Abstract

The mixed lineage leukemia (MLL) gene plays a critical role in epigenetic regulation of gene expression and is a frequent target of chromosomal translocations leading to leukemia. MLL plant homeodomain 3 (PHD3) is lost in all MLL translocation products, and reinsertion of PHD3 into MLL fusion proteins abrogates their transforming activity. PHD3 has been shown to interact with the RNA-recognition motif (RRM) domain of human nuclear Cyclophilin33 (CYP33). Here, we show that CYP33 mediates downregulation of the expression of MLL target genes HOXC8, HOXA9, CDKN1B, and C-MYC, in a proline isomerase-dependent manner. This downregulation correlates with the reduction of trimethylated lysine 4 of histone H3 (H3K4me3) and histone H3 acetylation. We have structurally characterized both the PHD3 and CYP33 RRM domains and analyzed their binding to one another. The PHD3 domain binds H3K4me3 (preferentially) and the CYP33 RRM domain at distinct sites. Our binding data show that binding of H3K4me3 to PHD3 and binding of the CYP33 RRM domain to PHD3 are mutually inhibitory, implying that PHD3 is a molecular switch for the transition between activation and repression of target genes. To explore the possible mechanism of CYP33/PHD3-mediated repression, we have analyzed the CYP33 proline isomerase activity on various H3 and H4 peptides and shown selectivity for two sites in H3. Our results provide a possible mechanism for the MLL PHD3 domain to act as a switch between activation and repression.
The MLL gene is involved in chromosomal rearrangements that result in a variety of acute leukemias, including de novo acute leukemia, therapy-induced acute myeloid leukemia, and most infant leukemia (1−6). Human MLL1 is a large protein consisting of 3969 amino acids containing multiple functional domains, including three AT-hook motifs, a CXXC zinc finger, two speckled nuclear localization signals, four PHD zinc fingers, a bromo domain, two FY-rich regions, a transcriptional activation domain, and a Su(var.)3-9, Enhancer of Zeste, Trithorax (SET) domain (7−9). MLL regulates expression of a large number of genes during embryonic development and hematopoiesis. Like its Drosophila homologue Trithorax (trx), MLL associates with chromatin and writes an epigenetic mark on lysine 4 of histone H3 (H3K4) through the methyltransferase activity of the C-terminal SET domain (9,10). Trimethylation on H3K4 (H3K4me3) is a global mark for transcriptional activation, and high levels of H3K4me3 are observed in transcriptionally active regions of MLL target genes (11,12).
MLL belongs to the Trithorax group (Trx-G) of proteins that are responsible for epigenetic memory of the transcriptionally open or active chromatin states in development and differentiation (13). Trx-G proteins antagonize silencing by the Polycomb group of proteins (Pc-G) (14). The balance of Trx-G and Pc-G functions to maintain a transcriptionally active or repressive state has to be elaborately regulated depending on the particular developmental stage. Direct interaction between MLL and Pc-G proteins, HPC2 and Bmi-1, has been reported, suggesting that physical cross-talk between Trx-G and Pc-G proteins is likely to be involved in the transition between activation and repression of target genes (15). It has been shown that nuclear Cyclophilin33 (CYP33) mediates this transition by binding to the third PHD finger (PHD3) of MLL (16). CYP33 contains an N-terminal RRM (RNA-recognition motif) domain and a C-terminal cyclophilin domain that has peptidyl-proline isomerase (PPIase) activity (17,18). The role of the PPIase activity in the biological function of CYP33 and the mechanism of how CYP33 modulates MLL function are still obscure.
PHD fingers are frequently found in chromatin-associated proteins, recognizing either methylated or unmethylated lysines of histone proteins (19−25). MLL has four PHD fingers whose function is mostly unknown. All the PHD fingers as well as the SET domain are absent in MLL chromosomal translocations found in leukemia. Recently, it was shown that reinsertion of PHD3 into MLL−AF9 and MLL−ENL fusion proteins abrogates immortalization of hematopoietic progenitor cells with a reduction in the level of expression of critical fusion protein targets such as HOXA9 or HOXC8, indicating that PHD3 has a crucial role in MLL function (26,27).
Herein, we show that CYP33 mediates downregulation of the expression of MLL target genes HOXC8, HOXA9, CDKN1B, and C-MYC, in a PPIase-dependent manner. This downregulation correlates with reduction of trimethylated lysine 4 of histone H3 (H3K4me3) and histone H3 acetylation. We have structurally characterized both the PHD3 and CYP33 RRM domains using NMR spectroscopy. Chemical shift perturbation analysis, Ala mutagenesis, and isothermal titration calorimetry have been used to show that H3K4me3 and CYP33 RRM bind at distinct sites on PHD3. A similar analysis with the RRM domain shows that PHD3 binds on a site that overlaps with the canonical RNA binding site on the CYP33 RRM domain. Our binding data show that binding of H3K4me3 to PHD3 and binding of the CYP33 RRM domain to PHD3 are mutually inhibitory, implying that PHD3 is a molecular switch for the transition between activation and repression of target genes. To explore the possible mechanism of CYP33/PHD3-mediated repression, we have analyzed CYP33 proline isomerase activity on various H3 and H4 peptides and shown selectivity for two sites in H3. Our results provide a possible mechanism for the MLL PHD3 domain to act as a switch between activation and repression.
Experimental Procedures
Protein and Peptide Preparation
DNA encoding MLL PHD3 [A1564−H1628 of human MLL (gi 56550039)] was cloned into the pGEX-4T-1 vector between the BamHI site and the NotI site. Recombinant PHD3 was expressed in Escherichia coli BL21 cells grown in LB or EMBL medium. Samples for NMR experiments were prepared by addition of appropriate isotopes to minimal medium. All the media used for PHD3 expression included 100 μM zinc chloride; 0.8 mM IPTG was used to induce expression when OD600 reached 0.6−0.7, and cells were subsequently grown for 4 h at 30 °C. The proteins were initially purified by affinity chromatography on a Glutathione Sepharose 4B column. The eluate was cleaved by thrombin to remove the GST tag and then dialyzed against 50 mM Tris (pH 8.0), 50 mM NaCl, 3 mM DTT, and 100 μM ZnCl2. Proteins were further purified by ion-exchange chromatography on a DEAE-Sepharose column. Full-length CYP33 (gi 4406227) and the RRM domain spanning residues T3−K83 were cloned into the pGEX-4T-1 vector between the BamHI site and the NotI site. The proteins were expressed in E. coli BL21 cells grown in LB medium or EMBL medium with appropriate isotope labeling. CYP33 RRM was induced by 0.8 mM IPTG at 37 °C. CYP33 was induced by 0.8 mM IPTG at 25 °C. Cells were allowed to grow for 4 h before being harvested. The proteins were initially purified by GST tag affinity chromatography. The eluate was cut with thrombin, dialyzed against 50 mM Tris (pH 8.0), 50 mM NaCl, and 1 mM DTT, and finally purified by ion-exchange chromatography using DEAE-Sepharose. All the mutants were generated using the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). Histone peptides with or without lysine methylation [H3 (residues 1−15 with un-, mono-, di-, or trimethylated K4), H3K9me3 (residues 1−15), H3K27me3 (residues 22−32), H3K36me3 (residues 31−41), H3K79me3 (residues 74−84), and H4K20me3 (residues 15−25)] for binding assays were synthesized by New England Peptide LLC (Gardner, MA), Biosynthesis (Lewisville, TX), and the Keck Facility at Yale University (New Haven, CT). A tyrosine was added to the end of the H3K4me1/2/3 peptides, and the concentration was determined by measuring the UV absorbance at 280 nm.
NMR Spectroscopy
Two-dimensional (2D) 15N−1H HSQC, three-dimensional (3D) HNCACB, 3D CBCA(CO)NH, 3D HNCO, and 3D HNHA spectra were acquired for backbone assignment. 2D 13C−1H HSQC, 3D CC(CO)NH-TOCSY, 3D HCC(CO)NH-TOCSY, and 3D HCCH-TOCSY spectra were acquired for side chain assignment. All NMR experiments were performed at 30 °C on a Varian 600 MHz instrument equipped with a cryogenic probe. All NMR samples were prepared in 25 mM potassium phosphate (pH 6.9), 50 mM NaCl, 1 mM DTT, 1 mM sodium azide, and 5% (v/v) D2O. Additional DTT (up to 3 mM) and 100 μM ZnCl2 were added to any NMR sample containing PHD3. Alignment for RDC measurements was determined with polyacrylamide gels (28) made of zwitterionic copolymers based on 50% content of a 1:1 APTMAC/acrylic acid mixture and 50% acrylamide; 9% of the initial concentration of the copolymer was used.
Structure Calculations
The structure calculations for PHD3 and CYP33 RRM were conducted with the same procedure unless otherwise specified. Interproton distance constraints were obtained from NOE cross-peak assignments in 3D NOESY spectra. Initial semiautomatic NOE assignment and structure calculation were performed with CYANA (29). After automatic NOE assignment, only unambiguous NOEs were selected, and ambiguous NOEs were manually examined and assigned. Assigned NOEs were then categorized into four groups: 1.8−2.8, 1.8−3.3, 1.8−4.2, and 1.8−5.5 Å for structure calculation. ϕ and ψ dihedral angle restraints were generated by TALOS prediction based on Cα, Cβ, C′, and N chemical shifts (30). 3JHNHA coupling constant restraints were calculated via comparison of the intensities of diagonal and cross-peaks in a 3D HNHA spectrum (31). Three sets of RDCs (1DHN, 1DNC′, and 1DC′Cα) were measured using TROSY-HNCO experiments (32). Initial values of the axial (Da) and rhombic (R) components of the alignment tensor were estimated from a histogram of RDC distribution, and final optimized Da and R values were obtained by iterative fitting of the measured RDCs to the preliminary structure using PALES (33). Zinc constraints have been used in the final structural refinement of PHD3 for zinc coordination by the side chains of seven cysteines and one histidine.
NMR Titration and Chemical Shift Perturbation Analysis
15N−1H HSQC spectra were recorded for probing interactions between PHD3 and histone peptides. For the H3K4 binding studies, 100 μM PHD3 was titrated with 100, 200, 400, 800, and 1600 μM histone H3K4me1/2/3. 15N−1H HSQC spectra were recorded at each titration point to calculate chemical shift changes of the PHD3 residues. To identify the CYP33 RRM-binding site on PHD3, [U-15N,13C,1H]PHD3 was mixed with unlabeled CYP33 RRM. Backbone chemical shifts were assigned using 2D HSQC, 3D HNCO, 3D HNCACB, and 3D CBCA(CO)NH spectra. Average chemical shifts from each spectrum were used to calculate chemical shift perturbation upon complex formation. Similarly, the complex of [U-15N,13C,1H]CYP33 RRM mixed with unlabeled PHD3 was used to calculate chemical shift perturbation of CYP33 RRM upon PHD3 binding. Combined chemical shift changes for each residue were calculated using the equation Δδtot = [(wNΔδN)2 + (wHΔδHN)2]1/2, where ΔδHN and ΔδN are the chemical shift differences in the HN and N resonances, respectively. Weighting factors (wN and wH) of 0.154 and 1 were used for N and H, respectively.
Isothermal Titration Calorimetry
ITC experiments were conducted on a VP-ITC MicroCalorimeter system at 25 °C. Protein samples were dialyzed against 25 mM potassium phosphate (pH 7.2), 50 mM NaCl, 1 mM DTT, and 100 μM ZnCl2 and then centrifuged and degassed prior to use. In each experiment, 8 μL aliquots of a titrant (CYP33 RRM or the H3K4me3 peptide) solution were added 30 times to PHD3. Data were corrected for dilution enthalpy and then were fit to a one-site binding model using Origin 7.0.
Peptidyl-Proline Isomerase Activity Assay
The PPIase assay was conducted as described previously (34). Pentapeptides with C-terminal pNA corresponding to H3P16, H3P30, H3P38, and H4P32 were purchased from Biosynthesis (Lewisville, TX). The peptides were dissolved in TFE containing 470 mM LiCl to achieve a final peptide concentration of 10 mM. Five milligrams of bovine chymotrypsin (Sigma-Aldrich) was dissolved in 10 mL of the buffer containing 50 mM Tris-HCl (pH 8.0) and 100 mM NaCl. The buffer solution with chymotrypsin was chilled at 4 °C, and 10 μL of peptide solution was added to 1 mL of a chilled buffer solution to initiate the reaction. The progress of the reaction was monitored by measuring the absorbance at 390 nm using a GenesysII spectrometer (Spectronics). For CYP33-treated samples, 40 nM CYP33 was added to the reaction buffer before peptides were added.
RT-PCR
R191A and F196A mutations in the PPIase active site were introduced by site-directed mutagenesis in the CYP33 coding sequence to create a PPIase dead CYP33 (P. Breslin, unpublished methods). The resulting polypeptide fused to GST and expressed from bacteria was tested for proline isomerase activity by chymotrypsin cleavage of a Suc-AAPF-pNA substrate. The first-order isomerization rate using the mutant protein was 5.7% of the one obtained for wild-type Cyp33 (J. Solanki and R. Schultz, unpublished observations). HEK293T cells were transfected with pCEP4 plasmid constructs expressing FLAG-tagged wild-type or the PPIase mutant CYP33 using Lipofectamine 2000 (Invitrogen). After 48 h, RNA was harvested using Trizol (Invitrogen) and reverse-transcribed using a High-Capacity cDNA Reverse Transcription kit (Applied Biosystems). Quantitative PCR was performed in triplicate using GoTaq qPCR Master mix (Promega) on an Applied Biosystem 7300 system. Expression levels of HOXC8, HOXA9, CDKN1B, and C-MYC cDNAs were normalized to β2M cDNA by the comparative Ct (cycle threshold) method.
Chromatin Immunoprecipitation
HEK293T cells were transfected with vector control or pCEP4-CYP33 using lipofectamine. After transfection for 48 h, cells were fixed in formalin and ChIP was performed against anti-histone H3, anti-H3K4me3, anti-acetylated histone H3 (acetylated at lysines 9 and 14), and IgG antibodies (Millipore), using the Magna ChIP kit from Upstate. The ChIP DNA was used as a template for quantitative PCR at the MLL target gene promoters. The fold occupancy of histone H3, H3K4me3, and H3 acetylation at MLL target gene promoters was normalized to input DNA levels.
Primer Sequences
RT-PCR: HOXC8 3′UTR (forward), 5′TGGAAACCTGAAGGAGATGTGGGT3′; HOXC8 3′UTR (reverse), 5′AAACAGCGAAGGAGAGGAAGGCAT3′; HOXA9 (forward), 5′TCCCACGCTTGACACTCACACTTT3′; HOXA9 (reverse), 5′AGTTGGCTGCTGGGTTATTGGGAT3′; CDKN1B (RT forward), 5′CCGGCTAACTCTGAGGACAC3′; CDKN1B (RT reverse), 5′CTTCTGAGGCCAGGCTTCTT3′; C-MYC (RT forward), 5′CCTACCCTCTCAACGACAGC3′; C-MYC (RT reverse), 5′CTCTGACCTTTGCCAGGAG3′; CYP33 (forward), 5′GTTTTCTGGGAAGACGCTTG3′; CYP33 (reverse), 5′ATTTGAGCGGGCCTTTTTAG3′; β2M (forward), 5′TGCTGTCTCCATGTTGATGTATCT3′; β2M (reverse), 5′TTGATTTTGGAGGGATCTCG3′. ChIP: HOXC8 promoter (forward), 5′AGTCTGAGGAATTCGCCTGGGCTGTTA3′; HOXC8 promoter (reverse), 5′CGCTCCTCACTGTCGGTAGGTAGA3′; CDKN1B promoter (forward), 5′CCGGCTAACTCTGAGGACAC3′; CDKN1B promoter (reverse), 5′ATACGCCGAAAAGCAAGCTA3′.
Results
Solution Structure of PHD3
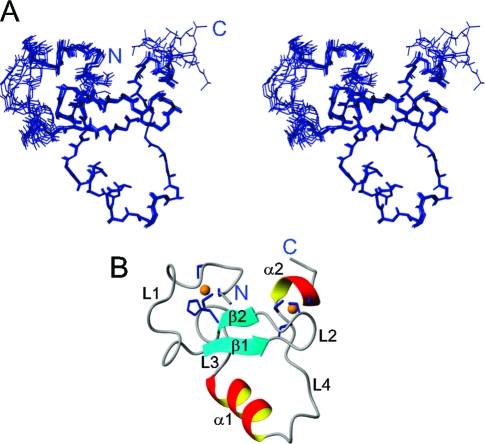
The solution structure of PHD3 was calculated using constraints obtained from multidimensional heteronuclear NMR spectroscopy. The constraints for structure determination and the structural statistics are summarized in Table 1 of the Supporting Information. NMR data and atomic coordinates have been deposited in the BioMagResBank and the RSCB Protein Data Bank. The final ensemble of 10 conformers of 100 calculated structures was selected by lowest energy with a backbone root-mean-square deviation (rmsd) of 0.54 Å. The solution structure suggests that the overall fold of PHD3 is similar to that of previously determined PHD fingers, coordinating two zinc atoms in one structural entity (Figure 1). Two zinc-binding clusters are formed by C1569, C1572, H1596, and C1599 and by C1588, C1591, C1621, and C1624. The structure consists of a small double-stranded antiparallel β-sheet, two short α-helices, and the four loops connecting them (Figure 1B). The β-sheet is located between two zinc clusters as is typically found in other PHD fingers. L1 is not well-defined in our NMR ensemble. 15N T1 and T2 relaxation time measurements showed that L1 is flexible in solution (data not shown).
Figure 1.
Solution structure of PHD3. (A) Stereoview of an ensemble of 10 conformers. (B) Ribbon representation of the lowest-energy structure showing secondary structural units. The side chains of the residues coordinating the two zinc atoms (orange) are colored blue.
PHD3 Selectively Binds H3K4me3
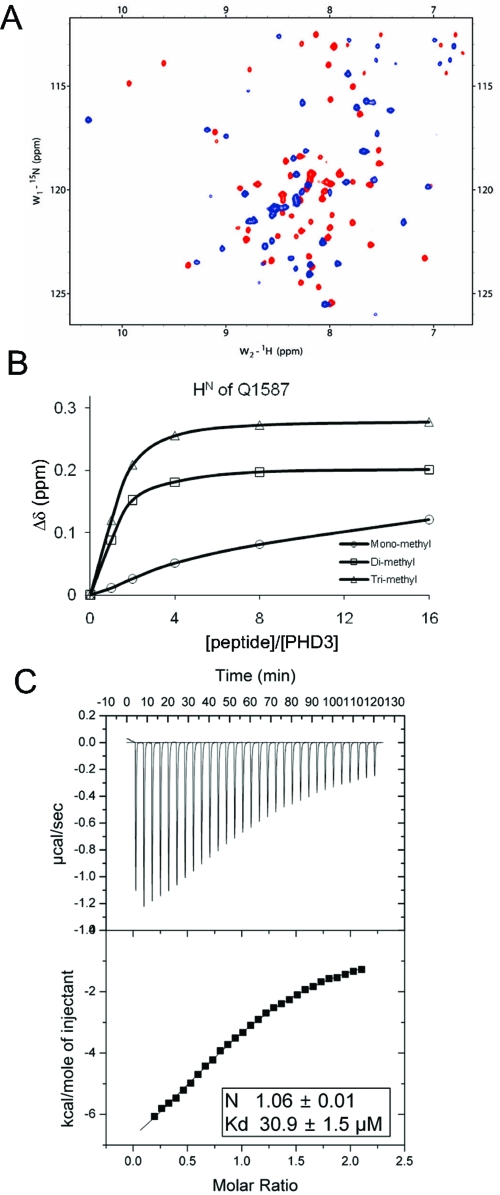
PHD3 is very similar in sequence and structure to PHD fingers that recognize a methylated lysine in histone tails (Figure 2). PHD fingers that recognize a methylated lysine have an invariant tryptophan on the β-sheet, whereas PHD fingers that bind exclusively to an unmethylated lysine do not have this tryptophan (Figure 2A). Recent structures of PHD finger and H3K4me3 complexes revealed that this tryptophan is critical for the recognition of methylated lysine (22,35). PHD3 also has a tryptophan, which is located at the same position as seen in known structures of PHD finger−H3K4me3 complexes (Figure 2B). Because the sequence and structural comparison strongly suggested that PHD3 might recognize a methylated lysine, the ability of PHD3 to recognize methylated lysine peptides was screened using a series of 15N−1H HSQC experiments. Synthetic histone peptides with trimethylation on H3 K4, H3 K9, H3 K27, H3 K79, and H4 K20 were titrated into PHD3 and monitored for chemical shift perturbation. Only the H3K4me3 (residues 1−15) peptide induced chemical shift changes of PHD3, whereas the others did not, indicating that PHD3 interacts with H3K4me3 (Figure 3A). Furthermore, PHD3 exhibited differential affinity for mono-, di-, and trimethylated H3K4, as observed in the PHD fingers already shown to bind H3K4me3. The difference in the magnitude of the chemical shift changes of a representative amide resonance (Q1587) on 15N−1H HSQC spectra upon titration with mono-, di-, and trimethylated H3K4 indicates that PHD3 preferentially binds to trimethylated H3K4 (Figure 3B). The binding stoichiometry and affinity of the PHD3−H3K4me3 interaction were determined by ITC. PHD3 binds H3K4me3 with a 1:1 stoichiometry and a Kd of ∼30 μM (Figure 3C). This binding preference of the MLL PHD3 domain was also very recently reported by others (36).
Figure 2.
Sequence and structural comparison of PHD fingers. (A) Sequence alignment of the MLL PHD3 finger with PHD fingers of chromatin-associating PHD fingers. Zinc-coordinating residues are colored orange, and the invariant tryptophan is colored red. AIRE and HBC80 bind to an unmethylated lysine 4 on histone H3 (20,43). ING2, Yng1, and BPTF bind to a methylated lysine 4 on histone H3 (22,35,44). (B) Structure comparison of the MLL PHD3 finger (left) with PHD fingers of BPTF (middle, PDB entry 2F6J) and ING2 (right, PDB entry 2G6Q). The backbone of histone H3 is colored blue, and H3K4me3 is colored violet. The side chain of the invariant tryptophan is colored red.
Figure 3.
PHD3 preferentially binds H3K4me3. (A) Overlay of 15N−1H HSQC spectra of PHD3 in the absence (red) and presence (blue) of H3K4me3. (B) Titration plot based on the chemical shift changes of the Q1587 HN resonance of PHD3. Chemical shift changes upon addition of mono-, di-, and trimethylated H3K4 were plotted as a function of the molar ratio. (C) ITC measurement of the binding of PHD3 to H3K4me3. Data from addition of 8 μL aliquots of 745 μM peptide to a solution of 65 μM PHD3 are shown.
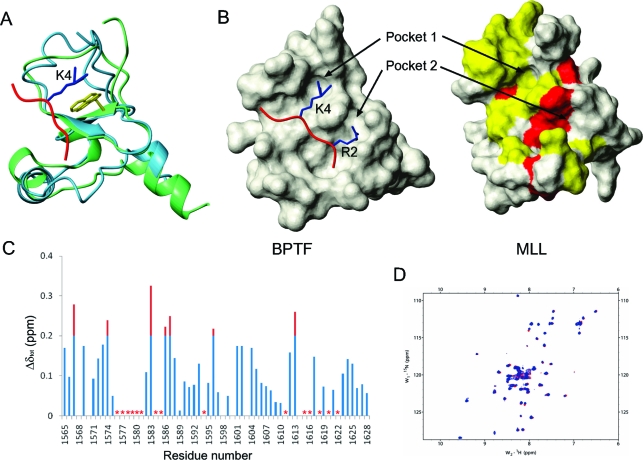
We next identified the H3K4me3 binding site on the basis of a structural comparison and chemical shift perturbation data. The structure of PHD3 was compared to the structure of the BPTF PHD−H3K4me3 complex (PDB entry 2F6J). The backbone atoms of the core region from K1584 to E1600 of PHD3 were fit to the core region of the BPTF PHD with a backbone rmsd of 0.93 Ǻ. The overall folds of the two PHD fingers are very similar, and the conserved tryptophan of each PHD superimposes well (Figure 4A). The BPTF PHD has an H3K4me3-docking groove with two pockets for the side chains of R2 and trimethylated K4 of H3. The same groove is found in the structure of PHD3, suggesting the binding site for H3K4me3 (Figure 4B). The chemical shift changes upon H3K4me3 binding also supported the binding site predicted by structural comparison. When PHD3 was titrated with H3K4me3, the backbone resonances of the residues that form the groove were broadened out or significantly shifted (Figure 4C), indicating that this region is involved in the interaction with H3K4me3. To confirm the binding site and that the conserved tryptophan is essential in recognizing H3K4me3, we mutated it to alanine and probed the interaction by NMR. No significant change in chemical shift was observed on the 15N−1H HSQC spectrum of PHD3-W1594A upon addition of H3K4me3, indicating that the interaction between the mutant and H3K4me3 is negligible (Figure 4D).
Figure 4.
Identification of the H3K4me3 recognition site on PHD3. (A) Superimposition of the MLL PHD3 finger with the BPTF PHD finger in complex with H3K4me3 (PDB entry 2F6J). The MLL PHD3 finger is colored blue, the BPTF PHD finger green, and the N-terminal H3 tail red. The side chain of the consensus tryptophan of each PHD finger is colored yellow. The side chain of the K4 of histone H3 is colored blue. (B) Surface comparison of the MLL PHD3 finger (right) with the BPTF PHD finger in complex with H3K4me3 (left). The N-terminal H3 tail is colored red, and the side chains of R2 and K4 are colored blue. Residues whose backbone resonance was broadened out (yellow) or significantly perturbed (red) in panel C are mapped on the structure of the MLL PHD3 finger. (C) Plot of chemical shift differences (Δδtot) of the backbone HN and N resonances of PHD3 between the free and H3K4me3-bound state. Asterisks indicate the residues whose resonances are broadened out. (D) Overlay of 15N−1H HSQC spectra of the W1594A mutant in the presence (blue) and absence (red) of H3K4me3.
Identification of Recognition Sites for the Interaction between PHD3 and CYP33 RRM
It was shown previously that PHD3 interacts with CYP33 RRM using a yeast two-hybrid assay, co-immunoprecipitation, and GST pull-down (16). To identify the binding site on PHD3 for CYP33 RRM, chemical shift perturbation of backbone resonances of PHD3 was analyzed upon titration with CYP33 RRM. We identified the residues at the binding interface by plotting the combined chemical shift changes of HN and N versus the residue number (Figure 5A) and mapping them onto the structure of PHD3 (Figure 5B). In particular, chemical shifts of residues in α1, α2, and L4 showed significant perturbations, indicating that this region is likely the site of interaction. Interestingly, the MLL PHD3 domain has a longer L4 than other chromatin-associating PHD fingers shown in Figure 2A, implying a functional role other than H3K4me3 binding. To confirm the importance of this loop for CYP33 RRM binding, we mutated two residues in L4, V1617, which showed the most significant change in chemical shift upon CYP33 RRM titration, and Y1619, an aromatic residue frequently found at protein−protein interfaces, to alanine and measured the binding affinity for the mutants using ITC. The V1617A mutation decreases the affinity by ∼6.2-fold (Figure 5C). The 15N−1H HSQC spectrum of V1617A showed that this mutation does not affect the structural stability of PHD3 (Figure S1 of the Supporting Information). The Y1619A mutation also showed decreased affinity, but the 15N−1H HSQC spectrum indicated that the impaired binding is probably caused by disruption of the overall fold (Figure S1 of the Supporting Information).
Figure 5.
Identification of the binding sites for the interaction between PHD3 and CYP33 RRM. (A) Plot of chemical shift changes (Δδtot) of the backbone HN and N resonances of PHD3 upon CYP33 RRM titration. Asterisks indicate the residues whose resonances are broadened out. (B) Residues whose backbone resonance were broadened out (yellow) or significantly perturbed (red) in panel A are mapped on the structure of PHD3. The side chain of V1617 is colored green. (C) ITC measurements of the binding of CYP33 RRM (left) to wild-type PHD3 and to the V1617A mutant (right). Wild-type PHD3 (62 μM) was titrated with 755 μM CYP33 RRM, and 98 μM V1617A was titrated with 1 mM CYP33 RRM. (D) Plot of chemical shift changes (Δδtot) of the backbone HN and N resonances of CYP33 RRM upon PHD3 titration. (E) Residues whose backbone resonances were significantly perturbed (red) in panel D are mapped on the structure of CYP33 RRM. The lowest-energy structure from calculation is shown as a ribbon-and-arrow diagram.
The binding site on CYP33 RRM for PHD3 was also identified. First, conventional NMR experiments were conducted to assign resonances of CYP33 RRM and determine the solution structure. The detailed structural data and NMR statistics for structure calculation are described in Table 2 of the Supporting Information. A crystal structure of the CYP33 RRM domain was also reported recently by others (37). By identifying residues that experienced large chemical shift perturbations upon complex formation, we identified the binding site on CYP33 RRM for PHD3 (Figure 5D). Many residues on the surface composed of the central β-sheet and the loop between β2 and β3 participate in PHD3 binding (Figure 5E). To assess the role of the residues in this region in recognition of PHD3, L39, K45, R47, F49, and F51 were mutated to alanine, and the affinities of the mutants for PHD3 were measured by ITC. Among them, L39A and F49A showed affinities decreased by ∼4.3- and ∼14-fold, respectively, without disrupting the structure as assessed by 15N−1H HSQC spectra (Figure S2 of the Supporting Information). In contrast, K45A and R47A did not abrogate the interaction, and the F51A mutation disrupts the structure of CYP33 RRM (data not shown). We also identified the RNA-binding site of CYP33 RRM by probing chemical shift perturbation upon titration of RNA (Figure S3 of the Supporting Information). Interestingly, the binding site of CYP33 RRM for PHD3 generally overlaps with the canonical RNA-binding site, suggesting that PHD3 competes with RNA. The biological relevance of RNA binding of CYP33 RRM and potential competition with PHD3 is an important area for future study.
CYP33 Represses Transcription of MLL Target Genes and Changes H3K4 Methylation and H3 Acetylation Levels
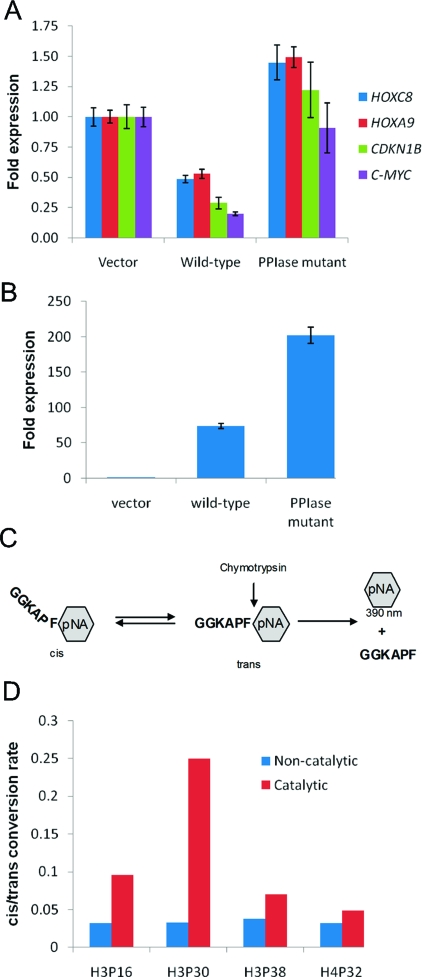
MLL regulates a number of genes, including HOX genes (e.g., HOXC8 and HOXA9) and non-HOX genes (e.g., CDKN1B and C-MYC) (38−41). To assess the general role of CYP33 on MLL target genes, the expression of HOXC8, HOXA9, CDKN1B, and C-MYC was monitored in HEK293T cells overexpressing CYP33. The expression level was quantified by real-time RT-PCR at 48 h after transfection of an empty vector or a CYP33-containing vector. High-level expression of CYP33 was confirmed by quantitative RT-PCR (Figure S4 of the Supporting Information). Expression levels of MLL target genes and CYP33 were normalized against β2M, whose expression is not influenced by CYP33. All MLL target genes tested here were markedly downregulated upon CYP33 expression, indicating that CYP33 negatively regulates the function of MLL (Figure 6A).
Figure 6.
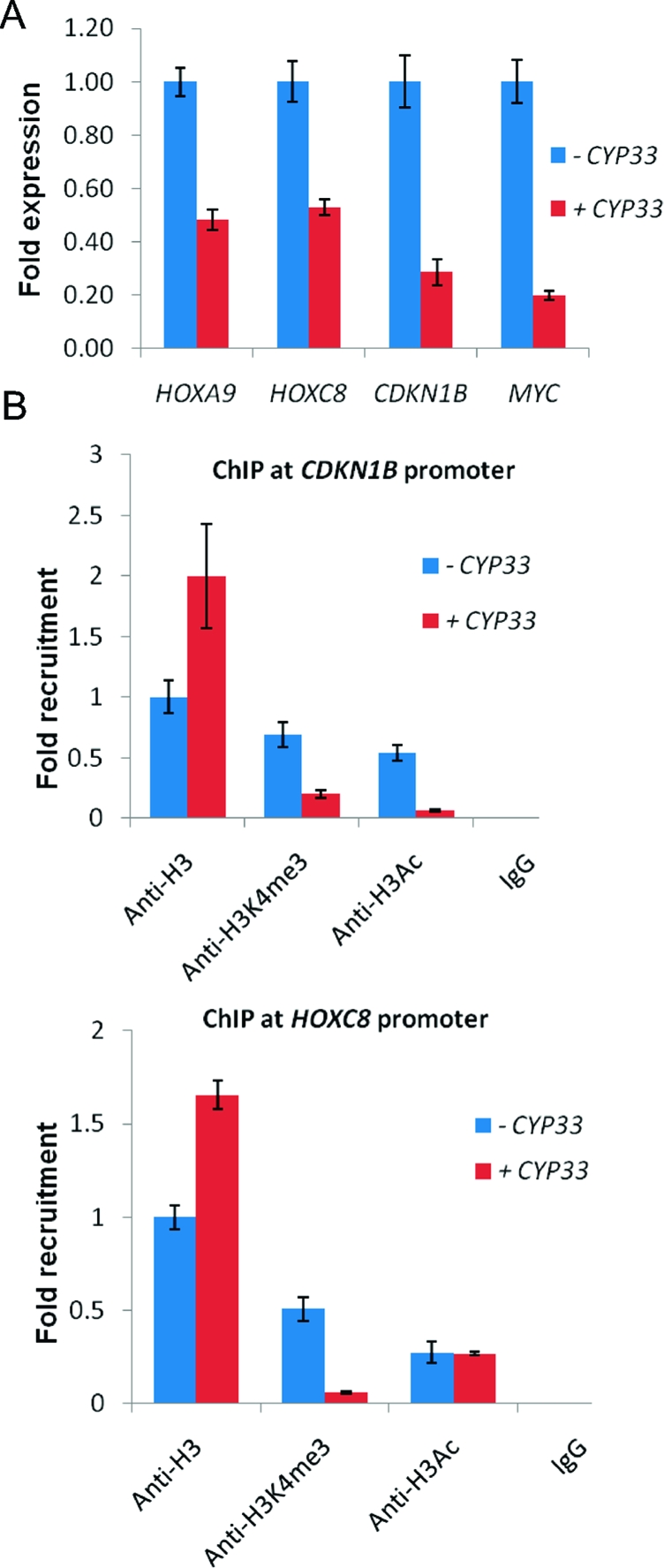
CYP33 represses transcription of MLL target genes and changes chromatin modification levels. (A) Expression of HOXA9, HOXC8, CDKN1B, and C-MYC in HEK293T cells transfected by an empty vector (blue) or CYP33-containing plasmid (red). (B) ChIP at promoters of CDKN1B (top) and HOXC8 (bottom) for probing the density of histone H3, the change in levels of H3K4me3, and the K9 and K14 acetylation of H3 (H3Ac) upon CYP33 overexpression. HEK293T cells were transfected with an empty vector (blue) or a CYP33-containing plasmid (red). IgG was used as a control.
We next examined the level of H3K4me3, which is an activation mark written by MLL, on MLL target genes. Chromatin immunoprecipitation experiments were performed 48 h after transfection of HEK293T cells with an empty vector or a CYP33-containing vector. CYP33 overexpression was confirmed by quantitative RT-PCR (Figure S4 of the Supporting Information). The H3K4me3 mark is reduced by CYP33 overexpression at the promoter regions of CDKN1B and HOXC8 (Figure 6B). In addition, the overall acetylation level of H3 on CDKN1B is also reduced by CYP33 (Figure 6B). The increase in H3 concentration corresponds to a higher nucleosome density on the CDKN1B and HOXC8 promoters after transcriptional silencing. Taken together, CYP33 overexpression leads to a change in histone modifications and chromatin structure consistent with a switch from gene activation to repression, resulting in downregulation of the expression of MLL target genes.
Proline Isomerization Activity Is Necessary for CYP33-Mediated Downregulation of MLL Target Genes
CYP33 has a cyclophilin domain that has PPIase activity as found in other cyclophilin family proteins (17). It has been previously reported that downregulation of HOXC8 by CYP33 overexpression can be inhibited by cyclosporin A, a general inihibitor of cyclophilins’ PPIase activity (15). To determine more specifically the role of the CYP33 PPIase activity in negative regulation of the transactivation function of MLL, a CYP33 mutant (R191A/F196A) that lacks PPIase activity was employed. The expression of HOXC8, HOXA9, CDKN1B, and C-MYC was quantified in HEK293T cells transfected with either the mutant or wild-type CYP33 expression vector. All four genes tested were no longer downregulated upon mutant CYP33 expression, even though the expression level of the mutant CYP33 was higher than for wild-type CYP33, indicating that the CYP33 PPIase activity is essential for repression of MLL target genes (Figure 7A,B).
Figure 7.
PPIase activity is necessary for CYP33-mediated repression. (A) The expression of MLL target genes was monitored by Q-RT-PCR after transfection of HEK293T cells with an empty vector, wild-type CYP33, and the CYP33-PPIase mutant. (B) Expression levels of the wild-type and PPIase mutant of CYP33 normalized to β2M. (C) Scheme for the chymotrypsin-coupled proline isomerase assay (45). (D) Comparison of the cis/trans conversion rates for four prolines in histone tails with (red) or without (blue) CYP33.
The in vivo substrates of CYP33 are not known. Because CYP33 is recruited to the multiprotein MLL complex via the PHD3−RRM interaction, possible substrates include MLL itself, a protein recruited to the MLL complex, and a histone. Recently, it was shown that the proline isomerase Fpr4 targets H3 P38 to alter the pattern of modifications at K36, thereby altering gene expression (42). On the basis of analogy to Fpr4, we tested PPIase activity of CYP33 on four prolines of histone tails: H3 P16, H3 P30, H3 P38, and H4 P32. The proline isomerization activity of proteins can be measured using a chymotrypsin-coupled assay with synthetic pentapeptides in which candidate prolines are followed by a phenyalanine and p-nitroanaline (pNA) group (Figure 7C). Similar to Fpr4, CYP33 shows little to no activity for H4P32 and high activity for H3P30 (Figure 7D). In contrast to Fpr4, CYP33 has activity for H3P16 and shows only very modest activity for H3P38. Thus, CYP33 does show proline isomerase activity on H3 histone tail prolines and displays substrate specificity that differs from that of Fpr4.
CYP33 RRM and H3K4me3 Binding to PHD3 Are Mutually Inhibitory
On the basis of binding site identification, PHD3 utilizes two separate surfaces for the binding of H3K4me3 and CYP33 RRM (Figure 8A). To confirm this, we have tested the binding of two mutants in PHD3 to both H3K4me3 and CYP33 RRM. W1594A, which impairs H3K4me3 binding, does not significantly affect the affinity for the RRM domain. ITC data showed that the Kd of the W1594A PHD3 for CYP33 RRM is 19.6 μM, which is ∼1.3-fold weaker than that of the wild type (14.7 μM) (Figure 8B). Similarly, V1617A PHD3, which abrogates RRM binding by ∼6.2-fold, does not significantly affect the affinity for H3K4me3 (Figure 8B). The Kd of V1617A PHD3 for the H3K4me3 peptide is 52.4 μM, which is ∼1.7-fold weaker than the wild-type value (30.9 μM).
Figure 8.
PHD3 uses two separate surfaces to bind H3K4me3 and CYP33 RRM, and the interactions are mutually inhibitory. (A) Two surfaces with a 180° difference on the MLL PHD3 domain serve for the H3K4me3-binding site (left) and for the RRM-binding site (right). The residues whose backbone resonances were broadened out (light blue or light green) or significantly shifted (dark blue or dark green) upon complex formation are displayed on the surface. (B) Binding constants of two PHD3 mutants measured by ITC. V1617A abrogates CYP33 RRM binding affinity ∼6.2-fold but does not significantly change the affinity for H3K4me3. W1594A lacks H3K4me3 binding but does not significantly change the affinity for CYP33 RRM. Note that these two residues are located on the separate surfaces of PHD3 shown in panel A. (C) Thermodynamic box showing the binding constants for each interaction. The binding constants were measured by ITC. PHD3 (62 μM) was saturated with 540 μM H3K4me3 and then titrated with 635 μM CYP33 RRM. PHD3 (65 μM) was saturated with 200 μM CYP33 RRM and then titrated with 745 μM H3K4me3.
With two binding sites on PHD3, it is possible that the two proteins bind with no influence on one another or that the binding of one can influence the binding of the other. If the PHD3 domain does indeed act as a switch, the latter behavior would be expected. In particular, we would predict that binding of one protein will inhibit the binding of the other. To test this, we have taken ITC measurements of the binding of H3K4me3 to a PHD3−CYP33 RRM complex and of CYP33 RRM to a PHD3−H3K4me3 complex. Consistent with our prediction, PHD3−H3K4me3 binding is significantly influenced by the presence of the RRM domain, resulting in an ∼5.7-fold increased Kd (Figure 8C). Similarly, PHD3 saturated with H3K4me3 was titrated with CYP33, in which case an ∼4.4-fold increase in the Kd for the PHD3−CYP33 RRM interaction was observed (Figure 8C). These data clearly show that the binding of the two partners, H3K4me3 and CYP33 RRM, is mutually inhibitory; i.e., increasing concentrations of CYP33 will displace PHD3 from H3K4me3.
Discussion
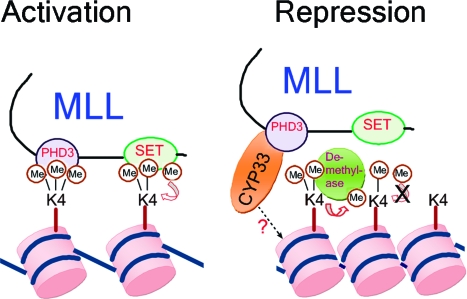
MLL plays a key role in controlling the expression of developmentally regulated genes. To do this, it must be able to strike a delicate balance between activation and repression. Our results suggest that the PHD3 domain of MLL acts as a key switch between activation and repression (see Figure 9). PHD3 binds both H3K4me3 and CYP33 on distinct surfaces on the structure of PHD3. This binding is mutually inhibitory, so increased levels of CYP33 will cause PHD3 to dissociate from H3K4me3, a key epigenetic mark for activation. Low levels of CYP33 would result in binding of PHD3 to the H3K4me3 mark. Although the specific role of the binding of MLL PHD3 to H3K4me3 remains to be elucidated, one possible scenario is that this binding is required for propagation of the H3K4me3 mark by the MLL methyltransferase activity. In this scenario, MLL could bind to trimethylated lysine 4 on a modified histone H3 and target an unmodified histone H3 molecule on the same or a neighboring nucleosome for trimethylation (see Figure 9). High levels of CYP33 would result in dissociation of PHD3 from H3K4me3, preventing the methylation of new histones and making the remaining H3K4me3 marks accessible to H3 K4 demethylases such as RBP2 and JARID1C, resulting in their demethylation. We have shown that increasing the level of CYP33 decreases the level of expression of MLL target genes and decreases the level of H3K4me3 at target gene promoters (Figure 6). In addition, increasing the level of CYP33 also increases the density of H3 at MLL target genes, consistent with a more dense, repressive chromatin state (Figure 6).
Figure 9.
Model for the role of MLL PHD3 in activation and repression. Binding of PHD3 to H3K4me3 protects this mark from demethylation and possibly facilitates the propagation of the H3K4me3 mark, all leading to activation. When the CYP33 concentration is high, PHD3 preferentially binds to CYP33, resulting in the release of H3K4me3 from the MLL complex, loss of SET domain-mediated H3K4 methylation, increased nucleosome density, and concomitant repression. In addition, the CYP33 proline isomerase may act on the histone tail to enhance the addition of epigenetic marks for repression.
The amount of CYP33 available to bind to MLL PHD3 may be regulated by its binding to RNA. This could provide a mechanism for regulating gene promoters by moving them to different nuclear compartments with a high RNA concentration (transcription factories) or a low RNA concentration (heterochromatin compartments), even in the presence of a constant average nuclear CYP33 concentration.
Our results also show that the proline isomerase activity of CYP33 is essential for its repressive function on MLL target genes. On the basis of analogy to Fpr4, which has been shown to isomerize histone tail residues to alter epigenetic modifications (42), we tested the ability of CYP33 to isomerize H3 and H4 tail peptides. Our results show significant activity for both H3P30 and H3P16 and demonstrate an altered histone tail specificity relative to Fpr4. This activity suggests a mechanism for the proline isomerase function of CYP33, namely that by means of isomerization of prolines in the histone tail, CYP33 can effect a change in the epigenetic modification profile on the H3 tail. The isomerization of one or more prolines could easily alter the activity of histone acetyl transferase (HAT), histone deacetylase (HDAC), histone methyl transferase, or histone demethylase enzymes at specific loci, resulting in this proposed alteration of the epigenetic profile. Alternatively, CYP33 may target prolyl-peptide bonds on MLL itself or on proteins that interact with MLL. Recruitment of HDAC1 to the repression domain of MLL is enhanced by CYP33 overexpression (15). These mechanisms are not mutually exclusive. It is possible that Cyp33 targets different peptide bonds in different proteins and contributes to repression of MLL target genes by more than one mechanism. Future efforts in this area will need to focus on testing this array of possibilities to deduce the specific mechanism.
Acknowledgments
We thank Peter Breslin and Wei Wei for providing the CYP33 mutant without PPIase activity.
Supporting Information Available
Tables describing structural statistics and additional figures. This material is available free of charge via the Internet at http://pubs.acs.org.
This work was supported by grants from the Leukemia and Lymphoma Society (SCOR 7006-05) to J.H.B. and National Cancer Institute (National Institutes of Health) Grant PO1 CA105049 to M.O.D.
Coordinates for the structures of the MLL PHD3 domain and the CYP33 RRM domain have been deposited in the Protein Data Bank as entries 2KYU and 2KYX, respectively. NMR experimental data for the MLL PHD3 domain and the CYP33 RRM domain have been deposited in the BioMagResBank as entries 16983 and 16989, respectively.
Footnotes
Abbreviations: BPTF, bromo domain and PHD transcription factor; CYP33, Cyclophilin33; H3K4me3, trimethylated lysine 4 of histone H3; ITC, isothermal titration calorimetry; MLL, mixed lineage leukemia; NMR, nuclear magnetic resonance; PHD, plant homeodomain; PHD3, third PHD finger of MLL; PPIase, peptidyl-proline isomerase; RRM, RNA-recognition motif; SET, Su(var.)3-9, Enhancer of Zeste, Trithorax.
Supplementary Material
References
- Chen C. S.; Sorensen P. H.; Domer P. H.; Reaman G. H.; Korsmeyer S. J.; Heerema N. A.; Hammond G. D.; Kersey J. H. (1993) Molecular rearrangements on chromosome 11q23 predominate in infant acute lymphoblastic leukemia and are associated with specific biologic variables and poor outcome. Blood 81, 2386–2393. [PubMed] [Google Scholar]
- Pui C. H.; Relling M. V.; Rivera G. K.; Hancock M. L.; Raimondi S. C.; Heslop H. E.; Santana V. M.; Ribeiro R. C.; Sandlund J. T.; Mahmoud H. H.; et al. (1995) Epipodophyllotoxin-related acute myeloid leukemia: A study of 35 cases. Leukemia 9, 1990–1996. [PubMed] [Google Scholar]
- Rowley J. D. (1993) Rearrangements involving chromosome band 11Q23 in acute leukaemia. Semin. Cancer Biol. 4, 377–385. [PubMed] [Google Scholar]
- Hunger S. P.; Tkachuk D. C.; Amylon M. D.; Link M. P.; Carroll A. J.; Welborn J. L.; Willman C. L.; Cleary M. L. (1993) HRX involvement in de novo and secondary leukemias with diverse chromosome 11q23 abnormalities. Blood 81, 3197–3203. [PubMed] [Google Scholar]
- Super H. J.; McCabe N. R.; Thirman M. J.; Larson R. A.; Le Beau M. M.; Pedersen-Bjergaard J.; Philip P.; Diaz M. O.; Rowley J. D. (1993) Rearrangements of the MLL gene in therapy-related acute myeloid leukemia in patients previously treated with agents targeting DNA-topoisomerase II. Blood 82, 3705–3711. [PubMed] [Google Scholar]
- Biondi A.; Cimino G.; Pieters R.; Pui C. H. (2000) Biological and therapeutic aspects of infant leukemia. Blood 96, 24–33. [PubMed] [Google Scholar]
- Zeleznik-Le N. J.; Harden A. M.; Rowley J. D. (1994) 11q23 translocations split the “AT-hook” cruciform DNA-binding region and the transcriptional repression domain from the activation domain of the mixed-lineage leukemia (MLL) gene. Proc. Natl. Acad. Sci. U.S.A. 91, 10610–10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birke M.; Schreiner S.; Garcia-Cuellar M. P.; Mahr K.; Titgemeyer F.; Slany R. K. (2002) The MT domain of the proto-oncoprotein MLL binds to CpG-containing DNA and discriminates against methylation. Nucleic Acids Res. 30, 958–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T.; Mori T.; Tada S.; Krajewski W.; Rozovskaia T.; Wassell R.; Dubois G.; Mazo A.; Croce C. M.; Canaani E. (2002) ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol. Cell 10, 1119–1128. [DOI] [PubMed] [Google Scholar]
- Milne T. A.; Briggs S. D.; Brock H. W.; Martin M. E.; Gibbs D.; Allis C. D.; Hess J. L. (2002) MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol. Cell 10, 1107–1117. [DOI] [PubMed] [Google Scholar]
- Ruthenburg A. J.; Allis C. D.; Wysocka J. (2007) Methylation of lysine 4 on histone H3: Intricacy of writing and reading a single epigenetic mark. Mol. Cell 25, 15–30. [DOI] [PubMed] [Google Scholar]
- Bernstein B. E.; Kamal M.; Lindblad-Toh K.; Bekiranov S.; Bailey D. K.; Huebert D. J.; McMahon S.; Karlsson E. K.; Kulbokas E. J. III; Gingeras T. R.; Schreiber S. L.; Lander E. S. (2005) Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120, 169–181. [DOI] [PubMed] [Google Scholar]
- Ingham P. W. (1998) Trithorax and the regulation of homeotic gene expression in Drosophila: A historical perspective. Int. J. Dev. Biol. 42, 423–429. [PubMed] [Google Scholar]
- Brock H. W.; van Lohuizen M. (2001) The Polycomb group: No longer an exclusive club?. Curr. Opin. Genet. Dev. 11, 175–181. [DOI] [PubMed] [Google Scholar]
- Xia Z. B.; Anderson M.; Diaz M. O.; Zeleznik-Le N. J. (2003) MLL repression domain interacts with histone deacetylases, the polycomb group proteins HPC2 and BMI-1, and the corepressor C-terminal-binding protein. Proc. Natl. Acad. Sci. U.S.A. 100, 8342–8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fair K.; Anderson M.; Bulanova E.; Mi H.; Tropschug M.; Diaz M. O. (2001) Protein interactions of the MLL PHD fingers modulate MLL target gene regulation in human cells. Mol. Cell. Biol. 21, 3589–3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H.; Kops O.; Zimmermann E.; Jaschke A.; Tropschug M. (1996) A nuclear RNA-binding cyclophilin in human T cells. FEBS Lett. 398, 201–205. [DOI] [PubMed] [Google Scholar]
- Zeng L.; Zhou Z.; Xu J.; Zhao W.; Wang W.; Huang Y.; Cheng C.; Xu M.; Xie Y.; Mao Y. (2001) Molecular cloning, structure and expression of a novel nuclear RNA-binding cyclophilin-like gene (PPIL4) from human fetal brain. Cytogenet. Cell Genet. 95, 43–47. [DOI] [PubMed] [Google Scholar]
- Baker L. A.; Allis C. D.; Wang G. G. (2008) PHD fingers in human diseases: Disorders arising from misinterpreting epigenetic marks. Mutat. Res. 647, 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F.; Collins R. E.; De Cegli R.; Alpatov R.; Horton J. R.; Shi X.; Gozani O.; Cheng X.; Shi Y. (2007) Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature 448, 718–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.; Fischle W.; Wang W.; Duncan E. M.; Liang L.; Murakami-Ishibe S.; Allis C. D.; Patel D. J. (2007) Structural basis for lower lysine methylation state-specific readout by MBT repeats of L3MBTL1 and an engineered PHD finger. Mol. Cell 28, 677–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.; Ilin S.; Wang W.; Duncan E. M.; Wysocka J.; Allis C. D.; Patel D. J. (2006) Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 442, 91–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi S. K.; Qiu C.; Bernstein E.; Li K.; Jia D.; Yang Z.; Erdjument-Bromage H.; Tempst P.; Lin S. P.; Allis C. D.; Cheng X.; Bestor T. H. (2007) DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448, 714–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X.; Hong T.; Walter K. L.; Ewalt M.; Michishita E.; Hung T.; Carney D.; Pena P.; Lan F.; Kaadige M. R.; Lacoste N.; Cayrou C.; Davrazou F.; Saha A.; Cairns B. R.; Ayer D. E.; Kutateladze T. G.; Shi Y.; Cote J.; Chua K. F.; Gozani O. (2006) ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 442, 96–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J.; Swigut T.; Xiao H.; Milne T. A.; Kwon S. Y.; Landry J.; Kauer M.; Tackett A. J.; Chait B. T.; Badenhorst P.; Wu C.; Allis C. D. (2006) A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442, 86–90. [DOI] [PubMed] [Google Scholar]
- Chen J.; Santillan D. A.; Koonce M.; Wei W.; Luo R.; Thirman M. J.; Zeleznik-Le N. J.; Diaz M. O. (2008) Loss of MLL PHD finger 3 is necessary for MLL-ENL-induced hematopoietic stem cell immortalization. Cancer Res. 68, 6199–6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muntean A. G.; Giannola D.; Udager A. M.; Hess J. L. (2008) The PHD fingers of MLL block MLL fusion protein-mediated transformation. Blood 112, 4690–4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cierpicki T.; Bushweller J. H. (2004) Charged gels as orienting media for measurement of residual dipolar couplings in soluble and integral membrane proteins. J. Am. Chem. Soc. 126, 16259–16266. [DOI] [PubMed] [Google Scholar]
- Guntert P. (2004) Automated NMR structure calculation with CYANA. Methods Mol. Biol. 278, 353–378. [DOI] [PubMed] [Google Scholar]
- Cornilescu G.; Delaglio F.; Bax A. (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13, 289–302. [DOI] [PubMed] [Google Scholar]
- Vuister G. W.; Bax A. (1993) Quantitative J correlation: A new approach for measuring homonuclear three-bond J(HNHa) coupling constants in 15N-enriched proteins. J. Am. Chem. Soc. 115, 7772–7777. [Google Scholar]
- Yang D.; Venters R. A.; Mueller G. A.; Choy W. Y.; Kay L. E. (1999) TROSY-based sequences for the measurement of 1HN-15N, 15N-13CO, 1HN-13CO, 13CO-13Ca and 1HN-13Ca dipolar couplings in 15N, 13C, 2H-labeled proteins. J. Biomol. NMR 14, 333–343. [Google Scholar]
- Zweckstetter M.; Bax A. (2000) Prediction of sterically induced alignment in a dilute liquid crystalline phase: Aid to protein structure determination by NMR. J. Am. Chem. Soc. 122, 2. [Google Scholar]
- Kofron J. L.; Kuzmic P.; Kishore V.; Colon-Bonilla E.; Rich D. H. (1991) Determination of kinetic constants for peptidyl prolyl cis-trans isomerases by an improved spectrophotometric assay. Biochemistry 30, 6127–6134. [DOI] [PubMed] [Google Scholar]
- Pena P. V.; Davrazou F.; Shi X.; Walter K. L.; Verkhusha V. V.; Gozani O.; Zhao R.; Kutateladze T. G. (2006) Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature 442, 100–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang P. Y.; Hom R. A.; Musselman C. A.; Zhu L.; Kuo A.; Gozani O.; Kutateladze T. G.; Cleary M. L. (2010) Binding of the MLL PHD3 Finger to Histone H3K4me3 Is Required for MLL-Dependent Gene Transcription. J. Mol. Biol. 400, 137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hom R. A.; Chang P. Y.; Roy S.; Musselman C. A.; Glass K. C.; Selezneva A. I.; Gozani O.; Ismagilov R. F.; Cleary M. L.; Kutateladze T. G. (2010) Molecular Mechanism of MLL PHD3 and RNA Recognition by the Cyp33 RRM Domain. J. Mol. Biol. 400, 145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson R. D.; Hess J. L.; Yu B. D.; Ernst P.; van Lohuizen M.; Berns A.; van der Lugt N. M.; Shashikant C. S.; Ruddle F. H.; Seto M.; Korsmeyer S. J. (1999) Mammalian Trithorax and polycomb-group homologues are antagonistic regulators of homeotic development. Proc. Natl. Acad. Sci. U.S.A. 96, 14372–14377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z. B.; Popovic R.; Chen J.; Theisler C.; Stuart T.; Santillan D. A.; Erfurth F.; Diaz M. O.; Zeleznik-Le N. J. (2005) The MLL fusion gene, MLL-AF4, regulates cyclin-dependent kinase inhibitor CDKN1B (p27kip1) expression. Proc. Natl. Acad. Sci. U.S.A. 102, 14028–14033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner S.; Birke M.; Garcia-Cuellar M. P.; Zilles O.; Greil J.; Slany R. K. (2001) MLL-ENL causes a reversible and myc-dependent block of myelomonocytic cell differentiation. Cancer Res. 61, 6480–6486. [PubMed] [Google Scholar]
- Sierra J.; Yoshida T.; Joazeiro C. A.; Jones K. A. (2006) The APC tumor suppressor counteracts β-catenin activation and H3K4 methylation at Wnt target genes. Genes Dev. 20, 586–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson C. J.; Santos-Rosa H.; Kouzarides T. (2006) Proline isomerization of histone H3 regulates lysine methylation and gene expression. Cell 126, 905–916. [DOI] [PubMed] [Google Scholar]
- Org T.; Chignola F.; Hetenyi C.; Gaetani M.; Rebane A.; Liiv I.; Maran U.; Mollica L.; Bottomley M. J.; Musco G.; Peterson P. (2008) The autoimmune regulator PHD finger binds to non-methylated histone H3K4 to activate gene expression. EMBO Rep. 9, 370–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna S. D.; Ilin S.; Rogers R. S.; Tanny J. C.; Lavender H.; Li H.; Baker L.; Boyle J.; Blair L. P.; Chait B. T.; Patel D. J.; Aitchison J. D.; Tackett A. J.; Allis C. D. (2006) Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol. Cell 24, 785–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer G.; Bang H.; Berger E.; Schellenberger A. (1984) Conformational specificity of chymotrypsin toward proline-containing substrates. Biochim. Biophys. Acta 791, 87–97. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.