Abstract
The presence of Campylobacter concisus in the saliva of healthy individuals and patients with inflammatory bowel disease (IBD) was examined. C. concisus was detected in 97% of the healthy individuals and 100% of the patients with IBD tested. The C. concisus culture positivity rate in younger children was significantly lower than that in the other age groups.
Campylobacter concisus is a Gram-negative, spiral/curved, and flagellated bacterium (12) that has been associated with gingival inflammation (7). Associations between C. concisus and diarrheal disease, as well as inflammatory bowel disease (IBD), also have been reported (1, 3, 10, 11, 14).
It is not clear whether the C. concisus strains detected in stool and intestinal biopsy specimens are the same as or different from oral C. concisus strains. In addition, the prevalence of C. concisus in the oral cavities of healthy individuals and patients with IBD has not been investigated.
In this study, we examined the prevalence of C. concisus in the saliva of healthy individuals and patients with IBD by using bacterial culture and PCR. In addition, the protein profiles of C. concisus strains isolated from saliva and intestinal biopsy specimens from a child with Crohn's disease (CD, one type of IBD) were compared.
Saliva samples were collected from 59 healthy individuals (33 male) 3 to 60 (mean ± standard deviation, 19.5 ± 15.4) years old and 18 patients (11 male) 8 to 55 (18.2 ± 10.3) years old with IBD, including 13 with CD and 5 with ulcerative colitis (UC). Both the patients and the controls reside in the Sydney area. The controls had no gum disease or gastrointestinal symptoms. None of the participants were receiving antibiotics at the time of sample collection.
Bacterial culture was performed within 3 h of saliva collection. Briefly, 6 μl of saliva was streaked onto agar plates prepared using blood agar base no. 2 supplemented with 6% defibrinated horse blood and vancomycin (10 μg/ml; Oxoid Limited, Hampshire, United Kingdom). Plates were incubated at 37°C under microaerophilic conditions generated by a Campylobacter gas system (Oxoid). Following 3 days of incubation, the mixed bacterial culture was filtered through a 0.65-μm nitrocellulose filter onto a fresh agar plate (6) and cultured for a further 2 days. Candidate bacterial colonies were subjected to 16S rRNA gene PCR using primers F27 and R1492 (5). All PCR products were sequenced.
All saliva samples (1 μl) were also subjected to a nested C. concisus PCR using primers F27 and R1492 for the initial amplification, followed by a C. concisus-specific PCR using primers Concisus F and Concisus R (8).
To compare the relative sensitivities of culture and C. concisus PCR for the detection of C. concisus, serial dilutions (1:10 to 1:1014) of C. concisus were prepared and subjected to both detection methods.
The protein profiles of six oral C. concisus strains and one biopsy C. concisus strain isolated from a child with CD were compared by using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (4).
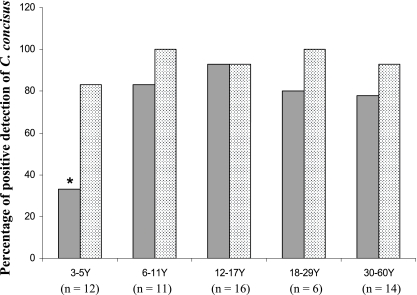
C. concisus was isolated from 75% (44/59) of the healthy controls. Four (33%) of the 12 children 3 to 5 years old were C. concisus culture positive, which was a significantly lower percentage than in the other age groups (Fisher's exact test, P < 0.01 compared to the group of patients 12 to 18 years old and P < 0.05 compared to the remaining groups) (Fig. 1). There was no gender difference in percent C. concisus culture positivity (females, 77% [20/26]; males, 73% [24/33]).
FIG. 1.
Detection of C. concisus by culture and PCR (dotted column) in saliva samples from healthy individuals in different age groups. The C. concisus culture positivity rate of children 3 to 5 years old was significantly lower than that of the other age groups (P < 0.01 compared to the group of patients 12 to 18 years old and P < 0.05 compared to the remaining groups).
C. concisus PCR was positive in 97% (57/59) of the healthy individuals, with no age-related differences (Fig. 1).
Comparison of the sensitivities in detecting C. concisus showed that the nested C. concisus PCR was 1,000 times more sensitive than the culture method.
C. concisus was isolated from 85% (11/13) of the patients with CD and 100% (5/5) of the patients with UC, percentages which are not statistically significantly different from that in age-matched controls. All 18 patients with IBD were positive for C. concisus by PCR.
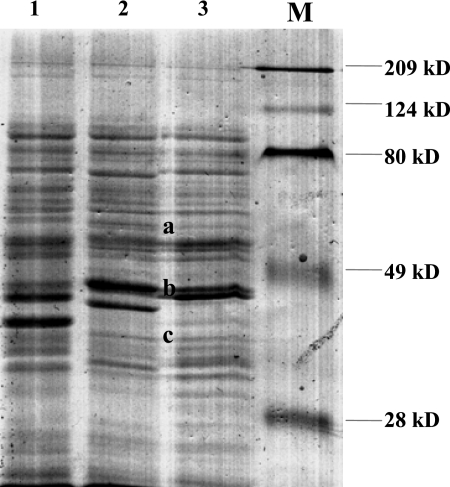
Five of the six oral C. concisus strains isolated from a child with CD showed identical protein profiles on SDS-PAGE (Fig. 2). Interestingly, the sixth oral C. concisus strain showed a pattern that was different from that of the other five oral strains but resembled the pattern of the biopsy strain isolated from the same child at a number of positions (Fig. 2, bands a to c).
FIG. 2.
Comparison of the SDS-PAGE profiles of six oral C. concisus strains and a biopsy C. concisus strain isolated from a child with CD. Profile 1: five of the six oral C. concisus strains showed this pattern. Profile 2: one of the six oral C. concisus strains showed this pattern. Profile 3 is the pattern of a biopsy C. concisus strain from the same child who yielded the isolates with profiles 1 and 2. Protein bands a to c are common to one oral C. concisus strain and the biopsy strain from the same child. Lane M, molecular size markers.
The high prevalence of C. concisus detected in saliva from healthy individuals suggests that C. concisus is part of the normal human oral microflora. Unexpectedly, we found that preschool children had significantly less C. concisus culture positivity than older subjects did (Fig. 1). Given that 83% of the children 3 to 5 years old were positive for C. concisus by PCR and the C. concisus PCR was 1,000 times more sensitive than culture in the detection of C. concisus, the lower culture positivity rate in children 3 to 5 years old likely reflects lower numbers of C. concisus bacteria in the saliva of these children. An earlier study examining the severity of gingival inflammation in individuals of different ages reported that gingivitis was less severe in children 4 to 6 years old than in teenage children and adults (9). Given that C. concisus has been associated with the onset of periodontal disease (7), increased numbers of C. concisus bacteria in the oral cavities of older children and adults may account for their increased severity of gingival inflammation. It is not clear why older children and adults harbor more C. concisus bacteria in their oral cavities. One possibility is that the subgingival space of permanent teeth provides conditions more suitable for C. concisus growth.
The prevalence of C. concisus in patients with IBD was the same as that of the age-matched controls. Protein analysis of the six oral C. concisus strains isolated from a child with CD revealed two different profiles, consistent with the heterogeneous nature of this species (2). Interestingly, one oral C. concisus strain had a number of protein bands in common with the biopsy C. concisus strain isolated from the same child. This result suggests that an individual may harbor multiple C. concisus strains in the oral cavity and it is possible that some of the oral C. concisus strains have colonized the lower gastrointestinal tract (GIT) and are involved in enteric diseases, including IBD. The previous isolation of C. concisus from esophageal, gastric, and intestinal biopsy specimens suggests that this bacterium is able to colonize and may cause mucosal inflammation in other parts of the GIT, in addition to the oral cavity (13, 14).
The high rate of C. concisus isolation from older children and adults in this study may also have relevance to IBD. The increased number of oral C. concisus bacteria in older children and adults may have permitted some oral virulent C. concisus strains to reach the threshold dose required to cause mucosal inflammation, which may have contributed to the increased incidence of IBD in adolescents and young adults.
In conclusion, this is the first study to examine the oral prevalence of C. concisus in healthy individuals of different age groups. Furthermore, results from this study suggest that further studies are required to investigate whether some oral C. concisus strains are involved in human IBD.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences of all of the C. concisus isolates obtained in this study were submitted to GenBank and assigned accession numbers GU255882 to GU255942.
Acknowledgments
Li Zhang thanks Albert Lastovica for his detailed e-mail conversation regarding the Cape Town method that was used, with modification, in this study for the isolation of C. concisus.
This work was supported by a Faculty Research Grant of the University of New South Wales and the Eli and Edythe L. Broad Foundation.
This work was approved by the Ethics Committees of the University of New South Wales and the South Eastern Sydney Area Health Service, Australia [HREC 06233/SESAHS(ES) 06/164, HREC 09237/SESIAHS 09/078, and HREC08335/SESIAHS(CHN)07/48].
Footnotes
Published ahead of print on 2 June 2010.
REFERENCES
- 1.Aabenhus, R., S. L. W. On, B. L. Siemer, H. Permin, and L. P. Andersen. 2005. Delineation of Campylobacter concisus genomospecies by amplified fragment length polymorphism analysis and correlation of results with clinical data. J. Clin. Microbiol. 43:5091-5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Engberg, J., S. L. W. On, C. S. Harrington, and P. Gerner-Smidt. 2000. Prevalence of Campylobacter, Arcobacter, Helicobacter, and Sutterella spp. in human fecal samples as estimated by a reevaluation of isolation methods for campylobacters. J. Clin. Microbiol. 38:286-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Engberg, J., D. D. Bang, R. Aabenhus, F. M. Aarestrup, V. Fussing, and P. Gerner-Smidt. 2005. Campylobacter concisus: an evaluation of certain phenotypic and genotypic characteristics. Clin. Microbiol. Infect. 11:288-295. [DOI] [PubMed] [Google Scholar]
- 4.Laemmli, U. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 5.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Ltd., Chichester, United Kingdom.
- 6.Lastovica, A. J. 2006. Emerging Campylobacter spp.: the tip of the iceberg. Clin. Microbiol. Newsl. 28:49-56. [Google Scholar]
- 7.Macuch, P. J., and A. C. R. Tanner. 2000. Campylobacter species in health, gingivitis, and periodontitis. J. Dent. Res. 79:785-792. [DOI] [PubMed] [Google Scholar]
- 8.Man, S. M., L. Zhang, A. S. Day, S. T. Leach, D. A. Lemberg, and H. Mitchell. 2010. Campylobacter concisus and other Campylobacter species in children with newly diagnosed Crohn's disease. Inflamm. Bowel Dis. 16:1008-1016. [DOI] [PubMed] [Google Scholar]
- 9.Matsson, L., and P. Goldberg. 1985. Gingival inflammatory reaction in children at different ages. J. Clin. Periodontol. 12:98-103. [DOI] [PubMed] [Google Scholar]
- 10.Newell, D. G. 2005. Campylobacter concisus: an emerging pathogen? Eur. J. Gastroenterol. Hepatol. 17:1013-1014. [DOI] [PubMed] [Google Scholar]
- 11.Snijders, F., E. J. Kuijper, B. de Wever, L. van der Hoek, S. A. Danner, and J. Dankert. 1997. Prevalence of Campylobacter-associated diarrhea among patients infected with human immunodeficiency virus. Clin. Infect. Dis. 24:1107-1113. [DOI] [PubMed] [Google Scholar]
- 12.Vandamme, P., F. E. Dewhirst, B. J. Paster, and S. L. W. On. 2005. Genus I. Campylobacter, p. 1147-1160. In G. M. Garrity, D. J. Brenner, N. R. Kriegand, and J. T. Staley (ed.), Bergey's manual of systematic bacteriology, Springer, New York, NY.
- 13.Vandamme, P., E. Falsen, B. Pot, B. Hoste, K. Kersters, and J. De Ley. 1989. Identification of EF group 22 campylobacters from gastroenteritis cases as Campylobacter concisus. J. Clin. Microbiol. 27:1775-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang, L., S. M. Man, A. S. Day, S. T. Leach, D. A. Lemberg, S. Dutt, M. Stormon, A. Otley, E. V. O'Loughlin, A. Magoffin, P. H. Y. Ng, and H. Mitchell. 2009. Detection and isolation of Campylobacter species other than C. jejuni from children with Crohn's disease. J. Clin. Microbiol. 47:453-455. [DOI] [PMC free article] [PubMed] [Google Scholar]