Abstract
The ability to detect HIV-2 and to discriminate between HIV-1 and HIV-2 infections was evaluated in 46 serum samples from Guinea-Bissau (GB) and Guinea-Conakry (GC) using serological tests and commercial (HIV-1) and in-house (HIV-2) real-time PCR assays. Samples were first identified as HIV-2 positive by Genie I/II assay in GB and GC. HIV positivity was detected in 44 of 46 samples by all screening and confirmatory assays. A diagnostic strategy based on Inno-LIA and HIV-1/2 RNA detection assays allowed accurate discrimination between HIV-1 and HIV-2 in 84% of single infections and confirmed 32% of double infections. In samples with double reactivity in the Inno-LIA test and no detection of both genomes, cross-reactivity likely hampered the identification of true double infections. In conclusion, the implementation of a diagnostic strategy, based on multiple specific serological tests and highly sensitive quantitative PCR assays, is recommended to ensure accurate HIV-2 diagnosis and appropriate therapy for individuals from areas in which the virus is endemic.
The occurrence of human immunodeficiency virus type 2 (HIV-2) infection is geographically restricted to West Africa, where the virus was first isolated from patients with AIDS originating from Cape Verde and Guinea-Bissau (2, 6).
HIV-2 infections have been also reported in European countries with socioeconomic relations with West Africa, such as France, United Kingdom, and Portugal (11, 18, 25). Although most of the HIV-2 infections in these countries occurred in patients originating from areas in which the virus is endemic (3, 22), the risk of HIV-2 transmission outside migrant populations in Europe could become relevant in the future, due to the increase of migration and international travel. This is particularly important for countries with high numbers of foreign citizens, like Italy (http://epp.eurostat.ec.europa.eu/cache/ITY_OFFPUB/KS-NK-06-008/EN/KS-NK-06-008-EN.PDF). In this context, a correct diagnosis of HIV-2 infection is crucial to ensure appropriate therapy and reduce the risk of transmission both in areas in which the virus is endemic and in areas in which it is not.
According to the Italian law, the use of HIV-2-specific diagnostic assays for the screening of blood donations became mandatory in 1992. HIV screening is recommended with HIV-1/2 antibody screening test followed by HIV-1/2 confirmatory test. Although sensitivity and specificity of screening assays have improved, the genetic variability of HIV still represents a challenge, in particular for early detection of infection. The standardization of screening assays for HIV-2 is also particularly difficult, as serum panels are rare and seroconversion samples are not available; moreover, screening assays usually include only one HIV-2 antigen in their format. As a consequence, variable sensitivity of fourth-generation screening assays for HIV-1 non-B subtypes and HIV-2 has been reported by different groups (16, 17, 19, 26). In addition, in countries in which the virus is not endemic, a correct serological diagnosis of HIV-2 infection may be missed, as adequate confirmation tools are not routinely used. The national program of external quality evaluation for the anti-HIV screening test in diagnostic laboratories indicated that an incorrect diagnosis of HIV-2-positive serum in Italy may be related to the use, on a daily routine, of HIV-1 confirmatory Western blots (5). The use of such confirmatory tests in regions in which the virus is not endemic may in fact lead to misclassification of HIV-2-infected individuals as HIV-1 positive. This is due to cross-reactivity between anti-HIV-2 antibodies and envelope glycoproteins of HIV-1. In this regard, it is important to note that HIV-2 subtype B sera react with the gp160 and gp120 HIV-1 glycoproteins, as well as with peptides from the gp41 immunodominant region of HIV-1 M and/or N strain (9). Diagnosis of HIV-2 infection is further complicated by the absence of specifically dedicated commercial assays for detection and/or monitoring of the level of HIV-2 RNA. The precise diagnosis of HIV-2 has implications for the choice of antiretroviral treatment, as HIV-2 strains are resistant to nonnucleoside reverse transcriptase and fusion inhibitors and are less sensitive in vitro to some protease inhibitors (10, 20).
The aim of the present study was to evaluate the ability to detect and discriminate between HIV-1 and HIV-2 infection in sera from areas in which the virus is endemic by diagnostic systems largely used in Italian blood transfusion services and clinical diagnosis. To address this issue, we analyzed serum samples collected in western African countries using screening and confirmatory serological assays and molecular methods.
MATERIALS AND METHODS
Patient samples.
Overall, 46 serum samples were collected from 37 individuals at the Drug Resource Enhancement against AIDS and Malnutrition (DREAM) center in Guinea-Bissau (GB) and 9 at the DREAM center in Guinea-Conakry (GC). All of them were identified, in local settings, as HIV-2 antibody positive by using a Genie I/II rapid assay. Demographic and clinical data were available for 34 individuals (25 from GB and 9 from GC). Of the patients, 17 had CD4+ cell counts above 300 per μl (mean ± standard deviation [SD], 517 ± 321; median, 350; range, 301 to 1,343 [values are per ml]) and 17 had values below 300/μl (mean ± SD, 198 ± 66; median, 216; range, 91 to 299 [values are per ml]). According to the Centers for Disease Control and Prevention (CDC) classification, three patients were at stage C, one was at stage B, and four were at stage A of infection; the stages for the other patients were unknown.
Of these samples, 28 were taken from patients under short-term antiretroviral treatment (less than 1 year) with a three-drug regimen, 6 were from untreated patients, and 12 were from patients of unknown treatment status. As described in detail in the next paragraphs, sera were tested by (i) two HIV-1/2 antibody/antigen combination assay systems, designed for large-scale screening, (ii) three different confirmatory assays, specific for HIV-1/2, HIV-1, and HIV-2, and (iii) commercial (HIV-1) and in-house (HIV-2) real-time PCR assays for genomic detection of HIV-1 and HIV-2 RNAs.
HIV screening assays.
All assays were performed following the manufacturers' package insert. The Architect HIV Ag/Ab Combo assay (Abbott, Diagnostic Division, Wiesbaden, Germany) is a chemiluminescent magnetic microparticle-based immunoassay used to determine the presence of HIV-1 p24 antigen and antibody to HIV-1 group M, HIV-1 group O, and HIV-2 by an automated, random access instrument. Genscreen Ultra HIV Ag-Ab (Bio-Rad Laboratories, Hercules, CA) is a microplate double-sandwich enzyme-linked immunosorbent assay used to determine the presence of HIV-1 p24 antigen and antibody to HIV-1 group M, HIV-1 group O, and HIV-2. Specimens with signal-to-cutoff (S/CO) values greater than or equal to 1.00 are considered reactive.
HIV antibody confirmatory assays.
HIV-positive samples were tested by Inno-LIA HIV I/II (Innogenetics N.V., Ghent, Belgium), New Lav Blot II (Bio-Rad Laboratories, Hercules, CA), and New Lav Blot I (Bio-Rad Laboratories, Hercules, CA) assays following the manufacturers' package inserts. The criteria for interpreting HIV-2 results were defined in accordance with the package inserts together with the World Health Organization (WHO) and Centers for Disease Control and Prevention (CDC) recommendations.
Nucleic acid amplification.
For HIV-1 RNA purification, the High Pure viral nucleic acid isolation kit was used with 500 μl of plasma, according to the manufacturer's instructions (Roche Diagnostics, Mannheim, Germany).
The amplification and detection were performed in an automated Cobas TaqMan 48 analyzer by using the hydrolysis probe technology with the Cobas TaqMan HIV-1 test. This reverse transcription (RT)-PCR assay targets a conserved region in HIV-1 gag and uses armored RNA as an internal standard for quantification to normalize for sample preparation and possible PCR inhibition. The viral loads of the specimens were calculated automatically with the AmpliLink software (Roche Diagnostics, Mannheim, Germany) by comparing the target signal with the quantification standard signal. The results were automatically reported in copies/ml.
HIV-2 RNA was extracted from 200 μl of plasma using silica columns provided with the QIAamp MinElute Virus Spin kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The assay, with a sensitivity of 100 IU/ml, was optimized with reference strains and validated further in an interlaboratory study organized by the National Institute for Biological Standards and Control (NIBSC) to evaluate two candidate international standards for an HIV-2 RNA nucleic acid amplification test (NAAT): strains CAM-2 and ROD. NIBSC is the United Kingdom's Official Medicines Control Laboratory (OMCL) and is the leading WHO International Laboratory for Standards (http://www.nibsc.ac.uk/default.aspx).
The proposed unit of 1,000 IU/ml for HIV-2 CAM-2 candidate material has been accepted by the WHO, which recommended it as the 1st International Standard for HIV-2 RNA (www.who.int/entity/bloodproducts/catalogue/SafeJan10.pdf). Three calibration samples were prepared using the HIV-2 CAM-2 standard at final concentrations of 1,000, 300, and 100 IU/ml. Every sample was spiked with 4 μl of RNA internal extraction control (PrimerDesign Ltd, United Kingdom) added to AVL buffer at the extraction step. Seven μl out of 60 μl of purified nucleic acid was amplified on a Light-Cycler V2.0 using Quantitect RT-PCR (Qiagen, Hilden, Germany) and specific HIV-2 primers and the TaqMan probe previously described (12), which recognize the long terminal repeat (LTR) region with low variability.
Concerning the crossing point values observed for the three diluted calibration standard samples, it was possible to make rapid estimates of the viral load, expressed as follows (semiquantitative): nonreactive if below the detection limit (<100 IU/ml), reactive (+) if between 100 and 300 IU/ml, reactive (++) if between 300 and 1,000 IU/ml, and reactive (+++) if over 1,000 IU/ml.
RNA extraction and PCR amplification.
Viral RNA was extracted from 140 μl of serum, using the QIAmp viral RNA extraction kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. Serum samples from healthy individuals were used as negative controls. RNA was reverse transcribed using the SuperScript II reverse transcriptase protocol (Invitrogen, Life Technologies, Carlsbad, CA). cDNA was amplified by nested PCR, using the FastStart high-fidelity PCR system (Roche Diagnostics, Mannheim, Germany). Primers for the first round of amplification were PDF1 (8) and LTR9574 (4). The first-round PCR conditions were 93°C for 10 min, followed by 10 cycles of denaturation at 93°C for 10 s, annealing at 55°C for 30 s, and extension at 68°C for 4 min. The subsequent 25 cycles were carried out under the same conditions, with incremental lengthening of the extension time (2-s cycle) and a last extension step at 72°C for 7 min. Five μl of first-round amplified product was used for nested PCR with the following primers (13): ENVA and ENVB. Nested PCR product (nt 7691 to 8425) encompassed the gp36 region of HIV-2 genome. The second-round PCR conditions were 95°C for 1 min, followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 45 s, and a last extension step at 72°C for 7 min. The nested PCR product was analyzed on a 2% agarose gel stained with ethidium bromide.
Sequence analysis.
The PCR product was purified using the High Pure PCR cleanup microkit (Roche Diagnostics, Mannheim, Germany) according to manufacturer's instructions. Sequencing of both strands was performed with primers ENVA and ENVB using the GenomeLab DTCS quick start kit (Beckman Coulter, Inc., Fullerton, CA). Sequencing reactions were run on an automated DNA sequencer (Beckman Coulter, Inc., Fullerton, CA).
Phylogenetic analysis.
The HIV-2 env sequence of patient 6 (HIV-2_ISS-6) was aligned and compared with reference sequences of the HIV-2 env gene (http://blast.ncbi.nlm.nih.gov/Blast.cgi) using CLUSTAL X (24); then the sequences were manually edited with the Bioedit program (14) and gaps were removed from the final alignment. Accession numbers of reference sequences (with reference sequence names in parentheses) retrieved from GenBank are as follows: AF170030 (A196226FR), AF170029 (A196328FR), AF170039 (A19649MLI), AF170038 (A196326MLI), AF170036 (A196310SN), AF170031 (A196205GH), and AF170034 (A196151MLI) for subtype A1; U05354 (A2CAM2GNB), AF170042 (A296308CV), U05358 (A2CAM6GNB), AF170041 (A296150CV), AF170044 (A296327CV), A03789 (A2RODSN), AF170043 (A296199CV), AF170050 (A296325FR), AF170045 (A296329GNB), AF170047 (A296330MAR), AJ238995 (A2CBL24GMB), AF170046 (A296324GNB), AF170049 (A296203CV), and AF170048 (A296206SE) for subtype A2; AF170052 (B97227COG), AF170054 (B97245IVC), AF170061 (B96306SN), AF170053 (B96339IVC), AF170059 (B96307MLI), U27200 (BEHOIVC), and A09998 (BD205GH) for subtype B; AF103818 (US_Marilyn) and AF115393 (CAM3) for simian immunodeficiency virus (SIV).
The phylogenetic tree was constructed using the PAUP package (23). We employed the general reversible model (TVM) of nucleotide substitution, incorporating maximum likelihood (ML) estimates of base composition and the shape parameter (α) of a gamma distribution (Γ) model of among-site rate variation (with eight rate categories), as it consistently gave much higher likelihood values using Modeltest v.3.7 implemented in PAUP (23). The maximum likelihood tree was estimated under this model using tree bisection-reconnection (TBR) branch swapping. The statistical robustness and reliability of the branching order within each phylogenetic tree were confirmed through a bootstrap analysis using 1,000 replicates for the neighbor-joining (NJ) tree and through the zero branch length test for the ML tree (23).
The tree was rooted using two simian immunodeficiency virus strains (AF103818 and AF115393) as an outgroup. The genetic distance between the HIV-2_ISS-6 isolate and the HIV-2 subtype A2 reference sequences was computed by Mega 4 software (15).
Nucleotide sequence accession number.
The nucleotide sequence obtained in this study was submitted to GenBank under accession number HM371426.
RESULTS
Serum samples collected from 37 subjects in Guinea-Bissau (GB) and 9 subjects in Guinea-Conakry (GC) were identified, in local settings, as HIV-2 antibody positive by a Genie I/II rapid assay. All 46 samples were sent to the Italian National Institute of Health for confirmation of HIV-1/2 positivity by serological and genomic assays. Demographic and clinical data were available for 34 individuals (25 from GB and 9 from GC) and are reported in Table 1.
TABLE 1.
Demographic and clinical data for 34 of 46 total patients
| Patient code | Countrya | Age (yr) | Sexc | No. of CD4 cells/μl | CDC stage | Therapy |
|---|---|---|---|---|---|---|
| 1 | GB | 50 | M | 350 | NAb | Yes |
| 2 | GB | 31 | F | 257 | NA | Yes |
| 3 | GB | 32 | F | 220 | NA | Yes |
| 4 | GB | 41 | F | 239 | NA | Yes |
| 5 | GB | 19 | F | 301 | NA | Yes |
| 6 | GB | 62 | M | 299 | NA | Yes |
| 7 | GB | 24 | F | 135 | NA | Yes |
| 10 | GB | 36 | M | 350 | NA | Yes |
| 11 | GB | 30 | F | 434 | NA | Yes |
| 13 | GB | 39 | F | 230 | NA | Yes |
| 15 | GB | 47 | M | 210 | NA | Yes |
| 16 | GB | 29 | F | 220 | NA | Yes |
| 20 | GB | 21 | F | 345 | NA | Yes |
| 21 | GB | 45 | F | 260 | NA | Yes |
| 22 | GB | 48 | M | 360 | NA | Yes |
| 24 | GB | 24 | F | 216 | NA | Yes |
| 28 | GB | 33 | M | 340 | NA | Yes |
| 29 | GB | 43 | F | 198 | NA | Yes |
| 30 | GB | 36 | F | 298 | NA | Yes |
| 31 | GB | 22 | F | 314 | NA | Yes |
| 32 | GB | 35 | M | 312 | NA | Yes |
| 33 | GB | 47 | F | 321 | NA | Yes |
| 34 | GB | 33 | F | 98 | NA | Yes |
| 35 | GB | 25 | F | 330 | NA | Yes |
| 36 | GB | 26 | F | 110 | NA | Yes |
| 52 | GC | 25 | F | 143 | 3 | Yes |
| 53 | GC | 58 | M | 91 | 3 | Yes |
| 54 | GC | 14 | F | 1343 | 1 | No |
| 65 | GC | 51 | F | 837 | 1 | No |
| 68 | GC | 45 | M | 461 | NA | Yes |
| 69 | GC | 46 | F | 408 | 2 | No |
| 71 | GC | 27 | M | 1136 | 1 | No |
| 74 | GC | 30 | F | 856 | 1 | No |
| 76 | GC | 35 | F | 143 | 3 | No |
GB, Guinea-Bissau; GC, Guinea-Conakry.
NA, not available.
M, male; F, female.
Screening and confirmation of HIV positivity.
The 46 samples were first tested with two antibody/antigen combination assays designed for large-scale screening of HIV-1/2: the fully automated Architect HIV Ag/Ab Combo system and the Genscreen Ultra HIV Ag/Ab assay. As shown in Table 2, HIV positivity was detected in 44 out of 46 patients by both screening assays. Analysis of the 46 samples by Inno-LIA HIV I/II confirmatory assay detected HIV positivity (HIV-1, HIV-2, or double infection) in the same 44 samples, showing a complete concordance (100%) with the screening assays.
TABLE 2.
Serological and genomic screening of 46 patients
| Patient code | Result of screening assayd |
Result of confirmatory assay |
Result of real-time PCR |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GS Ultra HIV Ag/Ab | HIV Ag/Ab Combo | INNO-LIA HIV 1/2a |
ImmunoBlotc |
HIV-1 RNA (copies/ml) | HIV-2 RNAb | |||||||
| HIV-1 |
HIV-2 |
HIV type | New LAV Blot I (HIV-1) | New LAV Blot II (HIV-2) | ||||||||
| p24 | gp41 | gp120 | gp36 | gp105 | ||||||||
| 6 | 7.049 | 159.66 | − | − | − | 3+ | 3+ | HIV-2 | HIV-1 | HIV-2 | − | ++ |
| 20 | 6.938 | 95.06 | 1+ | − | − | 3+ | 3+ | HIV-2 | HIV-1 | HIV-2 | − | + |
| 68 | 7.253 | 199.24 | − | − | − | 3+ | 3+ | HIV-2 | HIV-1 | HIV-2 | − | ++ |
| 65 | 7.092 | 144.35 | 3+ | − | − | 3+ | 3+ | HIV-2 | HIV-1 | HIV-2 | − | + |
| 2 | 7.846 | 153.93 | 2+ | +/− | − | 3+ | 3+ | HIV-2 | Ind. | HIV-2 | − | ++ |
| 47 | 2.313 | 57.87 | − | − | − | 3+ | 3+ | HIV-2 | Ind. | HIV-2 | − | + |
| 50 | 5.747 | 152.84 | − | − | − | 3+ | 3+ | HIV-2 | Ind. | HIV-2 | − | + |
| 71 | 7.542 | 182.73 | 1+ | − | − | 3+ | 3+ | HIV-2 | Ind. | HIV-2 | − | ++ |
| 69 | 7.863 | 115.15 | − | − | − | 3+ | 3+ | HIV-2 | Ind. | Ind. | − | + |
| 46 | 6.102 | 83.14 | 3+ | − | − | 3+ | 3+ | HIV-2 | HIV-1 | HIV-2 | − | − |
| 76 | 7.606 | 143.37 | 2+ | − | − | 3+ | 1+ | HIV-2 | Ind. | HIV-2 | − | − |
| 4 | 8.086 | 441.58 | 1+ | 2+ | 2+ | − | − | HIV-1 | NT | Ind. | 1,576,835 | − |
| 13 | 7.507 | 390.93 | 2+ | 2+ | 2+ | − | − | HIV-1 | NT | Ind. | 483,476 | − |
| 24 | 7.221 | 468.79 | 3+ | 3+ | 3+ | 3+ | 3+ | HIV-1/HIV-2 | HIV-1 | HIV-2 | 1,466,148 | + |
| 29 | 7.849 | 134.44 | 3+ | 3+ | 2+ | 3+ | 3+ | HIV-1/HIV-2 | HIV-1 | HIV-2 | 921 | + |
| 32 | 7.226 | 296.14 | 3+ | 3+ | 3+ | 2+ | 3+ | HIV-1/HIV-2 | HIV-1 | HIV-2 | 958,059 | + |
| 35 | 8.086 | 102.36 | 3+ | 3+ | 3+ | 3+ | 3+ | HIV-1/HIV-2 | HIV-1 | HIV-2 | 3,248 | + |
| 36 | 7.598 | 120.28 | 3+ | 2+ | 2+ | 2+ | 2+ | HIV-1/HIV-2 | HIV-1 | HIV-2 | 263,021 | + |
| 44 | 8.086 | 443.81 | 3+ | 3+ | 2+ | 2+ | 3+ | HIV-1/HIV-2 | HIV-1 | HIV-2 | 420 | + |
| 40 | 7.022 | 203.57 | 3+ | 3+ | − | 3+ | 3+ | HIV-1/HIV-2 | HIV-1 | HIV-2 | 340 | + |
| 42 | 7.501 | 441.88 | 3+ | 2+ | 1+ | 3+ | 3+ | HIV-1/HIV-2 | HIV-1 | HIV-2 | 157 | + |
| 53 | 7.528 | 147.09 | +/− | 2+ | 1+ | 3+ | 2+ | HIV-1/HIV-2 | HIV-1 | HIV-2 | 67 | ++ |
| 31 | 7.065 | 203.83 | 3+ | 3+ | − | 2+ | 3+ | HIV-1/HIV-2 | HIV-1 | HIV-2 | <47 | ++ |
| 54 | 7.313 | 153.97 | +/− | 2+ | +/− | 2+ | 2+ | HIV-1/HIV-2 | Ind. | HIV-2 | − | + |
| 38 | 7.089 | 484.83 | 3+ | 3+ | 2+ | 3+ | 2+ | HIV-1/HIV-2 | HIV-1 | HIV-2 | − | + |
| 39 | 7.067 | 256.36 | 3+ | 3+ | 1+ | 3+ | 3+ | HIV-1/HIV-2 | HIV-1 | HIV-2 | − | + |
| 41 | 7.531 | 348.78 | 3+ | 2+ | 1+ | 3+ | 3+ | HIV-1/HIV-2 | HIV-1 | HIV-2 | − | + |
| 5 | 7.515 | 478.82 | 3+ | 3+ | 2+ | 3+ | 3+ | HIV-1/HIV-2 | HIV-1 | HIV-2 | − | + |
| 10 | 7.485 | 382.58 | 1+ | 2+ | − | 3+ | 3+ | HIV-1/HIV-2 | HIV-1 | HIV-2 | − | ++ |
| 11 | 7.639 | 268.79 | 3+ | 3+ | 2+ | 3+ | 3+ | HIV-1/HIV-2 | HIV-1 | HIV-2 | − | + |
| 15 | 7.049 | 739.97 | 2+ | 2+ | 2+ | 2+ | 3+ | HIV-1/HIV-2 | HIV-1 | HIV-2 | − | + |
| 3 | 7.655 | 330.26 | 2+ | 2+ | 1+ | 3+ | 3+ | HIV-1/HIV-2 | HIV-1 | HIV-2 | − | +/− |
| 7 | 7.278 | 367.76 | 3+ | 2+ | 2+ | 2+ | 3+ | HIV-1/HIV-2 | HIV-1 | HIV-2 | − | +/− |
| 52 | 7.588 | 430.28 | 3+ | 2+ | 3+ | 2+ | − | HIV-1/HIV-2 | HIV-1 | Ind. | 500,000 | − |
| 43 | 7.658 | 724.69 | 3+ | 3+ | 3+ | 3+ | 2+ | HIV-1/HIV-2 | HIV-1 | HIV-2 | 5,300 | − |
| 21 | 7.496 | 358.88 | 2+ | 3+ | 3+ | 3+ | 3+ | HIV-1/HIV-2 | HIV-1 | HIV-2 | 142 | − |
| 45 | 7.852 | 184.42 | 3+ | 3+ | 2+ | 2+ | 2+ | HIV-1/HIV-2 | HIV-1 | HIV-2 | 830 | − |
| 28 | 7.588 | 678.89 | 3+ | 3+ | 2+ | 1+ | 1+ | HIV-1/HIV-2 | HIV-1 | HIV-2 | 226 | − |
| 33 | 7.528 | 512.33 | 3+ | 3+ | 3+ | 3+ | 3+ | HIV-1/HIV-2 | HIV-1 | HIV-2 | 6,172 | − |
| 1 | 7.512 | 551.83 | 2+ | 3+ | 3+ | 3+ | 3+ | HIV-1/HIV-2 | HIV-1 | HIV-2 | 9,308 | − |
| 30 | 7.081 | 785.93 | 2+ | 3+ | 3+ | 3+ | 3+ | HIV-1/HIV-2 | HIV-1 | HIV-2 | <47 | − |
| 34 | 7.512 | 629.18 | 3+ | 3+ | 3+ | 2+ | +/− | HIV-1/HIV-2 | HIV-1 | HIV-2 | <47 | − |
| 16 | 7.005 | 461.62 | 3+ | 3+ | 2+ | 2+ | 3+ | HIV-1/HIV-2 | HIV-1 | HIV-2 | − | − |
| 74 | 8.086 | 394.25 | 3+ | 3+ | 3+ | 2+ | +/− | HIV-1/HIV-2 | HIV-1 | HIV-2 | − | − |
| 22 | 0.278 | 0.12 | − | − | − | − | − | Neg. | NT | Neg. | − | − |
| 37 | 0.318 | 0.11 | − | − | − | − | − | Neg. | NT | Neg. | − | − |
Only reactivity to antigens of interest is reported.
+, 100 to 300 IU/ml; ++, 300 to 1,000 IU/ml; −, not reactive (<100 IU/ml).
NT, not tested; Neg., negative; Ind., indeterminate.
GS Ultra HIV Ag/Ab, Genscreen Ultra HIV Ag/Ab assay; HIV Ag/Ab Combo, Architect HIV Ag/Ab Combo.
The two negative samples were also not reactive by the New LAV Blot II confirmatory assay, and HIV-1/HIV-2 RNAs were undetectable by genomic assays in both samples (Table 2, patients 22 and 37).
Discrimination between HIV-1 and HIV-2 positivity by serological and genomic assays.
To assess HIV-2 positivity and discriminate between HIV-1 and HIV-2 infections, results from Inno-LIA HIV I/II, able to differentiate simultaneously between HIV-1 and HIV-2, were compared with results from the New LAV Blot II Western blot (WB) assay, specifically designed for the identification of antibodies to HIV-2. Selected samples with no clear-cut results were also tested by New LAV Blot I, specific for detection of HIV-1 antibodies. In addition, all samples were tested by commercial (HIV-1) and in-house (HIV-2) real-time PCR assays for genomic detection of HIV-1 and HIV-2 RNAs.
Among the 44 HIV-positive patients, the Inno-LIA assay identified 11 HIV-2-positive samples, 2 HIV-1-positive samples, and 31 samples that were HIV-1/HIV-2 dually reactive (“untypable” according to the manufacturers' instructions).
Overall, HIV-2 antibodies were detected by Inno-LIA in 42 out of 46 subjects, with a concordance rate with the Genie I/II assay of 91%. When further analyzed by HIV-2 WB (New LAV Blot II), 40 of 46 samples showed a banding pattern suggestive of HIV-2 infections, with a concordance rate of 87% with the Genie I/II assay. Forty samples were HIV-2 positive by both Inno-LIA and New LAV Blot II, while two samples showed discordant results (Table 2, samples 52 and 69). In particular, sample 52 was dually reactive by Inno-LIA but HIV-2 indeterminate by New LAV Blot II, while sample 69 was identified as HIV-2 positive by Inno-LIA but indeterminate by New LAV Blot II.
Analysis of sera by in-house real-time PCR showed detection of HIV-2 RNA in 27 out of 42 samples (64%) with positivity to anti-HIV-2 antibodies by Inno-LIA assay. In two samples, HIV-2 RNA levels were very close to the detection limit of the assay (Table 2, samples 3 and 7).
High concordance between results of Inno-LIA and HIV-1/HIV-2 genomic assays defined a clear pattern for 21 samples: 9 HIV-2 positive, 2 HIV-1 positive, and 10 HIV-1/HIV-2 dually reactive (Table 2, rows 1 to 9, rows 12 to 13, and rows 14 to 23, respectively), allowing, at least in these cases, reasonable identification of single and double infections.
To better confirm HIV-1 and HIV-2 infections, 31 HIV-1/HIV-2 double-positive samples by the Inno-LIA assay were also tested by New LAV Blot I, specific for detection of HIV-1 antibodies. Overall, HIV-1 positivity was detected in 30 of 31 subjects, with a concordance rate of 97%. Only sample 54 showed discordant results. In this case, a single reactivity to HIV-1 gp41 antigen in the Inno-LIA assay and a negative result in the HIV-1 NAAT were observed (Table 2, sample 54), leaving uncertainty about the diagnosis of HIV-1 infection. Overall, the data showed that the use of a diagnostic procedure based on Inno-LIA assay followed by HIV-1/HIV-2 genomic detection allows accurate discrimination between HIV-1 and HIV-2 in 84% of single infections (11 out of 13 samples) but proved to be crucial to confirm only 32% of double infections (10 out of 31 samples). In the latter patients, confirmation of the results with WB may be useful to define the diagnosis of double infections.
Performance of New LAV Blot I on samples with HIV-2 single infection.
HIV diagnosis in areas in which the virus is not endemic might be carried out by HIV-1 confirmatory assay only. Thus, the performance of the New LAV Blot I assay was evaluated on the nine identified HIV-2-positive samples as well as on two additional samples that, although HIV-2 RNA negative, showed HIV-2 positivity by both Inno-LIA and New LAV Blot II (Table 2, samples 46 and 76).
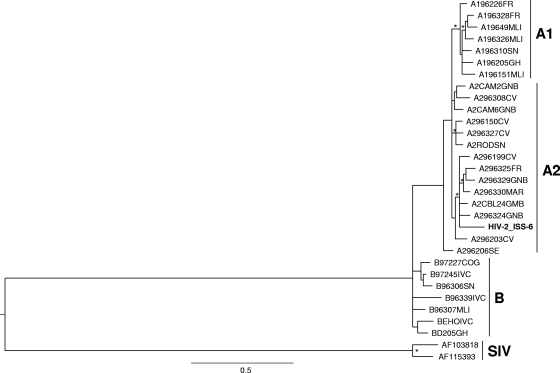
The New LAV Blot I assay identified a general cross-reactivity of these samples with the internal gag and pol proteins of HIV-1. However, five samples also showed cross-reactivity with the gp160 (Table 3, samples 6, 20, and 68) or gp41 (Table 3, samples 65 and 46) envelope protein of HIV-1 and were misdiagnosed as HIV-1-infected samples by this assay. Cross-reactivity with the immunodominant regions of HIV-1 envelope proteins was previously reported for HIV-2 subtype B. Amplification of the gp36 region (nt 7691 to 8425) for genotyping had to be carried out directly from viral RNA and was successfully achieved only from sample 6. Phylogenetic analysis of the obtained sequence (HIV-2_ISS-6) confirmed infection with HIV-2 subtype A2; the sequence clustered with the A296324GNB and A296329GNB sequences from Guinea- Bissau (Fig. 1). The mean genetic distance of the strains which clustered with the HIV-2_ISS-6 sequence confirmed the close relationship of our isolate with the strains from Guinea-Bissau and neighboring countries like Gambia (Table 4).
TABLE 3.
Reactivity of HIV-2-positive sera with HIV-1 antigens in Western blot
| Patient code | Reactivity |
Assay resulta | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
gag |
pol |
env |
|||||||||
| p17/18 | p24/25 | p40 | p55 | p34/31 | p52/51 | p68/66 | gp41 | gp120 | gp160 | ||
| 6 | − | 1+ | − | +/− | 3+ | − | +/− | − | − | 1+ | HIV-1 |
| 20 | − | 3+ | − | 2+ | +/− | +/− | 1+ | +/− | − | 2+ | HIV-1 |
| 68 | − | 2+ | − | 2+ | 1+ | − | 3+ | − | − | 2+ | HIV-1 |
| 65 | 3+ | 3+ | 3+ | 3+ | 3+ | − | 1+ | 2+ | − | − | HIV-1 |
| 2 | − | 3+ | − | 3+ | 3+ | 1+ | 2+ | − | − | +/− | Ind. |
| 47 | − | − | − | 2+ | − | − | − | − | − | − | Ind. |
| 50 | − | − | − | 2+ | − | − | − | − | − | − | Ind. |
| 71 | − | 2+ | − | 1+ | 1+ | − | − | − | − | − | Ind. |
| 69 | − | − | − | + | − | − | − | − | − | − | Ind. |
| 46 | − | 3+ | 2+ | 3+ | 2+ | +/− | +/− | 1+ | − | − | HIV-1 |
| 76 | 1+ | 3+ | − | 2+ | 1+ | − | 1+ | − | − | +/− | Ind. |
Ind., indeterminate.
FIG. 1.
Phylogenetic relationships of the HIV-2_ISS-6 isolate (in bold) to representatives of the HIV-2 subtypes A1, A2, B, and SIV (indicated on the right) retrieved from the HIV sequence database (see Materials and Methods). The maximum likelihood (ML) phylogenetic tree was generated from env nucleotide sequences with the Hasegawua, Kishino, and Yano (HKY-85) nucleotide substitution model under gamma distribution (branch lengths were estimated with the best fitting nucleotide substitution model according to a hierarchical likelihood ratio test). The symbol * along a branch represents significant statistical support for the clade subtending that branch (P = 0.001 in the zero-branch-length test; bootstrap support, 75%). The scale bar indicates 0.5% nucleotide sequence divergence. The letters to the right of the sequence name are the country codes for the origins of the patients: FR, France; MLI, Mali; SN, Senegal; GH, Ghana; GNB, Guinea-Bissau; CV, Cape Verde; MAR, Morocco; GMB, Gambia; SE, Sweden; COG, Congo; IVC, Ivory Coast.
TABLE 4.
Average genetic distance between the HIV-2_ISS-6 sequence and HIV-2 reference sequencesa
| Gp1 | Gp2 | Gp3 | Gp4 | |
|---|---|---|---|---|
| Gp1 | 0.135 | 0.121 | 0.143 | |
| Gp2 | 0.079 | 0.073 | ||
| Gp3 | 0.086 | |||
| Gp4 |
Sequences in each group: Gp1, HIV-2_ISS-6; Gp2, A296330MAR; Gp3, A296324GNB, A2CBL24GMB; Gp4, A296329GNB, A296325FR, A296199CV. Distances are given in genetic distance units (GDU).
DISCUSSION
In the present study, 46 samples from areas in which HIV-1/HIV-2 is endemic, identified in local settings as HIV-2 antibody positive by Genie I/II rapid assay, were analyzed by HIV-1/HIV-2 serological assays (screening and confirmation) and NAAT.
Overall, complete concordance in detecting generic HIV positivity between our screening and confirmatory assays was observed in 44 of the 46 study participants. However, all of these 46 samples were defined as HIV positive by Genie I/II rapid assay, strongly suggesting that nonspecific detection of HIV positivity by Genie I/II assay likely occurred in two of them.
For at least one patient (Table 1 and Table 2, sample 22), who was on anti-retroviral therapy because of a reduced number of CD4 cells, we could not exclude HIV infection with undetectable serum markers or, alternatively, improper serum storage.
Among all patients, only two (patients 52 and 69) showed discordant results using HIV-2 confirmatory tests.
In sample 52, Inno-LIA detected a clear reactivity to both HIV-1 envelope proteins but also to HIV-2 gp36 (Table 2). In addition, NAAT and HIV-2 WB assay could not exclude or confirm HIV-2 infection. The HIV-2 WB assay showed an indeterminate result due to reactivity to only one pol protein (p68) (data not shown). Thus, we could not definitively assess whether patient 52 had single HIV-1 infection (with serum showing cross-reaction on HIV-2 gp36 antigen) or double HIV-1/HIV-2 infection.
Sample 69 was clearly identified as HIV-2 positive by Inno-LIA, with a strong reactivity to gp36 and gp105 antigens, and the result was confirmed by genomic detection of HIV-2, but it was identified as indeterminate by HIV-2 WB. The indeterminate result was due to an uncommon serological pattern characterized by reactivity to only one envelope protein of HIV-2 (gp105/140) but no reactivity to the internal proteins (data not shown). Clearly, in this sample there was an apparent discrepancy between the lack of antibodies to gp36 in HIV-2 WB with respect to the Inno-LIA assay. Overall, the serological pattern of patient 69 suggests that synthetic peptide-based assays may be more sensitive and specific for a correct diagnosis of HIV-2 infection in uncommon samples.
For those samples with double reactivity in the Inno-LIA test, but in which only one or neither genome was detected (21 out of 31), identification of true double infections in this group may be hampered by cross-reactivity between HIV-1 and HIV-2 (1, 21). Consequently, a more complex diagnostic strategy which includes other discriminatory assays or new specific discriminatory assays should be carried out to ensure appropriate diagnosis of these cases. Among the double infections detected by Inno-LIA, sample number 54 showed strong reactivity to both HIV-2 envelope proteins but to only one of the two HIV-1 envelope antigens (Table 2, sample 54). Results from genomic and WB assays indicated a clear HIV-2 but not HIV-1 infection (the negative HIV-1 NAAT result was not due to anti-retroviral therapy; see Table 1). These findings suggest that this patient was infected by HIV-2 and showed antibodies cross-reacting on HIV-1 gp41 antigen.
Cross-reactivity with the immunodominant regions of HIV-1 envelope proteins was previously reported for HIV-2 subtype B, which is common in Mali and Ivory Coast (9). This may have implication for the correct diagnosis of HIV-2 in areas in which the virus is endemic and where adequate confirmation tools may not be used (3, 5). Our results indicate that five patients with single HIV-2 infection would have been misdiagnosed as HIV-1 positive if the HIV-1 WB confirmatory assay had been improperly used. Interestingly, sequence analysis showed that one of these patients was infected with HIV-2 subtype A2, suggesting the need for studies to clarify the impact of HIV-2 genotype variability on accuracy of serological diagnosis.
The issue of “migration and health” is a main health theme in developed countries. Improved surveillance in migrant populations is recommended, particularly to control the HIV epidemic and ensure appropriate therapy. The study of HIV-2 circulation in areas in which the virus is not endemic should be a small but significant part of surveillance programs, at least for countries, like Italy, with increasing numbers of migrants from Africa. To date, a single study, performed in Northern Italy, reported a prevalence of 10.2% for HIV-2 infection in a small population of HIV-infected patients from West Africa, suggesting the need for more extensive studies (7). On this basis, a concerted effort to improve the accuracy of HIV-2 diagnosis as well as standardization of an international HIV-2 quantitative PCR assay is needed.
Acknowledgments
We are grateful to Sabrina Tocchio for excellent editorial and secretarial assistance.
Footnotes
Published ahead of print on 23 June 2010.
REFERENCES
- 1.Andersson, S., Z. da Silva, H. Norrgren, F. Dias, and G. Biberfeld. 1997. Field evaluation of alternative testing strategies for diagnosis and differentiation of HIV-1 and HIV-2 infections in an HIV-1 and HIV-2-prevalent area. AIDS 11:1815-1822. [DOI] [PubMed] [Google Scholar]
- 2.Barin, F., S. M'Boup, F. Denis, P. Kanki, J. S. Allan, T. H. Lee, and M. Essex. 1985. Serological evidence for virus related to simian T-lymphotropic retrovirus III in residents of west Africa. Lancet 2:1387-1389. [DOI] [PubMed] [Google Scholar]
- 3.Barin, F., F. Cazein, F. Lot, J. Pillonel, S. Brunet, D. Thierry, F. Damond, F. Brun-Vézinet, J. C. Desenclos, and C. Semaille. 2007. Prevalence of HIV-2 and HIV-1 group O infections among new HIV diagnoses in France: 2003-2006. AIDS 21:2351-2353. [DOI] [PubMed] [Google Scholar]
- 4.Berry, N., K. Ariyoshi, O. Jobe, P. T. Ngum, T. Corrah, A. Wilkins, H. Whittle, and R. Tedder. 1994. HIV type 2 proviral load measured by quantitative polymerase chain reaction correlates with CD4+ lymphopenia in HIV type 2-infected individuals. AIDS Res. Hum. Retroviruses 10:1031-1037. [DOI] [PubMed] [Google Scholar]
- 5.Candido, A., P. Chionne, E. Madonna, C. Rovetto, E. Salvi, A. Caratelli, P. Piccinini, P. Verani, and M. Rapicetta. 2006. National program of external quality evaluation in diagnostic laboratories for HBsAg, anti-HCV and anti-HIV screening test. Activity and results 1994-2003. Rapporti ISTISAN, 06/3. Istituto Superiore di Sanità, Rome, Italy. (In Italian.)
- 6.Clavel, F., D. Guétard, F. Brun-Vézinet, S. Chamaret, M. A. Rey, M. O. Santos-Ferreira, A. G. Laurent, C. Dauguet, C. Katlama, C. Rouzioux, et al. 1986. Isolation of a new human retrovirus from West African patients with AIDS. Science 233:343-346. [DOI] [PubMed] [Google Scholar]
- 7.Costarelli, S., C. Torti, A. Rodella, F. Baldanti, S. Paolucci, G. Lapadula, N. Manca, E. Quiros-Roldan, I. Izzo, and G. Carosi. 2008. Screening and management of HIV-2-infected individuals in Northern Italy. AIDS Patient Care STDS 22:489-494. [DOI] [PubMed] [Google Scholar]
- 8.Damond, F., I. Loussert-Ajaka, C. Apetrei, D. Descamps, S. Souquière, A. Leprêtre, S. Matheron, F. Brun-Vézinet, and F. Simon. 1998. Highly sensitive method for amplification of human immunodeficiency virus type 2 DNA. J. Clin. Microbiol. 36:809-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Damond, F., C. Apetrei, D. L. Robertson, S. Souquière, A. Leprêtre, S. Matheron, J. C. Plantier, F. Brun-Vézinet, and F. Simon. 2001. Variability of human immunodeficiency virus type 2 (HIV-2) infecting patients living in France. Virology 280:19-30. [DOI] [PubMed] [Google Scholar]
- 10.Damond, F., F. Brun-Vezinet, S. Matheron, G. Peytavin, P. Campa, S. Pueyo, F. Mammano, S. Lastere, I. Farfara, F. Simon, G. Chene, and D. Descamps. 2005. Polymorphism of the human immunodeficiency virus type 2 (HIV-2) protease gene and selection of drug resistance mutations in HIV-2-infected patients treated with protease inhibitors. J. Clin. Microbiol. 43:484-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dougan, S., B. Patel, J. H. Tosswill, and K. Sinka. 2005. Diagnoses of HIV-1 and HIV-2 in England, Wales, and Northern Ireland associated with west Africa. Sex. Transm. Infect. 81:338-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferns, R. B., and J. A. Garson. 2006. Development and evaluation of a real-time RT-PCR assay for quantification of cell-free human immunodeficiency virus type 2 using a Brome Mosaic Virus internal control. J. Virol. Methods 135:102-108. [DOI] [PubMed] [Google Scholar]
- 13.Gao, F., L. Yue, D. L. Robertson, S. C. Hill, H. Hui, R. J. Biggar, A. E. Neequaye, T. M. Whelan, D. D. Ho, and G. M. Shaw. 1994. Genetic diversity of human immunodeficiency virus type 2: evidence for distinct sequence subtypes with differences in virus biology. J. Virol. 68:7433-7447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall, T. A. 1999. Bioedit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98 NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 15.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: Molecular Evolutionary Genetics Analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 16.Ly, T. D., L. Martin, D. Daghfal, A. Sandridge, D. West, R. Bristow, L. Chalouas, X. Qiu, S. C. Lou, J. C. Hunt, G. Schochetman, and S. G. Devare. 2001. Seven human immunodeficiency virus (HIV) antigen-antibody combination assays: evaluation of HIV seroconversion sensitivity and subtype detection. J. Clin. Microbiol. 39:3122-3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malm, K., M. von Sydow, and S. Andersson. 2009. Performance of three automated fourth-generation combined HIV antigen/antibody assays in large-scale screening of blood donors and clinical samples. Transfus. Med. 19:78-88. [DOI] [PubMed] [Google Scholar]
- 18.Matheron, S., G. Mendoza-Sassi, F. Simon, R. Olivares, J. P. Coulaud, and F. Brun-Vezinet. 1997. HIV-1 and HIV-2 AIDS in African patients living in Paris. AIDS 11:934-936. [PubMed] [Google Scholar]
- 19.Perry, K. R., S. Ramskill, R. P. Eglin, J. A. Barbara, and J. V. Parry. 2008. Improvement in the performance of HIV screening kits. Transfus. Med. 18:228-240. [DOI] [PubMed] [Google Scholar]
- 20.Reeves, J. D., and R. W. Doms. 2002. Human immunodeficiency virus type 2. J. Gen. Virol. 83:1253-1265. [DOI] [PubMed] [Google Scholar]
- 21.Rouet, F., K. D. Ekouevi, A. Inwoley, M. L. Chaix, M. Burgard, L. Bequet, I. Viho, V. Leroy, F. Simon, F. Dabis, and C. Rouzioux. 2004. Field evaluation of a rapid human immunodeficiency virus (HIV) serial serologic testing algorithm for diagnosis and differentiation of HIV type 1 (HIV-1), HIV-2, and dual HIV-1-HIV-2 infections in West African pregnant women. J. Clin. Microbiol. 42:4147-4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soriano, V., P. Gomes, W. Heneine, A. Holguin, M. Doruana, R. Antunes, et al. 2000. Human immunodeficiency virus type 2 (HIV-2) in Portugal: clinical spectrum, circulating subtypes, virus isolation, and plasma viral load. J. Med. Virol. 61:111-116. [PubMed] [Google Scholar]
- 23.Swofford, D. L. 1999. PAUP: Phylogenetic Analysis Using Parsimony (and Other Methods), version 4.0 b2a. Sinauer Associates, Inc., Sunderland, MA.
- 24.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. Clustal W: improving sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valadas, E., L. França, S. Sousa, and F. Antunes. 2009. 20 years of HIV-2 infection in Portugal: trends and changes in epidemiology. Clin. Infect. Dis. 48:1166-1167. [DOI] [PubMed] [Google Scholar]
- 26.Weber, B., L. Gürtler, R. Thorstensson, U. Michl, A. Mühlbacher, P. Bürgisser, R. Villaescusa, A. Eiras, C. Gabriel, H. Stekel, S. Tanprasert, S. Oota, M. J. Silvestre, C. Marques, M. Ladeira, H. Rabenau, A. Berger, U. Schmitt, and W. Melchior. 2002. Multicenter evaluation of a new automated fourth-generation human immunodeficiency virus screening assay with a sensitive antigen detection module and high specificity. J. Clin. Microbiol. 40:1938-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]