to their colonization status at two sampling occasions. The CFU of strains which were detected only after overnight enrichment were not considered here. Boxes show median, quartile, and extreme values.
FIG. 2.
Genetic diversity among carriage isolates of S. aureus. Each section represents one isolate. Sections with the same color represent the same cluster as defined by AFLP analysis. Two isolates which did not fall within any of the AFLP clusters were genotyped by MLST. The diameters of the circles correspond to the numbers of isolates.
Microevolution: spa and clfB loci.
There were 53 participants whose strains were identical by AFLP on the two sampling occasions (Table 3). Among these carriers, in seven cases a mutation in either the clfB or spa locus was detected (Table 4). In five cases (10%), this resulted in a change in spa type. Most mutations (six) were deletions, and one was point mutation.
TABLE 4.
Mutations in clfB and spa genes in strains which were identical by AFLP on the two sampling occasions
| Locus | Participant no. | Parameter | Value for samplinga: |
Mutation | |
|---|---|---|---|---|---|
| I | II | ||||
| clfB | 373 | clfB_500 | 26 | 406 | Deletion of one 18-bp repeat (TCGGATTCAGATAGCGAT) |
| spa | 62 | spa_500 | 126 | 366 | Deletion outside spa repeats |
| spa type | t2485 | t2485 | (loss of domain B5 | ||
| spa repeats | 15-12-16-02-16-17-24-24 | 15-12-16-02-16-17-24-24 | containing primer spa | ||
| 1113) | |||||
| 63 | spa_500 | 88 | 405 | Deletion of six repeats | |
| spa type | t091 | t2343 | |||
| spa repeats | 07-23-21-17-34-12-23-02-12-23 | 07-23-21-23 | |||
| 76 | spa_500 | 37 | 198 | Point mutation | |
| spa type | t015 | t050 | |||
| spa repeats | 08-16-02-16-34-13-17-34-16-34 | 08-16-02-16-34-34-17-34-16-34 | |||
| 101 | spa_500 | 37 | 371 | Deletion of one repeat | |
| spa type | t015 | t1078 | |||
| spa repeats | 08-16-02-16-34-13-17-34-16-34 | 08-16-02-16-34-13-17-34-34 | |||
| 118 | spa_500 | 158 | 404 | Deletion of one repeat | |
| spa type | t4025 | t029 | |||
| spa repeats | 09-02-16-34-13-13-17-34-16-34-34 | 09-02-16-13-13-17-34-16-34-34 | |||
| 402 | spa_500 | 196 | 370 | Deletion of five repeats | |
| spa type | t678 | t350 | |||
| spa repeats | 09-02-16-34-42-17-34-16-34-42-17-34-16-34 | 09-02-16-34-42-17-34-16-34 | |||
Boldface added to facilitate comparison of spa repeats.
DISCUSSION
A large number of studies have examined longitudinal carriage of Staphylococcus aureus (reviewed in reference 18), but only a handful employed genotyping methods precise enough to monitor the change in bacterial strain identity and sample sizes large enough to draw more general conclusions. Our data show that strain replacement among adult carriers of S. aureus occurred quite infrequently, and after approximately 9 months, only 7 out of 89 carriers acquired a strain different from the original one. These results confirm the conclusions of earlier, smaller studies, although direct comparison is difficult, primarily because these earlier studies included only individuals who were carriers. Nevertheless, the overall picture is similar. In the first study (16), one-third (11/31) of the carriers had the same strain after approximately 19 months. In the second study (18), 18% of the carriers (3/17) still harbored the same strain after 8 years.
Rare strain replacement and infrequent cocolonization of one human host with multiple strains (12) imply that one strain is usually able to completely monopolize its host, probably by competitive exclusion (1, 13). Interference due to agr types, as has been proposed by Massey et al. (9), cannot explain our results; if this were important, we would have observed cocolonization and replacement with different strains belonging to the same agr types (9). It is possible that the only chance for other strains to take over arises when the abundance of the resident strain drops. Data from an artificial human inoculation study (10) clearly showed that carriers come into contact with new S. aureus strains quite frequently. This suggests that low effective transmission rates in S. aureus are due to interference rather than to a simple lack of transfer opportunity. Massey et al. (9) proposed that lower transmission of S. aureus (compared to other human-colonizing staphylococci such as S. epidermidis) was the main evolutionary factor behind an evolutionary increase in S. aureus virulence. Among-strain interference is a likely cause of low effective transmission rates of S. aureus and thus potentially the ultimate reason for the evolutionary increase of its virulence. The link between interference and evolutionary increase of virulence needs to be elucidated.
Removal of a resident strain can open the way to colonization by a different (alien) strain. This might be highly relevant for hospitalized patients, who are often receiving antimicrobial drugs and at the same time are at increased risk of encountering strains more resistant than their resident strain (i.e., MRSA). Moreover, comparison of our data with those of Wertheim et al. (20) suggests that hospitalized patients exchange strains much more frequently than healthy individuals. The role of increased instability of nasal carriage in the hospital environment and its role in nosocomial acquisition of MRSA should be further investigated.
The mechanism of competitive exclusion among S. aureus strains is not known, but two artificial human inoculation studies (10, 17) strongly suggest the role of a human host factor. After a round of decolonization and artificial recolonization with a mixture of strains, the former carriers tended to become recolonized with their resident strain, in particular those who had been repeatedly found to be colonized before (persistent carriers). This suggests that a match between a bacterial strain and human host is needed, but so far the characteristics of host and bacteria which play a role are unknown. This is supported by results of Sakwinska et al. (12), who did not find any evidence that host characteristics such as hospitalization, antibiotic exposure, age, gender, etc., were associated with any particular genotype of S. aureus. Here, we found no evidence that participants' characteristics affected the loss or acquisition of colonization status, but the number of individuals was limited, possibly precluding finding any differences.
The change in colonization status was quite frequent, with 30% (27/89) of the previous carriers having eliminated the colonization with S. aureus in approximately 9 months and similar number of noncarriers becoming colonized. Given the rare occurrence of strain replacement, it is likely that a significant proportion of the noncolonization represents detection failure due to a decrease of CFU below the detection limit or, possibly, a change in physiological niche preventing detection.
We have observed no changes in the genotype composition of carried S. aureus strains in a time span of 9 months. Such changes have been documented in S. aureus (5, 15), but the interval was longer. Moreover, the most pronounced change was due to a swift expansion of one strain, USA300. The question whether the success of newly arising strains such as USA300 is due to their ability to colonize previously uncolonized persons or rather to successful competition with resident strains remains open.
We have found similar mutation rates in the spa locus as reported in earlier studies (7, 8), with a molecular clock of 91 months (six mutational changes in 544 months). On the other hand, only one mutation in the clfB locus was found, which might indicate its higher stability. Laboratories which use these loci for genotyping should be aware of this mutation rate. The potential functional impact of mutations in these loci remains obscure.
To elucidate the role of nasal carriage of S. aureus in infection, it is necessary to understand the colonization dynamics of healthy people. This study provides such a reference point. Individual bacterial strains persisted for a long time in the carriers, but mutations in evolutionarily labile loci occurred very often. Lower bacterial counts were associated with increased probability of eliminating the colonization.
Acknowledgments
This work was supported by a Marie Heim Vögtlin grant (PMPDA-106195) from the Swiss National Science Foundation to O.S.
We thank all the volunteers for their participation. Many thanks go to Marc Robinson-Rehavi and Sebastién Moretti for bioinformatic help with spa typing.
Footnotes
Published ahead of print on 2 June 2010.
REFERENCES
- 1.Aly, R., H. I. Maibach, H. R. Shinefield, A. Mandel, and W. G. Strauss. 1974. Bacterial interference among strains of Staphylococcus aureus in man. J. Infect. Dis. 129:720-724. [DOI] [PubMed] [Google Scholar]
- 2.Elphinstone, M. S., G. N. Hinten, M. J. Anderson, and C. J. Nock. 2003. An inexpensive and high-throughput procedure to extract and purify total genomic DNA for population studies. Mol. Ecol. Notes 3:317-320. [Google Scholar]
- 3.Feil, E. J., J. E. Cooper, H. Grundmann, D. A. Robinson, M. C. Enright, T. Berendt, S. J. Peacock, J. M. Smith, M. Murphy, B. G. Spratt, C. E. Moore, and N. P. J. Day. 2003. How clonal is Staphylococcus aureus? J. Bacteriol. 185:3307-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Felsenstein, J. 2004. PHYLIP (Phylogeny Inference Package) version 3.6. Department of Genome Sciences, University of Washington, Seattle, WA.
- 5.Gorwitz, R. J., D. Kruszon-Moran, S. K. McAllister, G. McQuillan, L. K. McDougal, G. E. Fosheim, B. J. Jensen, G. Killgore, F. C. Tenover, and M. J. Kuehnert. 2008. Changes in the prevalence of nasal colonization with Staphylococcus aureus in the United States, 2001-2004. J. Infect. Dis. 197:1226-1234. [DOI] [PubMed] [Google Scholar]
- 6.Harmsen, D., H. Claus, W. Witte, J. Rothganger, H. Claus, D. Turnwald, and U. Vogel. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41:5442-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kahl, B. C., A. Mellmann, S. Deiwick, G. Peters, and D. Harmsen. 2005. Variation of polymorphic region C of the protein A gene during persistent airway infection of cystic fibrosis patients reflects two independent mechanisms of genetic change in Staphylococcus aureus. J. Clin. Microbiol. 43:502-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuhn, G., P. Francioli, and D. S. Blanc. 2007. Double-locus sequence typing using clfB and spa, a fast and simple method for epidemiological typing of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 45:54-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Massey, R. C., M. J. Horsburgh, G. Lina, M. Hook, and M. Recker. 2006. The evolution and maintenance of virulence in Staphylococcus aureus: a role for host-to-host transmission? Nat. Rev. Microbiol. 4:953-958. [DOI] [PubMed] [Google Scholar]
- 10.Nouwen, J., H. Boelens, A. van Belkum, and H. Verbrugh. 2004. Human factor in Staphylococcus aureus nasal carriage. Infect. Immun. 72:6685-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peacock, S. J., A. Justice, D. Griffiths, G. D. I. de Silva, M. N. Kantzanou, D. Crook, K. Sleeman, and N. P. J. Day. 2003. Determinants of acquisition and carriage of Staphylococcus aureus in infancy. J. Clin. Microbiol. 41:5718-5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sakwinska, O., G. Kuhn, C. Balmelli, P. Francioli, M. Giddey, V. Perreten, A. Riesen, F. Zysset, D. S. Blanc, and P. Moreillon. 2009. Genetic diversity and ecological success of Staphylococcus aureus strains colonizing humans. Appl. Environ. Microbiol. 75:175-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shinefield, H. R., J. C. Ribble, and M. Boris. 1971. Bacterial interference between strains of Staphylococcus aureus, 1960 to 1970. Am. J. Dis. Child. 121:148-152. [DOI] [PubMed] [Google Scholar]
- 14.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tenover, F. C., S. McAllister, G. Fosheim, L. K. McDougal, R. B. Carey, B. Limbago, D. Lonsway, J. B. Patel, M. J. Kuehnert, and R. Gorwitz. 2008. Characterization of Staphylococcus aureus isolates from nasal cultures collected from individuals in the United States in 2001 to 2004. J. Clin. Microbiol. 46:2837-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Belkum, A., N. H. R. Eriksen, M. Sijmons, W. Van Leeuwen, M. VandenBergh, J. Kluytmans, F. Espersen, and H. Verbrugh. 1997. Coagulase and protein A polymorphisms do not contribute to persistence of nasal colonisation by Staphylococcus aureus. J. Med. Microbiol. 46:222-232. [DOI] [PubMed] [Google Scholar]
- 17.van Belkum, A., N. J. Verkaik, C. P. de Vogel, H. A. Boelens, J. Verveer, J. L. Nouwen, H. A. Verbrugh, and H. F. L. Wertheim. 2009. Reclassification of Staphylococcus aureus nasal carriage types. J. Clin. Microbiol. 199:1820-1826. [DOI] [PubMed] [Google Scholar]
- 18.van den Bergh, M. F. Q., E. P. F. Yzerman, A. van Belkum, H. A. M. Boelens, M. Sijmons, and H. A. Verbrugh. 1999. Follow-up of Staphylococcus aureus nasal carriage after 8 years: redefining the persistent carrier state. J. Clin. Microbiol. 37:3133-3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.von Eiff, C., K. Becker, K. Machka, H. Stammer, and G. Peters. 2001. Nasal carriage as a source of Staphylococcus aureus bacteremia. N. Engl. J. Med. 344:11-16. [DOI] [PubMed] [Google Scholar]
- 20.Wertheim, H. F. L., M. C. Vos, A. Ott, A. van Belkum, A. Voss, J. Kluytmans, P. H. J. van Keulen, C. Vandenbroucke-Grauls, M. H. M. Meester, and H. A. Verbrugh. 2004. Risk and outcome of nosocomial Staphylococcus aureus bacteraemia in nasal carriers versus non-carriers. Lancet 364:703-705. [DOI] [PubMed] [Google Scholar]