Summary
The Saccharomyces cerevisiae Yap1p and Skn7p transcription factors collaborate in the activation of oxidative stress response (OSR) genes. Although Yap1p and Skn7p oxidative stress response elements (YRE, OSRE) have been characterized and identified in some OSR genes, many OSR genes lack such elements. In this study, the complex, oxidative responsive, CCP1 promoter was used as a model to investigate the cis-acting elements responsible for activation by oxidative stress. In addition to consensus YRE and OSRE sequences, novel Yap1p and Skn7p binding sites were identified in the CCP1 promoter. These new sites were found to mediate Yap1p-and Skn7p-dependent activation of OSR genes including TSA1 and CTT1 previously thought to lack Yap1p and Skn7p binding sites. The novel YREs and OSREs were found to be enriched in the promoter regions of a set of 179 OSR genes. The widespread existence of novel Yap1p and Skn7p binding sites strongly suggest that direct binding of Yap1p and Skn7p is responsible for activation of many more OSR genes than previously believed.
Introduction
Aerobic organisms are continuously challenged by the formation of reactive oxygen species (ROS) that arise from an incomplete reduction of molecular oxygen during respiration. ROS are responsible for a wide range of intracellular damage to DNA, proteins and cellular structures (Moradas-Ferreira et al., 1996). Saccharomyces cerevisiae has multiple mechanisms for responding to oxidative stress and maintaining a reduced intracellular environment. The results of global gene expression experiments identified 579 genes that are upregulated in response to treatment with 0.32 mM H2O2 (20 min) (Gasch et al., 2000). This rapid and widespread genomic response suggests transcriptional regulation plays a major role in the adaptive response to oxidative stress. In S. cerevisiae, the transcription factors Yap1p and Skn7p regulate a subset of those genes directly (Morgan et al., 1997; Lee et al., 1999a).
Yap1p is a member of the c-jun family of proteins that, together with fos, comprise the AP1 transcription factor of higher eukaryotes. Yap1p contains a basic leucine zipper (bZIP) DNA-binding domain at the N-terminus. Yap1p was first identified as a functional homologue of mammalian AP-1 on the basis of its ability to bind to an AP-1 response element (ARE, TGACTAA) from SV40 and to activate transcription from an ARE-driven reporter gene (Harshman et al., 1988). Fernandes et al. (1997) showed that the preferred Yap1p binding site is TTACTAA, consisting of two inverted TTA half-sites. The Yap1p response element (YRE) differs at one position from the original SV40 AP-1 binding site TGACTAA, and at two positions from the optimal mammalian AP-1 and yeast GCN4 binding site TGACTCA (Fernandes et al., 1997).
The role of Yap1p in the oxidative stress response (OSR) is apparent from the hydrogen peroxide sensitive phenotype of yap1 null mutants (Schnell et al., 1992). A YRE was first identified in the promoters of TRX2 encoding thioredoxin (Kuge and Jones, 1994) and GSH1 encoding γ-glutamylcysteine synthetase (Wu and Moye-Rowley, 1994). Many more Yap1p target genes have as been identified, and the consensus YRE is reported to be TT/GAC/ GTAA (Toone and Jones, 1999). However, many known Yap1p target genes do not have a YRE conforming to this consensus in their promoter regions (Krems et al., 1996).
SKN7 encodes a response regulator containing a receiver domain typical of two-component signal transduction systems and similarity to the DNA-binding domain of heat shock factor (Hsf1p) (Brown et al., 1993; Morgan et al., 1995). A phospho-accepting aspartate residue (D427) in the receiver domain is required for activation of Skn7p through the SLN1-SKN7 pathway (Ketela et al., 1998; Li et al., 1998). Expression of cell wall and cell cycle genes are dependent on Skn7p D427 indicating the involvement of the SLN1-SKN7 pathway in these processes (Brown et al., 1993; 1994; Morgan et al., 1995; Li et al., 1998; Raitt et al., 2000).
In addition to its role in the SLN1 pathway, the Skn7 protein is required for the yeast OSR (Krems et al., 1996). The oxidative stress function of Skn7p is independent of the receiver domain D427 (Morgan et al., 1997). The OSR gene, TRX2, encoding thioredoxin, is directly regulated by the Skn7p and Yap1p transcription factors (Morgan et al., 1997). Both proteins bind directly to the TRX2 promoter to induce the expression of TRX2 in response to oxidant. The Skn7p binding site in TRX2 was localized to a region adjacent to a consensus YRE, which includes a GGCTGGC sequence later identified (GGCCGGC) in the promoter region of GPX2 as the Skn7p oxidative stress response element (OSRE) that mediates expression in response to oxidative stress (Tsuzi et al., 2004).
A repeated sequence in the promoter of OCH1 that is bound by Skn7p and mediates responsiveness to SLN1 activating (sln1*) alleles was previously identified (Li et al., 2002). The so-called ‘sln1* response element’ (SSRE) in the OCH1 promoter (ATTTGGC n1–2 GGC) is not responsive to oxidative stress. Interestingly, the Skn7p binding sites in TRX2, GPX2 and OCH1 share the GGC repeat motif, which may be an important feature of Skn7p binding sites.
In addition to Yap1p and Skn7p, additional transcription factors, such as Msn2p, Msn4p and Hsf1p, are known to contribute to the OSR of specific genes (Martinez-Pastor et al., 1996). The induction of CTT1 (Schuller et al., 1994) and AHP1 by hydrogen peroxide, for example, requires the Msn2/4 transcription factors and the STRE (stress response) binding sites in addition to Yap1p and Skn7p (Winderickx et al., 1996; Lee et al., 1999a; Rep et al., 2001). Interestingly, there are no known YREs or OSREs in the promoters of these genes. The TSA1 promoter likewise lacks YREs, although Yap1p and Skn7p are known to bind to it (Lee et al., 1999a). These results suggest the possible existence of novel YRE and OSRE sequences in addition to known elements.
The S. cerevisiae CCP1 gene encoding mitochondrial cytochrome c peroxidase, is a Yap1p- and Skn7p-dependent OSR gene (Charizanis et al., 1999). The OSR of CCP1 can not be fully explained by known Yap1p and Skn7p binding sites in the CCP1 promoter. In this study, analysis of the CCP1 promoter revealed novel binding sites involved in regulating the OSR. The presence of these sites in OSR genes previously thought to lack Yap1p and Skn7p elements helps to unify mechanisms of OSR gene regulation.
Results
Localization of the OSR elements in the CCP1 promoter
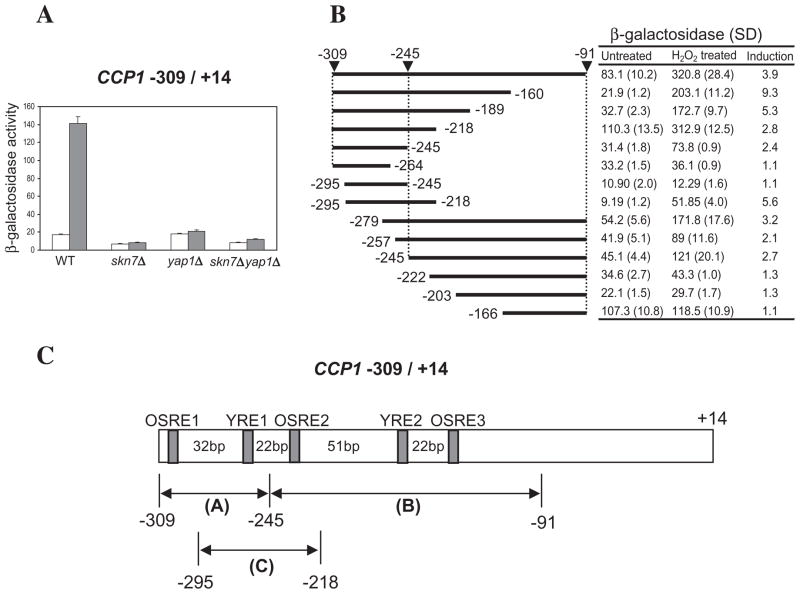
The OSR gene, CCP1, is induced by hydrogen peroxide in a Yap1p- and Skn7p-dependent manner (Lee et al., 1999a). Sequences between −309 and +14 of the CCP1 promoter are sufficient for this response as an in-frame fusion of CCP1 −309 to +14 with the Escherichia coli lacZ gene also exhibited Skn7p and Yap1p induction in response to hydrogen peroxide (Fig. 1A).
Fig. 1.
Induction of CCP1 by hydrogen peroxide maps to three upstream regions and is Skn7p- and Yap1p-dependent.
A. Wild type (WT) (JF1565), skn7Δ (JF1904), yap1Δ (JF2272) and skn7Δyap1Δ (JF2312) strains carrying the −309 to +14 CCP1–lacZ reporter plasmid were untreated (white bars) or treated with 0.3 mM H2O2 (shaded bars) for 30 min.
B. Localization of the oxidative stress response elements in the CCP1 promoter. Deletions of the CCP1−309 to −91-OCH1TATA–lacZ reporter were introduced into the skn7Δ strain, JF1904, carrying the SKN7 expression plasmid, pSL232. Transformants were untreated or treated with 0.3 mM H2O2 for 30 min. β-Galactosidase activities are given in Miller units (Miller, 1972) and are the average activities of three or four independent transformants. SD, standard deviation.
C. Two Yap1p response elements (YRE1, YRE2) and three Skn7p response elements (OSRE1, OSRE2, OSRE3) identified in this study are shown in the model of the CCP1 promoter. Distances between elements are shown in bp.
CCP1 −309 to −91 fused to minimal promoter sequences from the OCH1 gene (Li et al., 2002) likewise directed an OSR (Fig. 1B) that was Skn7p- and Yap1p-dependent (data not shown) indicating that regulatory features of the CCP1 gene are not dependent on any specific TATA box. To localize the regulatory elements responsible for an oxidative response, deletions of the functional CCP1−309 to −91-OCH1TATA–lacZ reporter were tested for activation following oxidant treatment. Three separate regions (−309 to −245, −245 to −91 and −295 to −218; A, B and C, respectively, in Fig. 1C) of the CCP1 promoter were capable of independently mediating induction by hydrogen peroxide.
Analysis of the OSR elements in CCP1 −245 to −91 (B) fragment
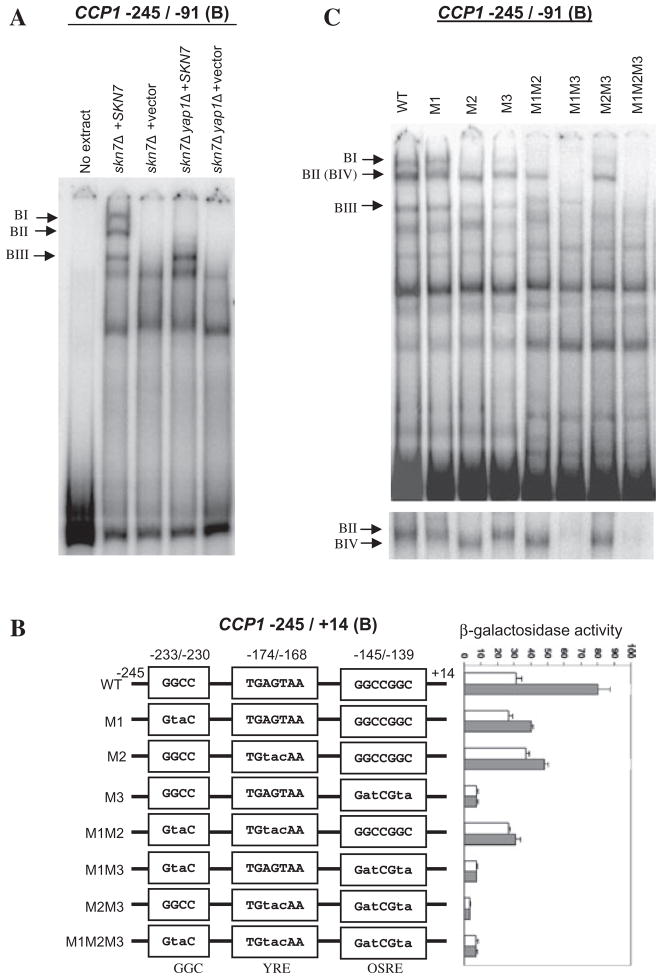
Inspection of fragment B revealed the presence of YRE and OSRE consensus sequences (labelled YRE2 and OSRE3 in Fig. 1C). Using electrophoretic mobility shift assays three Yap1p- or Skn7p-dependent complexes were detected (Fig. 2A). Two complexes (BI and BII) were dependent on the presence of both Yap1p and Skn7p, and the third complex (BIII) was dependent on Skn7p only.
Fig. 2.
Identification of three elements between −245 and −91 (fragment B) of CCP1 that contribute to the oxidative stress response.
A. Electrophoretic mobility shift assays showing the formation of Skn7p- and Yap1p-dependent complexes with CCP1 promoter fragment B. Extracts prepared from skn7Δ (JF1904) and skn7Δyap1Δ (JF2312) strains carrying the SKN7 plasmid pSL232 (Li et al., 1998) or empty vector, pRS315, were mixed with labelled probe corresponding to −245 to −91 (fragment B) of the CCP1 promoter. Complexes were separated by non-denaturing polyacrylamide gel electrophoresis at 200 V for 6 h. Skn7p- and Yap1p-dependent complexes are labelled to the left.
B. The sequence of the GGC-containing sequence (−233/−230), the potential YRE (−174/−168) and OSRE (−145/−139) are shown in three separate boxes in the line drawing. The sequence changes in mutants M1, M2 and M3 are indicated by lower case letters. Each fragment was fused to the lacZ gene in pSEYC102 (gift from S. Moye-Rowley, University of Iowa) and the activity of each reporter construct was assayed in the skn7Δ strain (JF1904) carrying the SKN7 expression plasmid, pSL232 (Li et al., 1998). Strains were untreated (white bars) or treated with 0.3 mM H2O2 (shaded bars). β-Galactosidase activity is given in Miller units (Miller, 1972) and is the average of three or four independent transformants. Error bars represent the standard deviation of the mean.
C. Electrophoretic mobility shift assays using fragments containing wild-type or mutated (M1, M2, M3) CCP1 promoter sequences. Extracts were from the skn7Δ strain (JF1904) carrying the SKN7 expression plasmid, pSL232 (Li et al., 1998). Skn7p- and Yap1p-dependent complexes are labelled to the left. The image shown at the bottom was from a gel run for 6 h to resolve complexes II and IV.
Interestingly, results of β-galactosidase assays (Fig. 1B) showed a reduction in OSR from 2.7- to 1.3-fold using a CCP1 −222 to −91 reporter, even though the putative YRE and OSRE are still present. No known cis-acting elements are present in the 24 bp between −222 and −245; however, the presence of a GGC motif seemed potentially important given its relationship to the previously published GGCC/TGGC OSRE (Tsuzi et al., 2004). To examine the role of the upstream GGC-containing sequence, mutations were generated in all three possible elements: the YRE, the OSRE and the GGC-containing sequence. As expected, mutations in the putative YRE or OSRE reduced the OSR (Fig. 2B, M2 and M3). Mutation of the upstream GGC sequence (M1) also reduced the oxidative stress responsiveness from 2.6- to 1.5-fold, suggesting the importance of the GGC motif for a maximal response (Fig. 2B, M1). The roles of the putative YRE, OSRE and the GGC sequences in the formation of the Yap1p- and Skn7p-dependent DNA–protein complex were investigated by electrophoretic mobility shift assay (Fig. 2C). Four complexes were evident on various fragment B probes. Formation of complex BI required the presence of both the YRE and the OSRE and is presumed to contain both Skn7p and Yap1p. Prolonged electrophoresis revealed that the complex labelled BII is actually two closely migrating complexes, BII and BIV, seen in different lanes (Fig. 2C). Formation of complex BII required a YRE and either the OSRE or the GGC element, while the presence of complex BIV was restricted to probes lacking the YRE but retaining either the OSRE or GGC element (Fig. 2C, bottom). Based on these observations complex BIV is presumed to contain Skn7p but not Yap1p, and BII is presumed, like BI, to contain both proteins. Although no effect of the GGC mutation was observed using probes containing the M1 mutation alone, probes containing both GGC and OSRE mutations (Fig. 2C, M1, M3) eliminated complex BII. The effect of the GGC mutation in the presence of an OSRE mutation and not in the presence of a YRE mutation (Fig. 2C, M1, M2) indicates that the functions of the GGC motif and the OSRE sequences are partially redundant. In summary, analysis of fragment B revealed a conventional YRE, a conventional OSRE and a novel OSRE.
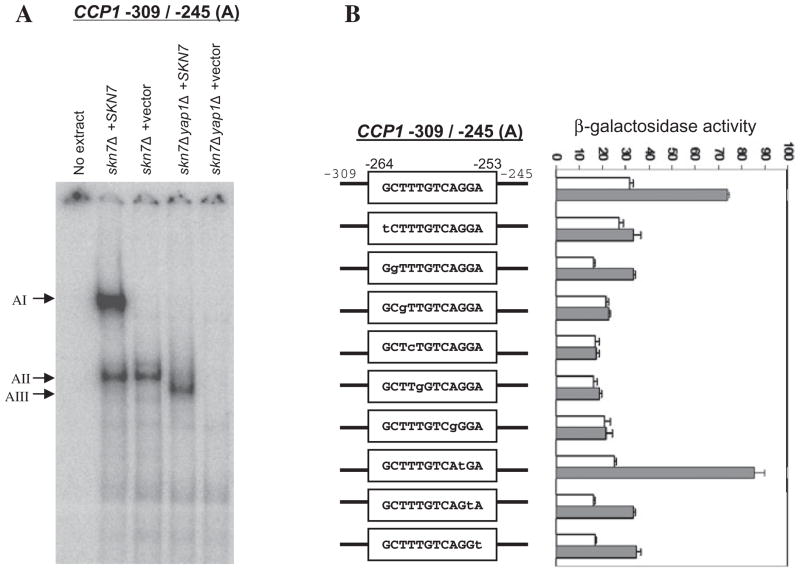
Identification of a novel YRE in −309 to −245 (A) CCP1 sequence
The Yap1p- and Skn7p-dependent OSR in CCP1 −309 to −245 (A) is more complex. Although this fragment lacks known Yap1p and Skn7p binding sites, two Yap1p- or Skn7p-dependent complexes were identified with a fragment A probe (Fig. 3A). Complex AI was dependent on both Yap1p and Skn7p and Complex AII was dependent on Yap1p but not Skn7p. In strains lacking Yap1p, a faster migrating Skn7p-dependent species, AIII, was observed. The fact that the Yap1p complex, AII, was observed in the presence of Skn7p while the Skn7p complex, AIII, was observed only in the absence of Yap1p suggests that the presence of Yap1p drives Skn7p into a higher-order complex (AI).
Fig. 3.
Identification of a novel YRE in the −309 to −245 region (fragment A) of the CCP1 promoter.
A. Electrophoretic mobility shift assays showing the formation of Skn7p- and Yap1p-dependent complexes with CCP1 promoter fragment A. Extracts prepared from skn7Δ (JF1904) and skn7Δyap1Δ (JF2312) strains carrying the SKN7 plasmid pSL232 (Li et al., 1998) or empty vector, pRS315, were mixed with labelled probe (fragment A) of the CCP1 promoter. Complexes were separated by non-denaturing polyacrylamide gel electrophoresis at 200 V for 2 h. Skn7p- and Yap1p-dependent complexes are labelled to the left.
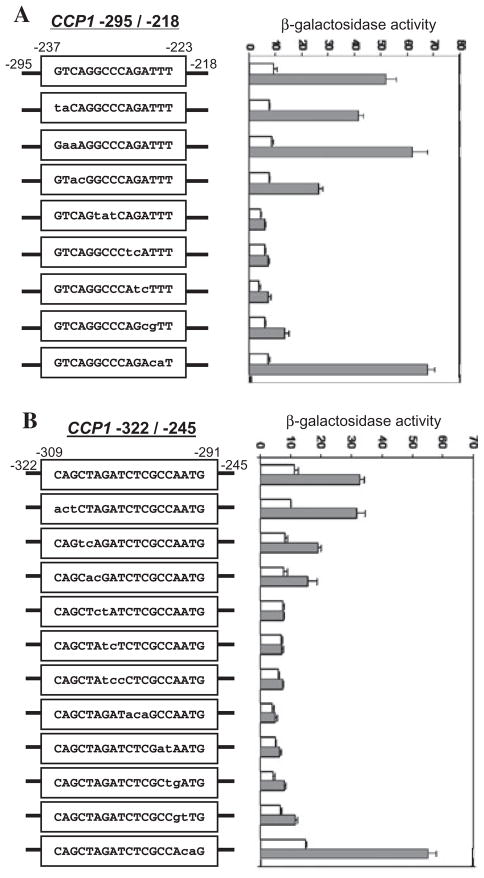
B. Activities of wild type and mutant CCP1−309 to −245-OCH1TATA–lacZ reporters. Reporters were introduced into the skn7Δ strain (JF1904) containing the SKN7 expression plasmid, pSL232. Wild type and mutated sequences between −237 and −223 within the −309 to −245 region of the CCP1 promoter are shown in boxes in the line drawing. Transformants were untreated (white bars) or treated with 0.3 mM H2O2 (shaded bars). β-Galactosidase activity is represented in Miller units (Miller, 1972) and is the average of three or four independent transformants. Error bars are the standard deviation of the mean.
The OSR elements in this sequence mapped to the two ends of fragment A as reporters consisting of CCP1 −309 to −264 and −295 to −245 were not oxidative stress responsive (Fig. 1B). Site-directed mutations in the CCP1−309 to −245-OCH1TATA–lacZ reporter identified the importance of the 7 bp core sequence (TTTGTCA) between −262 and −256 for a normal OSR (Fig. 3B). The complement of TTTGTCA (TGACAAA), differs from the YRE, TGACTAA only at the fifth base pair (from T to A). Mutations within the TGACAAA sequence abolished the OSR, whereas mutations outside of the TGACAAA motif were less important (Fig. 3B).
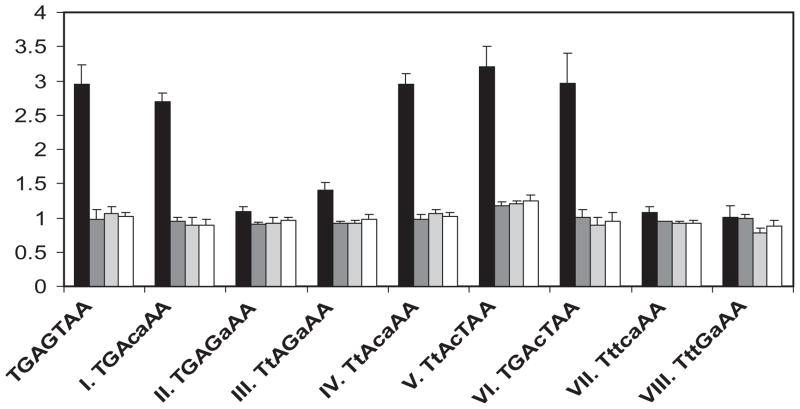
Identification of YRE variants
The functionality of a TGACAAA sequence having an A rather than a T at position 5 was evaluated by substituting it for the consensus YRE (TGAGTAA) in CCP1 fragment B. The activity of this variant was indistinguishable from that of the original CCP1 promoter sequence (Fig. 4, variant I), and like the original sequence, variant I activity was dependent on the presence of both Skn7p and Yap1p. As the consensus YRE sequence, TT/GAC/GTAA, is flexible in the second and the forth positions, additional variants were constructed to determine whether the second and the forth base pair remain flexible in the context of a variant I YRE. TTACAAA (variant IV) was found to be as active as the consensus YRE and as variant I (Fig. 4), while TTAGAAA (variant III) was only partially active, and TGAGAAA (variant II) exhibited no detectable OSR. The results indicate that the second position in a variant I YRE can be either T or G, but the fourth position has a strong preference for C.
Fig. 4.
Identification of different YRE variants. The consensus YRE (TGAGTAA) located within the −245 to +14 region of the CCP1 promoter was replaced by the different potential YRE sequences as shown below the bar graph. Reporter plasmids were introduced into the wild type (JF1565, black bars), skn7Δ (JF1904, dark grey bars), yap1Δ (JF2272, light grey bars) and skn7Δ yap1Δ (JF2312, white bars) strains. Transformants were untreated or treated with 0.3 mM H2O2 and the average fold induction by hydrogen peroxide for each of the four strains is shown as four consecutive bars for each sequence. Error bars are the standard deviation of the mean.
The existence of two TTA inverted half-sites is an important characteristic of the optimal YRE, TTACTAA. One of the half-sites can be substituted by TGA (Fernandes et al., 1997). However, when the two half-sites of YRE are simultaneously substituted with TGA to generate TGAC/GTCA, the sequence binds a different AP-1 protein, Gcn4p (Ellenberger et al., 1992). In variant I, one of the YRE half sites is TTT. To determine whether the symmetric site TTTC/GAAA consisting of two identical inverted half-sites (TTT) is still a functional YRE, the consensus YRE (TGAGTAA) in the CCP1 promoter (−245 to +14) was replaced with TTTCAAA or TTTGAAA. These substitutions completely eliminated the OSR (variants VII and VIII in Fig. 4).
Identification of novel OSREs in the CCP1 promoter
Although the upstream GGC motif in fragment B was not found to function with YRE2 to mediate oxidative stress (M3 in Fig. 2B), it was nonetheless found to work together with YRE1 in fragment C (−295 to −218). To determine the precise sequence of the OSRE in fragment C, site-directed mutations between −237 and −223 were evaluated. Mutations within the sequence GGCCCAGA eliminated the OSR while mutations outside the sequence had only minor effects on the response (Fig. 5A). Hence the GGC motif with OSRE-like function in fragment B is part of the larger motif, GGCCCAGA.
Fig. 5.
Identification of two novel OSREs in the CCP1 promoter. The original sequence and mutant derivatives are shown in boxes to the left. CCP1-OCH1TATA–lacZ reporter plasmids were introduced into the skn7Δ strain (JF1904) containing the SKN7 expression plasmid, pSL232. Transformants were untreated (white bars) or treated with 0.3 mM H2O2 (shaded bars). β-Galactosidase activity is represented in Miller units (Miller, 1972) and is the average of three or four independent transformants for each strain. Error bars show the standard deviation of the mean.
A. CCP1 sequence −237 to −223.
B. CCP1 sequence −309 to −291.
The −309 to −295 region was also required for oxidative stress responsiveness (Fig. 1B). A series of mutations covering this region were constructed within a CCP1 −322 to −245 reporter. The sequence GGCGAGATCT (the complement of −304 to −295) was found to be important for activity (Fig. 5B).
Validation of OSRE and YRE function in CCP1
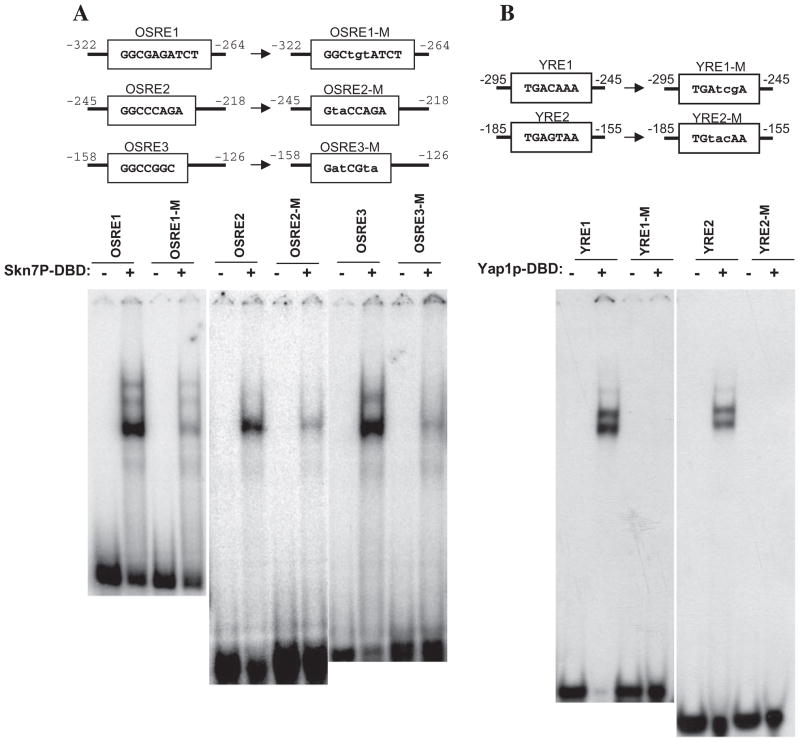
In summary, analysis of the CCP1 promoter revealed two putative Yap1p binding sites and three putative Skn7p binding sites (Fig. 1C). At least one YRE and one OSRE were present in each fragment capable of mediating an OSR implying that binding of Skn7p and Yap1p to a variety of cis-acting elements may be required for the function of these transcription factors in the CCP1 promoter. To evaluate whether Skn7p and Yap1p can bind directly to the CCP1 promoter, electrophoretic mobility shift experiments were carried out with recombinant Skn7p and Yap1p proteins. Binding of recombinant Skn7p to each OSRE and of recombinant Yap1p to each YRE was readily detected indicating that each of these sequences can be recognized by the respective proteins (Fig. 6). Skn7p and Yap1p binding was diminished when probes contained mutations in each of the five elements, consistent with the conclusion that the novel elements, OSRE1 and OSRE2 and YRE1 identified by reporter analysis, do in fact represent alternative Skn7p and Yap1p binding sites.
Fig. 6.
Electrophoretic mobility shift assays showing direct binding of Skn7p and Yap1p to the five oxidative stress response elements identified in CCP1. The size of the oligonucleotide probes and the sequences corresponding to Skn7p or Yap1p response elements in the CCP1 promoter (−322 to −264, −245 to −218, −158 to −126, −275 to −245 and −185 to −155) and mutant derivatives are shown. Five micrograms of purified Skn7p-DBD (A) or Yap1p-DBD (B) were used in each binding reaction. Complexes were separated by non-denaturing polyacrylamide gel electrophoresis at 200 V for 2 h.
The contribution of each element to CCP1 regulation in the native promoter was evaluated by RNA analysis of strains carrying a CCP1 expression plasmid including the entire CCP1 coding region and upstream sequences to −756 into which single mutations and combinations of mutations had been introduced. Transcript levels were determined at various times following oxidative stress treatment. In this analysis, individual mutations in OSRE1, OSRE2 and OSRE3 had modest effects on induction; pairs of mutations in the OSREs had more substantive effects on induction and a strain with mutations in all three OSREs were virtually non-inducible (Fig. 7A). Individual and paired mutations in the YREs, in contrast, retained substantial inducibility at early times, thus revealing a YRE-independent component to the CCP1 OSR. As induction was nonetheless entirely Yap1p-dependent (Fig. 7B), these results suggest that Yap1p regulation of CCP1 is not strictly dependent on the presence of YREs.
Fig. 7.
Northern (RNA) hybridization analysis of transcript levels in wild type and regulatory mutants of CCP1 following oxidant treatment.
A. Log-phase cultures of the ccp1 deletion strain, JF2374 (WT) carrying the CCP1 expression plasmid, pXH1811 or its derivatives (pXH1812-pXH1820) all of which include the CCP1 ORF as well as sequences out to −756 were treated (5–120′) or left untreated (0) with 0.3 mM H2O2 for the indicated times. Mutations in each construct are indicated by the diagram to the left.
B. Log-phase cultures of the ccp1 deletion strains, JF2374 (WT), JF2375 (skn7Δ), JF2376 (yap1Δ) and JF2377 (skn7Δyap1Δ) carrying the CCP1 expression plasmid, pXH1811 were treated (5–120′) or left untreated (0) with 0.3 mM H2O2 for the indicated times.
Equal loading of RNA in each lane in (A) and (B) was assured by examining ethidium bromide staining and by hybridization to PGK1 as a normalization probe (not shown).
YRE and OSRE variants mediate activation in other OSR genes
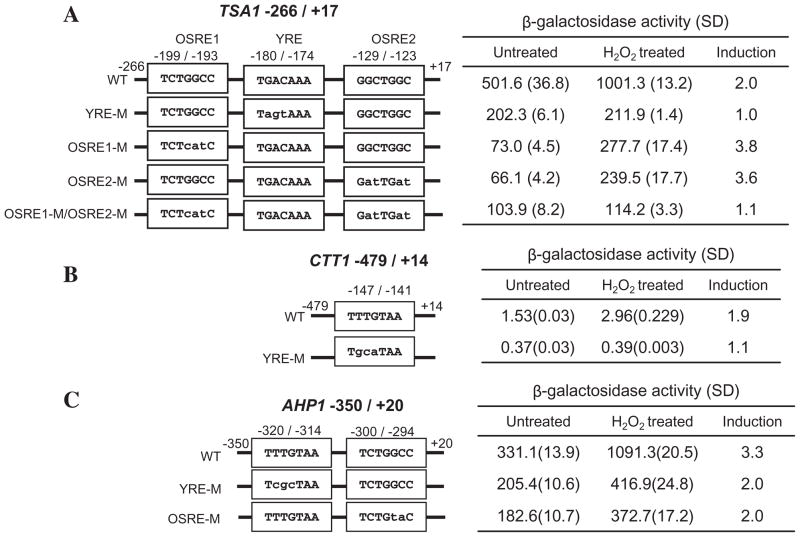
Yap1p response element (YRE) and OSRE variants were found in OSR genes in addition to CCP1 (Table 1). For example, the TSA1 promoter lacks a consensus YRE, but contains a variant I YRE (TGACAAA, −180 to −174), a new OSRE, GGCCAGA (−199 to −193) as well as a previously reported OSRE (GGCTGGC, −129 to −123) (Tsuzi et al., 2004). Mutation of the variant I YRE from TGACAAA to TAGTAAA abolished the OSR. Likewise, the TSA1 OSR was completely abolished when both OSREs were simultaneously mutated (OSRE1-M/OSRE2-M) (Fig. 8A).
Table 1.
Oxidative stress response elements characterized in this study.
| Element | Source | Sequence |
|---|---|---|
| YRE1 | CCP1 −256 to −262 | TGACAAA |
| YRE2 | CCP1 −174 to −168 | TGAGTAA |
| YRE | TSA1 −180 to −174 | TGACAAA |
| YRE | CTT1 −141 to −147 | TTACAAA |
| YRE | AHP1 −320 to −314 | TTACAAA |
| OSRE1 | CCP1 −295 to −304 | GGCGAGATCT |
| OSRE2 | CCP1 −233 to −226 | GGCCCAGA |
| OSRE3 | CCP1 −145 to −139 | GGCCGGC |
| OSRE | TSA1 −199 to −193 | GGCCAGA |
| OSRE | TSA1 −129 to −123 | GGCTGGC |
| OSRE | AHP1 −300 to −294 | GGCCAGA |
Fig. 8.
Identification of oxidative stress response elements in TSA1, AHP1 and CTT1.
A. The sequences of potential oxidative stress response elements (YRE, TGACAAA; OSRE1, GGCCAGA; OSRE2, GGCTGGC) found within the −266 to +17 region of the TSA1 promoter are shown in boxes in the line drawing. The wild-type sequence and mutant derivatives (YRE-M, OSRE1-M and OSRE2-M and OSRE1-M/OSRE2-M) were fused to the lacZ gene and each reporter plasmid introduced into JF1565. Transformants were untreated or treated with 0.3 mM H2O2.
B. Identification of Yap1 response elements in the −479 to +14 region of the CTT1 promoter. A single potential YRE (TTACAAA) seen in the CTT1 sequence was mutated as shown in the figure.
C. Identification of Yap1 and Skn7p response elements in the −350 to +20 region of the AHP1 promoter. One potential YRE (TTACAAA) and one potential OSRE (GGCCAGA) within this region of AHP1 were mutated. β-Galactosidase activity is represented in Miller units (Miller, 1972) and is the average of three or four independent transformants for each strain.
SD, standard deviation of the mean.
The CTT1 promoter also lacks a consensus YRE sequences but has a variant IV YRE. Reporters containing CTT1 −479 to +14 mediated a Yap1p- and Skn7p-dependent OSR that was significantly reduced when TTACAAA (from −147 to −141) was mutated to TTATGCA (Fig. 8B). Hence, TTACAAA is required for the CTT1 OSR. Finally, although a consensus YRE is present at −484 of the AHP1 promoter (Lee et al., 1999b), the region between −350 and +20 was found to exhibit a Yap1p and Skn7p OSR. This region lacks consensus YRE or OSRE sequences, but contains a variant IV YRE (TTACAAA, −320 to −314) and a novel OSRE (GGCCAGA, −300 to −294). Mutations in either the TTACAAA YRE-like element or the putative OSRE, GGCCAGA reduced oxidative stress induction from threefold to twofold (Fig. 8C). The residual OSR induction of AHP1 may be due to nearby STREs (Rep et al., 2001).
YRE and OSRE are enriched among OSR genes in S. cerevisiae
Statistical analysis of the distribution of YRE and OSRE in the promoters (−500 to −100) of S. cerevisiae genes was carried out using the pattern-matching program (SGD, http://db.yeastgenome.org/cgi-bin/PATMATCH/nph-patmatch). To determine whether consensus and novel YREs and OSREs are over-represented in the promoters of OSR genes, the frequency of these elements in the promoters of a set of 179 genes induced at 10 min and 20 min of H2O2 treatment (Gasch et al., 2000) was compared with the frequency of the same elements in the promoters of the 5888 hypothetical and known genes of the yeast genome. The results (Table 2B) show that the classical YRE (TKASTAA) is enriched (P < 0.0001) in OSR genes (37%) relative to their presence in the genome (18%). Of 179 OSR genes, only 66 possess one or more classical YREs. However 110 of the 179 OSR genes contained either a consensus or novel YRE. Taken together, the OSR gene set is highly enriched for the presence of YREs in the promoter (Table 2B). The simultaneous presence of a consensus YRE and a consensus OSRE is found in only six out of 179 (3%) OSR gene promoters. However, the simultaneous presence of any YRE and OSRE (including: GGCNGGC, GGCNNGGC, GGC NAGA, GGCNNAGA) was found in 37 of 179 or 28% of OSR gene promoters (Table 2A). Hence the newly recognized YRE and OSRE sequences increase the number of genes in this OSR set whose regulation is likely to be mediated by direct binding of Yap1p and Skn7p.
Table 2.
The oxidative stress gene set is enriched for YRE and OSRE sequences.
| Elementsa | Examples | Element containing genes | Total genesb | P-valuec (Fisher’s exact test) | |
|---|---|---|---|---|---|
| A. | YRE | 2022 91 |
5888 179 |
< 0.0001** | |
| OSRE | 1766 69 |
5888 179 |
0.0165* | ||
| YRE + OSRE | 594 37 |
5888 179 |
0.0019** | ||
| B. | TKASTAA | Consensus | 1071 66 |
5888 179 |
< 0.0001** |
| TKACAAA | CCP1, TSA1, CTT1, AHP1 | 1160 44 |
5888 179 |
0.0861 | |
| C. | YRE + GGCNNGGC | CTT1 | 127 12 |
5888 179 |
0.0008** |
| YRE + GGCNGGC | TSA1, CCP1 | 114 12 |
5888 179 |
0.0003** | |
| YRE + GGCNNAGA | CCP1 | 251 13 |
5888 179 |
0.0372* | |
| YRE + GGCNAGA | TSA1, AHP1, CCP1 | 220 14 |
5888 179 |
0.0097** | |
YRE, TKASTAA and TKACAAA; OSRE, GGCNGGC, GGCNNGGC, GGCNAGA, GGCNNAGA.
5888, total number genes in genome; 179, total genes induced by H2O2 at 10 and 20 min oxidative stress gene set (Gasch et al., 2000).
P-value from Fisher’s exact test (two-tailed) with
significant,
highly significant.
Discussion
The CCP1 gene encodes a mitochondrial cytochrome c peroxidase that uses hydrogen peroxide as a terminal electron acceptor for mitochondrial respiration (Williams and Stewart, 1976; Verduyn et al., 1991). CCP1 expression is induced by hydrogen peroxide and its importance in the removal of hydrogen peroxide generated as a byproduct of respiratory metabolism is evident from the hydrogen peroxide sensitivity of the deletion mutant (Charizanis et al., 1999; Lee et al., 1999a; Kwon et al., 2003). Ccp1p was also isolated in a screen for mutants that fail to activate Skn7p-dependent OSR and is therefore a candidate for the sensor responsible for the oxidative stress activation of Skn7p. This function of Ccp1p is independent of its peroxidase activity, as a point mutant in which electron flux between cytochrome c and cytochrome c peroxidase was reduced 104-fold nonetheless retained the ability to activate Skn7p (Charizanis et al., 1999). The complexity of the CCP1 promoter may be required for the multiple functions of Ccp1p in the OSR.
A new class of YRE
Due to the amino acid sequence similarity in the bZIP domain, AP-1 family members are expected to have related recognition sites. Fernandes et al. (1997) showed that the preferred Yap1p binding site is the sequence TTACTAA, which contains two inverted TTA half-sites, and differs from the AP-1 binding site (ARE, TGACTCA) at two positions. Native YREs were found in some Yap1p target genes and the consensus YRE sequence was defined as TT/GAC/GTAA having at least one TTA half-site. Analysis of the CCP1 promoter has led to the identification of a new class of YRE (TT/GACAAA) in which the TTA half-site is dispensable (e.g. variant 1 in Fig. 4). In the novel YRE, the second base pair remains flexible (G or T); however, the fourth base pair becomes inflexible. Substitution of C with G at position 4 significantly reduced oxidative stress responsiveness (compare variants I and II in Fig. 4). Molecular modelling suggests that the Yap1p-specific residues make novel contacts and cause physical constraints at the second and the sixth base pairs that may account for the distinct DNA-binding specificities of Yap1p and the other AP-1 proteins (Ellenberger et al., 1992; Fernandes et al., 1997). Characterization of the novel YRE suggests that the other base pairs in the element may also be involved in Yap1p recognition.
Although the activation of many OSR genes requires the presence of Yap1p and Skn7p, only a small fraction possess previously characterized Yap1p and Skn7p binding sites. The discovery of novel YREs and OSREs increases the size of the gene set known to be under the direct regulation of Yap1p and Skn7p. In one study, 14 out of 32 Yap1p target genes identified (Lee et al., 1999a) lacked a consensus YRE motif within 1 kb of the translation start site. The identification of new YRE variants accounts for 11 of the 14 genes in that study. TSA1 was previously shown to be a Yap1p and Skn7p target gene, but the absence of a YRE was thought to indicate possible indirect regulation (Lee et al., 1999a). The YRE variant (TGACAAA) identified in the CCP1 promoter was also found in TSA1 promoter, and the function of the YRE in TSA1 was confirmed both by reporter assays (Fig. 8) and by DNA binding studies (data not shown). Likewise, the existence of the functional YRE variant (TTACAAA) in CTT1 (Fig. 8) suggests that Yap1p may directly bind the CTT1 promoter.
A new class of OSRE
In the CCP1 promoter three OSREs were identified, only one of which is identical to the OSRE previously defined in GPX2 (Tsuzi et al., 2004). The precise sequences of the two new OSREs as defined by site-directed mutagenesis and reporter assays was found to be GGCCCAGA and GGCGAGATCT. Each site is capable of working with a nearby YRE to mediate an OSR. The heretofore unrecognized flexibility in Skn7p binding sites suggests that the Yap1p- and Skn7p-dependent OSR pathway is a more general mechanism in regulating the OSR in S. cerevisiae than previously appreciated. The two new OSREs in CCP1 are characterized by the presence of one GGC and one AGA triplet. This differs from the published OSRE (GGCC/TGGC) which is characterized by a set of direct GGC repeats. The importance of the AGA triplet in Skn7p binding and activation will require further analysis; however, the observation that this sequence is important for the OSR, and can be bound both by recombinant Skn7p (Fig. 6) and by Skn7p in extracts (data not shown) is consistent with a physiological role for this new sequence. Recombinant Skn7p was previously shown to bind to heat shock elements (HSEs) (Raitt et al., 2000), but the HSE-like elements in the CCP1 promoter were tested and found to be irrelevant to the OSR elements (data not shown).
Although the novel OSREs account for the activation of many Yap1p- and Skn7p-dependent OSR genes, there are still genes that appear to lack such sites suggesting the potential existence of additional Skn7p response element variants. For example, no OSREs were found in the CTT1 promoter, although a Yap1p- and Skn7p-dependent band can be detected by electophoretic mobility shift analysis (EMSA) (data not shown). In this case, the sequence GGCTAGGC, similar to one of the previously characterized Skn7p binding sites (SSRE-B, GGCTGGCC) in the SLN1-SKN7 target gene, OCH1 (Li et al., 2002), may be important. Although the presence of two base pairs between the two GGC motifs in this sequence differs from the OSRE3 sequence in CCP1 which has a single base pair between two GGC motifs, the sequence nonetheless binds Skn7p (Li et al., 2002) and may be another OSRE variant. The CTT1 (SSRE-B) sequence and additional novel OSRE sequences are further predicted by the analysis of their frequencies in the promoters of OSR genes relative to their frequencies in the genome. Table 2C shows that the sequence GGCNNGGC in CTT1 is significantly over-represented (P = 0.0008) among OSR genes compared with the genome, consistent with its possible function as an OSRE.
As expected, the GGCNGGC sequence found in TSA1 and CCP1 is also over-represented (P = 0.0003) among YRE containing OSR genes. Finally, the GGCNAGA sequence found in CCP1 (OSRE1), AHP1 and TSA1, and the GGCNNAGA sequence found in CCP1 (OSRE2) are both enriched among OSR genes (GGCNAGA, P = 0.0097) (GGCNNAGA, P = 0.0372).
Conservation of novel elements across fungal species
As bona fide regulatory elements are expected to be conserved across closely related fungi, orthologous sequences from Saccharomyces bayanus, S. mikatae and S. paradoxus were aligned with the S. cerevisiae CCP1 sequence to examine the conservation of the novel YRE and OSRE elements. Sequence identity in the region spanning −100 and −400 in the four species was 33%, while sequence identity within the sites ranged from 50% to 83% with the exception of YRE2 which was not conserved. Interestingly, when S. mikatae was removed from the alignment, total sequence identity among the three species was 42% and sequence identities within each of the five sites, including YRE2 ranged from 62% to 87%. The YRE and OSRE sites identified in CTT1 and AHP1 were similarly conserved.
Alterations in Skn7p–DNA complexes by Yap1p
Activation of the Yap1p- and Skn7p-dependent OSR genes involves the simultaneous presence of a YRE and OSRE. Fragment B of CCP1 contains one YRE (YRE2) and two OSREs (OSRE2 and OSRE3) (Fig. 1). Interestingly, the presence of a YRE in the promoter appeared to interfere with the formation of the Skn7p-dependent DNA–protein complex, BIV, perhaps promoting the formation of a Yap1p/ Skn7p DNA–protein complex (BII) instead (Fig. 2). Similar results were seen with fragment A. The Skn7-dependent complex, AIII, could be detected only in the absence of Yap1p. The loss of Skn7p–DNA complexes in the presence of Yap1p may be an explanation for the absence of SLN1-SKN7 responsiveness in many OSR genes.
CCP1 regulation
The number and types of regulatory elements participating in the OSR of CCP1 is unusually complex. Fragment A contains non-consensus elements OSRE1 (GGCGAGATCT) and YRE1 (TGACAAA) while fragment B contains consensus elements as well as an upstream non-consensus OSRE. Interestingly, the two fragments behave differently in EMSA experiments. The non-consensus sites in fragment A allow formation of a Yap1p complex (AII) in the absence of Skn7p, whereas fragment B promotes Yap1p binding only in the presence of Skn7p. Analysis of the intact CCP1 promoter suggests that the OSREs play a more important role in the CCP1 OSR than do the YREs as the OSR is abolished in promoters lacking all three OSREs but remains robust despite the loss of both YREs (Fig. 7A). Although we can not rule out the possibility of additional non-consensus YREs, our extensive computational and functional analysis of this promoter, together with the complete absence of induction in strains lacking either Skn7p or Yap1p (Fig. 7B), causes us to favour a model in which the Yap1p transcription factor may be recruited to the CCP1 promoter by Skn7p.
Experimental procedures
Strains and media and yeast techniques
The strains used in this work include the wild-type strain JF1565 (MATα his3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2Δ202 canR cyhR ) and isogenic derivatives JF1904 (skn7Δ::TRP1), JF2272 (yap1Δ) and JF2312 (yap1Δ skn7Δ::TRP1). The CCP1 gene in genome was disrupted in JF1565, JF1904, JF2272 and JF2312 to produce JF2374, JF2375, JF2376 and JF2377 (ccp1Δ::KAN) respectively. Media were prepared as described by Sherman et al. (1986). Synthetic complete medium (SC-aa) lacks the specified amino acid (e.g. SC-leucine). Yeast transformation was performed using Frozen-EZ yeast Transformation II kit (ZYMO RESEARCH, USA).
Plasmids
Construction of lacZ reporter genes involved the amplification of the target DNA sequences by polymerase chain reaction (PCR) using forward and reverse primers designed to contain EcoRI and BamHI sites respectively. The PCR products were digested with EcoRI and BamHI, and cloned into the lacZ plasmids pSEYC102 or pSL1156 (Li et al., 2002). Plasmid pSEYC102 is a CEN URA3 plasmid with a 3.3 kb lacZ insert lacking the first five amino acids of the lacZ ORF (gift from S. Moye-Rowley, University of Iowa). Plasmid pSL1156 was created by insertion of the OCH1TATA sequence (−154 to +26) without any native upstream activation sequences upstream to lacZ gene in pSEYC102 (Li et al., 2002). Promoter sequences that included a TATA box region were directly fused upstream to lacZ reporter gene in pSEYC102, while those promoter sequences lacking a TATA box region were placed upstream of the OCH1TATA–lacZ fusion gene in pSL1156. The CCP1 expression plasmid pXH1811 was produced by introducing CCP1 sequence from −756 to +1336 into the CEN HIS3 plasmid, PRS303 at EcoRI and XhoI sites. Details of plasmid construction and oligonucleotide sequences are provided in Table S1.
Site-directed mutagenesis of DNA sequences
Mutations were introduced by direct amplification of target DNA with mutagenic PCR primers or by using the Quick-Change™ Site-Directed Mutagenesis Kit (Stratagene, USA) with minor modification. For site-directed mutagenesis using the QuickChange protocol, DNA fragments to be mutagenized were first cloned into pBluescript II SK (+/–). Mutagenic primers containing the desired mutation were extended using Pfu Turbo DNA Polymerase. The resulting nicked circular strands were used in transformation of DH5α supercompetent cells (Invitrogen, USA) following digestion of the parental strands with DpnI. To introduce mutations into the CCP1 expression plasmid pXH1811, different mutated CCP1 promoter sequences were amplified from the pBluescript II SK (+/–) constructs. These PCR fragments were used as primers to introduce mutations into pXH1811 using site-directed mutagenesis.
β-Galactosidase assay
Yeast cultures for β-galactosidase assays were grown in SC media at 30°C and harvested by centrifugation at about 2 × 107 cells ml−1. Before the cultures were harvested, the cultures were incubated in the absence or in the presence of 0.3 mM H2O2 for 30 min. Extracts were prepared using glass bead lysis and cleared by centrifugation (Yu and Fassler, 1993). Extract concentration was determined using the Bio-Rad protein assay. Activities were calculated in Miller units (Miller, 1972). The final data are expressed as the average of three or four independent transformants.
Expression and purification of Skn7p and Yap1p proteins
A 3× myc tag was PCR-amplified with the primers myc-F (5′-GCTATAAAGCTTTAACTAGTGCGGCCGCTACT-3′) and myc-R (5′-GCTATACTCGAGGGCCGCTCTGAGCAAAAG-3′) and inserted into pET-28a(+) (Novagen, USA) at HinDIII and XhoI restriction sites. The BamHI–SalI-digested PCR product amplified with the primers SKN7-3F (5′-GGGATCCCATAT GAGCTTTTCCACCATAAA-3′) and skn7+583R (5′-GCT TAGTCGACCTTTCCTTTGCGCCGGAATTTT-3′), and then inserted into the same pET-28a(+) in frame with both the upstream 6× histidine tag and the downstream 3× myc tag. The DNA sequence encoding the Yap1p DNA binding domain (+1 to +658) was amplified with the primers YAP1+1F (5′-ACAAGATCTATGAGTGTGTCTACCGCCAAG-3′) and YAP1+658R (5′-ACTGTCGACGTGTCTGATTTATAGGCAT AG-3′), and then digested with BglII and SalI. The digested PCR product was cloned into the pET-28a(+) at the BamHI–SalI sites in frame with both the upstream 6× histidine tag and the downstream 3× myc tag. E. coli cultures containing the expression plasmids were grown to optical density at 600 nm (OD600) of 1 at 37°C and then shifted to 16°C. IPTG was added to 0.4 mM and the culture allowed to incubate for 20 h. Cells were harvested by centrifugation, resuspended in binding buffer and subjected to 10 freeze/thaw cycles to break open the cells. The Skn7p-DBD and Yap1p-DBD proteins were isolated using the His-Bind kit (Novagen, USA). Purified proteins were examined on a 10% 29:1 acrylamide/ bisacrylamide SDS gel. The protein concentration was determined with the Bio-Rad protein assay buffer.
Electophoretic mobility shift analysis
Preparation of yeast cell extracts, protein–DNA-binding reactions and electrophoretic fractionation of complexes were performed as described previously (Yu and Fassler, 1993) with minor modifications. DNA probes were generated by PCR amplification or directly synthesized and end-labelled with T4 polynucleotide kinase and [γ-32P]-ATP. The binding reaction system (20 μl) included 10 μg of protein extract, 0.5 μg of poly(dI-dC) and 0.5–1 ng (5–20 cp) of DNA in EMSA buffer (25 mM Tris-HCl, pH 7.5, 50 mM NaCl, 2 mM EDTA, 7 mM MgCl2, 10% glycerol). When the purified protein Skn7p-DBD or Yap1p-DBD was used in EMSA, the binding reaction system contained 0.5 μg of purified protein, 25 ng μl−1 poly(dI-dC), 2 μg μl−1 CHAPS {3-(3-Cholamidopropyl)dimethylamino]-1-propanesulphonate}, 0.25% BSA and 0.5–1 ng (5–20 cp) of DNA in EMSA buffer. Reactions were incubated at 37°C for 20 min. The reaction mixes were resolved on 4% non-denaturing polyacrylamide (29:1 acrylamide:bisacrylamide) in 0.5× TBE (89 mM Tris, 89 mM borate, 2.4 mM EDTA, pH 8.0) following electrophoresis at 200 V for 2 h or 6 h where extra resolution was required. Gels were dried, exposed and subject to phosphoimage analysis.
Northern analysis
Log-phase cultures grown at 30°C were harvested and resuspended in TES buffer (10 mM Tris-HCl, 10 mM EDTA, 0.5% SDS). Total RNA samples were isolated using hot acid phenol. Twenty micrograms of each total RNA sample were loaded onto denaturing agarose gels (1.2%), and transferred onto nylon membranes. CCP1 probe was labelled with [α-32P]-dATP (Prime-It II Kit, Stratagene). Hybridyzation was carried out in PerfectHyb solution (Sigma) at 68°C overnight. Quantification was performed by phosphoimage analysis.
Supplementary Material
Acknowledgments
We thank Scott Moye-Rowley, Bob Deschenes and Orna Carmel-Harel for critical reading of this manuscript. This work was supported by the National Institutes of Health (GM56719).
Footnotes
The following supplementary material is available for this article online:
Table S1. Plasmids and oligonucleotides used in this study.
This material is available as part of the online article from http://www.blackwell-synergy.com
References
- Brown JL, North S, Bussey H. SKN7, a yeast multicopy suppressor of a mutation affecting cell wall beta-glucan assembly, encodes a product with domains homologous to prokaryotic two-component regulators and to heat shock transcription factors. J Bacteriol. 1993;175:6908–6915. doi: 10.1128/jb.175.21.6908-6915.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JL, Bussey H, Stewart RC. Yeast Skn7p functions in a eukaryotic two-component regulatory pathway. EMBO J. 1994;13:5186–5194. doi: 10.1002/j.1460-2075.1994.tb06849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charizanis C, Juhnke H, Krems B, Entian KD. The mitochondrial cytochrome c peroxidase Ccp1 of Saccharomyces cerevisiae is involved in conveying an oxidative stress signal to the transcription factor Pos9 (Skn7) Mol Gen Genet. 1999;262:437–447. doi: 10.1007/s004380051103. [DOI] [PubMed] [Google Scholar]
- Ellenberger TE, Brandl CJ, Struhl K, Harrison SC. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein–DNA complex. Cell. 1992;71:1223–1237. doi: 10.1016/s0092-8674(05)80070-4. [DOI] [PubMed] [Google Scholar]
- Fernandes L, Rodrigues-Pousada C, Struhl K. Yap, a novel family of eight bZIP proteins in Saccharomyces cerevisiae with distinct biological functions. Mol Cell Biol. 1997;17:6982–6993. doi: 10.1128/mcb.17.12.6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, et al. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harshman KD, Moye-Rowley WS, Parker CS. Transcriptional activation by the SV40 AP-1 recognition element in yeast is mediated by a factor similar to AP-1 that is distinct from GCN4. Cell. 1988;53:321–330. doi: 10.1016/0092-8674(88)90393-5. [DOI] [PubMed] [Google Scholar]
- Ketela T, Brown JL, Stewart RC, Bussey H. Yeast Skn7p activity is modulated by the Sln1p-Ypd1p osmosensor and contributes to regulation of the HOG pathway. Mol Gen Genet. 1998;259:372–378. doi: 10.1007/s004380050824. [DOI] [PubMed] [Google Scholar]
- Krems B, Charizanis C, Entian KD. The response regulator-like protein Pos9/Skn7 of Saccharomyces cerevisiae is involved in oxidative stress resistance. Curr Genet. 1996;29:327–334. doi: 10.1007/BF02208613. [DOI] [PubMed] [Google Scholar]
- Kuge S, Jones N. YAP1 dependent activation of TRX2 is essential for the response of Saccharomyces cerevisiae to oxidative stress by hydroperoxides. EMBO J. 1994;13:655–664. doi: 10.1002/j.1460-2075.1994.tb06304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon M, Chong S, Han S, Kim K. Oxidative stresses elevate the expression of cytochrome c peroxidase in Saccharomyces cerevisiae. Biochim Biophys Acta. 2003;1623:1–5. doi: 10.1016/s0304-4165(03)00151-x. [DOI] [PubMed] [Google Scholar]
- Lee J, Godon C, Lagniel G, Spector D, Garin J, Labarre J, Toledano MB. Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J Biol Chem. 1999a;274:16040–16046. doi: 10.1074/jbc.274.23.16040. [DOI] [PubMed] [Google Scholar]
- Lee J, Spector D, Godon C, Labarre J, Toledano MB. A new antioxidant with alkyl hydroperoxide defense properties in yeast. J Biol Chem. 1999b;274:4537–4544. doi: 10.1074/jbc.274.8.4537. [DOI] [PubMed] [Google Scholar]
- Li S, Ault A, Malone CL, Raitt D, Dean S, Johnston LH, et al. The yeast histidine protein kinase, Sln1p, mediates phosphotransfer to two response regulators, Ssk1p and Skn7p. EMBO J. 1998;17:6952–6962. doi: 10.1093/emboj/17.23.6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Dean S, Li Z, Horecka J, Deschenes RJ, Fassler JS. The eukaryotic two-component histidine kinase Sln1p regulates OCH1 via the transcription factor, Skn7p. Mol Biol Cell. 2002;13:412–424. doi: 10.1091/mbc.01-09-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Pastor MT, Marchler G, Schuller C, Marchler-Bauer A, Ruis H, Estruch F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE) EMBO J. 1996;15:2227–2235. [PMC free article] [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- Moradas-Ferreira P, Costa V, Piper P, Mager W. The molecular defences against reactive oxygen species in yeast. Mol Microbiol. 1996;19:651–658. doi: 10.1046/j.1365-2958.1996.403940.x. [DOI] [PubMed] [Google Scholar]
- Morgan BA, Bouquin N, Merrill GF, Johnston LH. A yeast transcription factor bypassing the requirement for SBF and DSC1/MBF in budding yeast has homology to bacterial signal transduction proteins. EMBO J. 1995;14:5679–5689. doi: 10.1002/j.1460-2075.1995.tb00255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan BA, Banks GR, Toone WM, Raitt D, Kuge S, Johnston LH. The Skn7 response regulator controls gene expression in the oxidative stress response of the budding yeast Saccharomyces cerevisiae. EMBO J. 1997;16:1035–1044. doi: 10.1093/emboj/16.5.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raitt DC, Johnson AL, Erkine AM, Makino K, Morgan B, Gross DS, Johnston LH. The Skn7 response regulator of Saccharomyces cerevisiae interacts with Hsf1 in vivo and is required for the induction of heat shock genes by oxidative stress. Mol Biol Cell. 2000;11:2335–2347. doi: 10.1091/mbc.11.7.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep M, Proft M, Remize F, Tamas M, Serrano R, Thevelein JM, Hohmann S. The Saccharomyces cerevisiae Sko1p transcription factor mediates HOG pathway-dependent osmotic regulation of a set of genes encoding enzymes implicated in protection from oxidative damage. Mol Microbiol. 2001;40:1067–1083. doi: 10.1046/j.1365-2958.2001.02384.x. [DOI] [PubMed] [Google Scholar]
- Schnell N, Krems B, Entian KD. The PAR1 (YAP1/SNQ3) gene of Saccharomyces cerevisiae, a c-jun homologue, is involved in oxygen metabolism. Curr Genet. 1992;21:269–273. doi: 10.1007/BF00351681. [DOI] [PubMed] [Google Scholar]
- Schuller C, Brewster JL, Alexander MR, Gustin MC, Ruis H. The HOG pathway controls osmotic regulation of transcription via the stress response element (STRE) of the Saccharomyces cerevisiae CTT1 gene. EMBO J. 1994;13:4382–4389. doi: 10.1002/j.1460-2075.1994.tb06758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks JB. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1986. pp. 163–167. [Google Scholar]
- Toone WM, Jones N. AP-1 transcription factors in yeast. Curr Opin Genet Dev. 1999;9:55–61. doi: 10.1016/s0959-437x(99)80008-2. [DOI] [PubMed] [Google Scholar]
- Tsuzi D, Maeta K, Takatsume Y, Izawa S, Inoue Y. Regulation of the yeast phospholipid hydroperoxide glutathione peroxidase GPX2 by oxidative stress is mediated by Yap1 and Skn7. FEBS Lett. 2004;565:148–154. doi: 10.1016/j.febslet.2004.03.091. [DOI] [PubMed] [Google Scholar]
- Verduyn C, van Wijngaarden CJ, Scheffers WA, van Dijken JP. Hydrogen peroxide as an electron acceptor for mitochondrial respiration in the yeast Hansenula polymorpha. Yeast. 1991;7:137–146. doi: 10.1002/yea.320070207. [DOI] [PubMed] [Google Scholar]
- Williams PG, Stewart PR. The intramitochondrial location of cytochrome c peroxidase in wild-type and petite Saccharomyces cerevisiae. Arch Microbiol. 1976;107:63–70. doi: 10.1007/BF00427868. [DOI] [PubMed] [Google Scholar]
- Winderickx J, de Winde JH, Crauwels M, Hino A, Hohmann S, Van Dijck P, Thevelein JM. Regulation of genes encoding subunits of the trehalose synthase complex in Saccharomyces cerevisiae: novel variations of STRE-mediated transcription control? Mol Gen Genet. 1996;252:470–482. doi: 10.1007/BF02173013. [DOI] [PubMed] [Google Scholar]
- Wu AL, Moye-Rowley WS. GSH1, which encodes gamma-glutamylcysteine synthetase, is a target gene for yAP-1 transcriptional regulation. Mol Cell Biol. 1994;14:5832–5839. doi: 10.1128/mcb.14.9.5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Fassler JS. SPT13 (GAL11) of Saccharomyces cerevisiae negatively regulates activity of the MCM1 transcription factor in Ty1 elements. Mol Cell Biol. 1993;13:63–71. doi: 10.1128/mcb.13.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.