Abstract
Misfolding and subsequent self-assembly of protein molecules into various aggregates is a common molecular mechanism for a number of important human diseases. Curing protein misfolding pathologies and designing successful drugs for the inhibition or reversal of protein aggregation depends on understanding the peculiarities of the misfolding process. Protein aggregation is a very complex process characterized by a remarkable polymorphism, where soluble amyloid oligomers, amyloid fibrils and amorphous aggregates are found as final products. This polymorphism is associated with the existence of multiple independent and competing assembly pathways leading to aggregation. Regardless of the aggregation mechanism, soluble oligomers are inevitably formed during the self-association process. Some of these oligomers are now considered to be major initiators of the neurodegenerative cascades of corresponding diseases. However, not all oligomers are equally harmful, and several amyloidogenic proteins have been shown to form nontoxic oligomers, some of which were efficient fibrillation inhibitors. Unfortunately, the information on the structural properties of soluble oligomers and the mechanisms of their formation, interconversion and toxicity is sparse. This review provides an overview of some topics related to soluble oligomers and represents several illustrative examples of toxic, nontoxic, productive and off-pathway amyloid oligomers.
Introduction
Many biologically active proteins act as specific oligomers. Structural proteins assemble into sophisticated supramolecular complexes that play various roles in a cell’s life. The formation of such functional oligomers and supramolecular complexes is tightly controlled and regulated. On the other hand, protein misfolding and subsequent uncontrolled (or unwanted) self-aggregation are known pathogens, which are now considered as potential driving forces for the development of a number of human diseases [1–6]. In fact, pathogenic proteinaceous deposits are at the heart of several so-called conformational diseases, such as Alzheimer’s disease (AD), Parkinson’s disease (PD), diffuse Lewy bodies disease, Lewy bodies variant of AD, dementia with Lewy bodies, multiple system atrophy, Hallervorden–Spatz disease, light chain-associated amyloidosis, light chain deposition disease, amyloidosis associated with hemodialysis, Huntington disease, spinal and bulbar muscular atrophy, spinocerebellar ataxia, neuronal intranuclear inclusion disease, Creutzfeld–Jacob disease, Gerstmann–Straussler–Schneiker syndrome, fatal familial insomnia and Kuru. These, and many other diseases, originate from the conversion of soluble and harmless protein into stable, ordered, filamentous protein aggregates, commonly referred to as amyloid fibrils, which can accumulate in a variety of organs and tissues. At least 21 different proteins have been recognized as causative agents of these conformational diseases [7]. Protein aggregation in general, and amyloid fibrillation in particular, is a highly selective molecular self-assembly process. As a result, proteinaceous deposits found in different diseases predominantly contain aggregated forms of a specific causative protein, unique for a given disorder. This raises the question of what drives the transformation of a biologically active soluble protein into a pathogenic misfolded conformation with high self-aggregation potential. Some of the possible mechanisms include [8]: an intrinsic propensity of some proteins to assume a pathological conformation, which becomes evident either with aging (e.g. normal α-synuclein in sporadic forms of PD and other synucleinopathies [9], and normal transthyretin in patients with senile systemic amyloidosis [10]) or as a result of unnaturally and persistently high cellular or plasma concentrations (e.g. triplication of a normal α-synuclein gene in some familial forms of PD [11–13], accumulation of β2-microglobulin in patients undergoing long-term hemodialysis [14], locally high insulin concentrations at the injection sites because of the slow release of insulin from the injection site [15]); the point amino acid mutations in causative proteins (e.g. familial forms of AD and PD, various hereditary amyloidoses); the genetic expansion of a CAG repeat in ORFs of genes encoding corresponding proteins (e.g. Huntington disease, spinal and bulbar muscular atrophy and spinocerebellar ataxia); the abnormal post-translational modifications of the causative proteins (e.g. hyperphosphorylation of tau protein in AD); the proteolytic cleavage of the precursor protein (e.g. β-amyloid precursor protein in AD); the exposure to some environmental agents that can bring about pathogenic conformational changes in the causative proteins (e.g. structural changes induced by pesticides, herbicides or heavy metals in PD-related protein α-synuclein, structural consequences of oxidative damage, etc.). These and other mechanisms can act independently, additively, or even synergistically.
The accumulation of protein deposits is commonly associated with severe cellular degeneration at the deposition places, the precise mechanisms of which remain elusive [16]. It is not clear whether amyloid fibrils trigger the cellular degeneration or simply represent highly visible side products of the cellular disruption process. However, it has been established that protein misfolding/aggregation and cellular degeneration are coupled. As it was nicely summarized in a recent review [17], there are several potential mechanisms of such cytotoxicity originating from protein deposition. These include: the disruption of the tissue architecture and functions promoted by the invasion of the extracellular space of organ by amyloids [8,18]; the destabilization of intracellular and extracellular membranes by oligomers, the formation of which may precede or coincide with the appearance of amyloid fibrils [19,20]; the apoptotic cell death and receptor-mediated toxicity triggered by the oligomer interaction with various neuronal receptors [21]; the oligomer-mediated impairment of the presynaptic P/Q-type calcium currents [22]; the impaired maturation of autophagosomes to lysosomes mediated by the oligomer accumulation [23]; the dysfunction of autophagy, a lysosomal pathway for degrading organelles and proteins [24]; the oxidative damage-induced disruption of the cell viability promoted by the incorporation of redox metals into amyloid fibrils and the subsequent generation of reactive oxygen species [25–29]; the general disorganization of cellular protein homeostasis associated with the exhaustion of the cell defense mechanisms, such as a chaperone system [30,31]; proteasome inhibition [32]; the loss of crucial protein function(s) and/or the gain of toxic function(s).
All this explains why the problems of misfolding, aggregation and amyloid fibril formation have gained considerable attention from researchers. An intriguing recent development in this field is the immerging recognition of the multiple unique roles that soluble amyloid oligomers (which are oligomeric but soluble states of amyloidogenic proteins) play both as crucial precursors of amyloid fibrils and as independent toxic agents. Despite these facts, information on the structural properties of soluble oligomers and the mechanisms of their formation and interconversion is sparse, and the understanding of the molecular mechanisms of their toxicity remains mostly elusive. This review provides an overview of some topics related to these issues.
Template-dependent and template-independent mechanisms of amyloid fibril formation
It is now recognized that amyloid fibrillation is a highly dynamic process that represents the most dramatic consequence of protein misfolding and takes place in parallel with, or as an alternative to, physiological folding. The amyloid fibril is a relatively recent discovery, but seems to be the universal state of a polypeptide chain, as the number of proteins shown to form such structures in vitro is constantly increasing [2]. Despite different chemical natures of causative proteins, amyloid fibrils of different origins have a rather similar (but not identical!) morphology, consisting of two to six unbranched protofilaments 2–5 nm in diameter associated laterally or twisted together to form fibrils with a 4–13 nm diameter [2] and displaying many common properties, including a core cross-β-sheet structure in which continuous β-sheets are formed with β-strands running perpendicular to the long axis of the fibrils [33]. Apart from their characteristic appearance in the electron microscopy or atomic force microscopy (AFM) images (often observed as long twisted rope-like structures), amyloids are easily recognizable by their apple-green birefringence under a polarized light microscope after staining with a specific fluorescent dye (Congo red). Although fibrillation of various proteins produces fibrils with generally similar morphology, the phenomenon of amyloid fibrillar polymorphism has been recently recognized: fibrillation of a single amyloidogenic protein may result in the appearance of multiple forms of amyloid fibrils, depending on their induction conditions [17,34–38]. Such a polymorphism is probably due to the existence of multiple independent and competing assembly pathways leading to the amyloidogenesis [17,37,38].
In addition to the amyloid fibrils discussed above, proteins can self-assemble to form several other types of aggregate, e.g. soluble oligomers and amorphous aggregates. Amorphous aggregates are typically formed faster than fibrils. There is no special conformational prerequisite for amorphous aggregation to occur, and many destabilized and partially unfolded proteins precipitate out of solution in a form of amorphous aggregate. On the other hand, fibrillation requires special conditions that promote the formation of the specific amyloidogenic conformations [39]. The choice between the three aggregation pathways, fibrillation, amorphous aggregate formation and oligomerization is determined by the amino acid sequence and by the peculiarities of protein environment.
Fibrils are proposed to be formed in template-dependent and template-independent manners (reviewed in [17]). In template-dependent fibrillation, interaction with a pre-existing template brings about conformational changes in an amyloidogenic protein, promoting its accommodation to the template with the subsequent exposure of the interactive regions for the consecutive self-assembly [40]. Here, the template is taken in a broad essence, as almost any conformational species involved in the amyloid fibrillation (i.e. altered monomeric conformation, oligomeric forms, immature fibrils, protofibrils and fragments of fibrils) could play this role.
In template-independent fibrillation, the amyloidogenic conformations are formed spontaneously, in the absence of a template. After being formed, such an amyloidogenic self-interactive conformer favors the self-assembly process, which eventually leads to amyloid fibril formation [17]. The mentioned conformational transition from a soluble, biologically active form to the amyloidogenic species is a great illustration of the protein misfolding concept. Obviously, such a misfolding process can be triggered by a multitude of extrinsic and intrinsic factors. For example, the α-synuclein fibrillation was shown to be dramatically accelerated under any conditions favoring the transformation of this natively unfolded protein into the amyloidogenic form characterized as a partially folded monomeric conformation resembling the premolten globule state [9,41]. It is clear that in reality only the very early stages of amyloid fibril formation, the formation of monomeric amyloidogenic species and their assembly into the first amyloidogenic oligomers, are template independent. Once formed, such an amyloidogenic oligomer will immediately start acting as a template, promoting sequential addition of monomers through induced conformational transitions.
Even the longest journey begins with the first step: oligomerization as the inevitable step of protein aggregation
Recently, in an excellent review, Morris et al. [42] provided an outstanding summary of major models proposed for the description and analysis of the protein aggregation kinetics and mechanisms. The authors distilled the enormous literature on protein aggregation (as of March 2010, there were more than 53–500 papers in PubMed discussing various aspects of this phenomenon) down to several major classes of kinetic mechanisms. In this review, an interested reader can find thoughtful and deep analyses and comparisons of various protein aggregation models, together with the formalisms proposed for the quantitative description of aggregation kinetics [42]. Protein aggregation is an irreversible nucleated (or autocatalytic) process that resembles a condensation reaction, as it typically occurs only above a critical concentration. Typically, protein fibrillation is described by a sigmoidal curve and is considered to be a nucleation–polymerization reaction where the monomer addition steps are assumed to be thermodynamically unfavorable until a critical nucleus is formed (i.e. during the nucleation stage). However, aggregation is a thermodynamically favorable process during the polymerization stage (i.e. after the critical nucleus formation). The critical nucleus is defined as the least thermodynamically stable species in solution, which is the oligomer of minimal size capable of initiating further growth [42]. The nucleus can also be defined as the aggregate size after which the association rate exceeds the dissociation rate for the first time [43]. In addition to the homogeneous nucleation, heterogeneous nucleation (or seeding) can also take place on the surface of existing polymers [44]. Furthermore, aggregation can be further accelerated by the fragmentation of existing aggregates [45,46]. For some proteins with specific distributions of polar and hydrophobic residues (e.g. for Aβ1–40 peptide), fibrillation can start only above a certain critical micelle concentration at which the peptide micelles are formed. The formation of these micelles represents a crucial step, as fibrils nucleate inside them and then grow by irreversible binding of monomers to fibril ends [47].
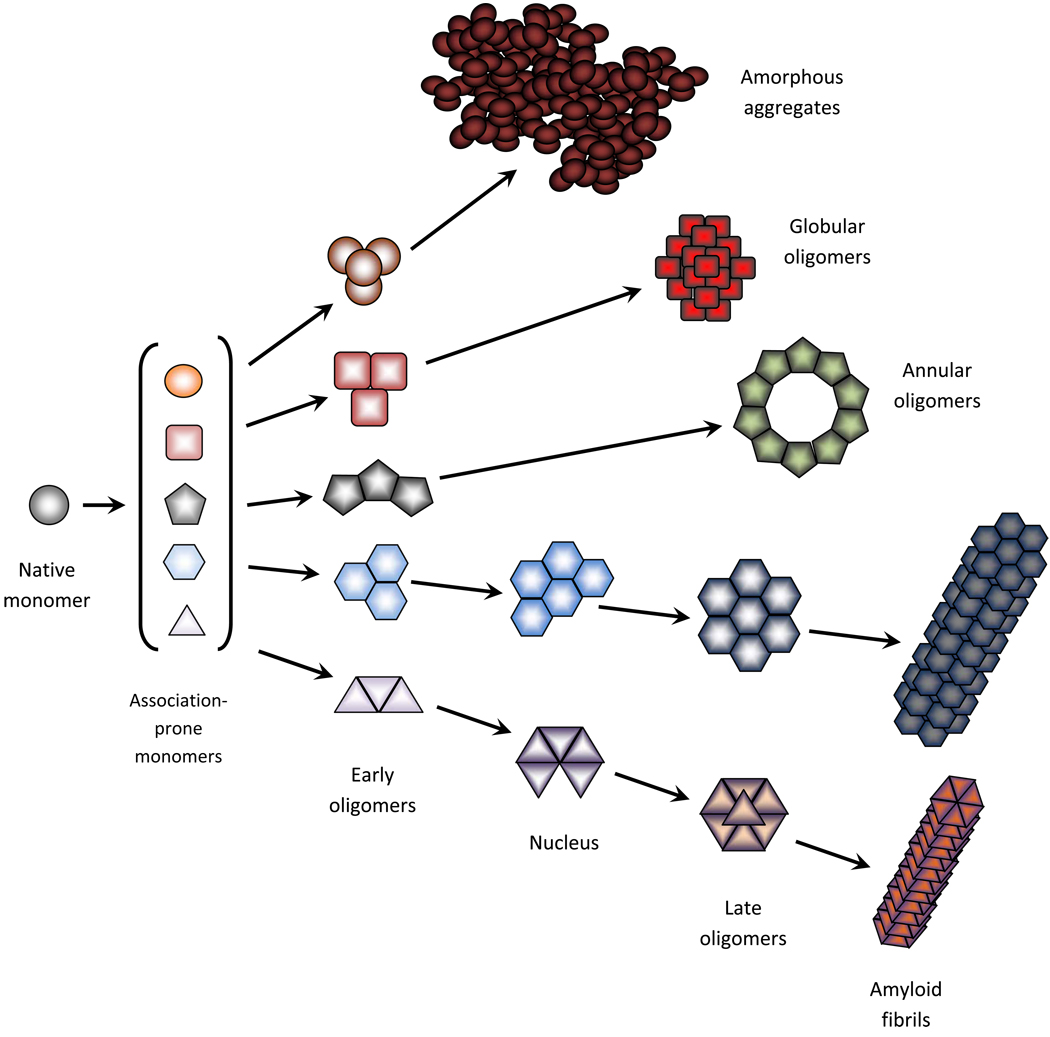
All the models mentioned above were developed to describe an aggregation process that obviously starts with a monomeric protein and ends with the aggregate formation. Figure 1 represents an idealized model of amyloid fibril formation and clearly shows that fibrillation is a directed process with a series of consecutive steps, including the formation of several different oligomers. In this model, various oligomers are comprised of structurally identical monomers and the formation of these oligomers constitutes productive steps in the fibrillation pathway. However, aggregation is known to induce dramatic structural changes in the aggregating protein. Therefore, monomers at different aggregation stages are not identical. In addition, recent studies have clearly shown that a given protein can self-assemble into various aggregated forms, depending on the peculiarities of its environment. In fact, the typical aggregation process only rarely results in the appearance of a homogeneous product where at the end of the reaction only one aggregate species (amyloid fibrils, amorphous aggregates or soluble oligomers) is present. More often, heterogeneous mixtures of various aggregated forms are observed. Furthermore, each aggregated form can have multiple morphologies and monomers comprising morphologically different aggregated forms can be structurally different. All this suggests that aggregation is not a simple reaction, but a very complex process with multiple related and unrelated pathways that can be connected or disjoined. However, regardless of the model or pathway considered, the appearance of a large aggregate inevitably involves the formation of some small oligomeric species.
Fig. 1.
A schematic, oversimplified representation of the protein self-association process. The formation of multiple association-prone monomeric forms generates multiple aggregation pathways. There are three major products of the aggregation reaction – amorphous aggregates (top pathway), morphologically different soluble oligomers (pathways second and third from the top) and morphologically different amyloid fibrils (bottom two pathways). Two types of soluble oligomer (spheroidal and annular) and two morphologically different amyloid fibrils are shown. Changes in color reflect potential structural changes within a monomer taking place at each elementary step. In reality, the picture is more complex and more species can be observed. Interconversions between various species at different pathways are also possible.
MysteriO’s: illustrative examples of toxic, nontoxic, productive and off-pathway oligomers
Recent studies have indicated that small oligomeric species are potentially more cytotoxic than mature fibrils, which can be considered as products of detoxification [19,20,48]. In fact, clinical manifestations of amyloidosis-related neurodegenerative diseases often precede a detectable accumulation of the fibrillar protein aggregates. In AD, the number of senile plaques in the affected region of the brain was shown to be poorly correlated with the local extent of neuron death or synaptic loss, or with cognitive impairment. On the other hand, some patients with abundant amyloid deposits do not show any neurodegenerative symptoms, suggesting that mature amyloid fibrils do not cause the onset of amyloidosis-related neurodegenerative diseases. Furthermore, a robust correlation between the soluble Aβ oligomer levels and the extent of synaptic loss and severity of cognitive impairment has been established [20,21,49–53]. Therefore, soluble oligomers are very important players in protein aggregation and in the related cytotoxicity. The term ‘soluble oligomer’ is used here to describe any nonmonomeric form of an amyloidogenic protein that is soluble in aqueous solutions and remains in solution after high-speed centrifugation, indicating that it is not an insoluble fibrillar or aggregated species. Several illustrative examples of these mysterious species are briefly described below.
Aβ oligomers
Among various amyloidogenetic proteins, oligomerization and its potential consequences are well documented for natural and synthetic Aβ peptides. Because various aspects of Aβ oligomer formation and toxicity were considered in a recent review by Sakono & Zako [49], only some key observations are presented below. Several different oligomeric forms, ranging from dimers to 24-mers and to higher relative molecular mass soluble species, have been reported for natural and synthetic Aβ peptides [49,50,54,55]. These oligomers are highly diverse in respect of their structure, size and shape (see Fig. 2). For example, natural Aβ oligomers with a wide-ranging relative molecular mass distribution (from <10 to >100 kDa) have been found in the AD brain [56]. This structural and morphological diversity is believed to be responsible for the diversity of biological effects ascribed to these oligomers and for the related complexity of AD pathology [20,21,49,50,54,55]. For example, the analysis of soluble fractions of human brain and amyloid plaque extracts revealed the presence of SDS-stable dimers and trimers, suggesting that these oligomers could play a crucial role as the fundamental building blocks in the formation of larger oligomers or insoluble amyloid fibrils [57–59]. Cultured cells have also been shown to secrete the similarly sized Aβ oligomers, which have been shown to inhibit long-term potentiation in vitro [60,61]. In vitro studies have revealed that Aβ dimers were three times more toxic than monomers, and that Aβ tetramers were 13 times more toxic, clearly supporting the concept of the high toxicity of low relative molecular mass Aβ oligomers [62]. The levels of the Aβ*56 oligomeric form (which corresponded to the SDS-stable Aβ nonamers and dodecamers) in the brain of the β-amyloid precursor protein transgenic Tg2576 mice were shown to be correlated with memory deficits in this animal model [63]. Purified dodecamers were shown to induce a significant fall-off in the spatial memory performance of wild-type rats, suggesting that nonamers and dodecamers can be associated with deleterious effects on cognition. A morphological analysis revealed a great shape variability in Aβ soluble oligomers. For example, small Aβ globular oligomers (5 nm in diameter), referred to as Aβ-derived diffusible ligands (ADDLs), were frequently found in Hams-F12 medium [64]. These ADDLs were shown to strongly interact with the dendritic arbors of cultured neurons, to cause neuronal cell death and to block long-term potentiation. Furthermore, an analysis of the soluble brain extracts using ADDL-specific antibody established the presence of ADDLs in human AD brain, suggesting that the formation and existence of ADDLs in the human AD brain can cause disease [64]. The largest globular Aβ assemblies are amylospheroids, which are highly neurotoxic, off-pathway, spheroidal structures with diameters of 10–15 nm [65]. Finally, various annular Aβ oligomers, with relative molecular mass ranging from 150 to 250 kDa and with an outer diameter of 8–12 nm and an inner diameter of 2.0–2.5 nm, have also been described [66,67]. Such doughnut-like oligomers are preferentially formed from mutant Aβ (such as those carrying the Arctic mutation). These annular Aβ oligomers can act on the nonspecific amyloid pores, which structurally resemble pores formed by the bacterial cytolytic β-barrel pore-forming toxins, and which could be responsible for the Aβ-associated cytotoxicity [66].
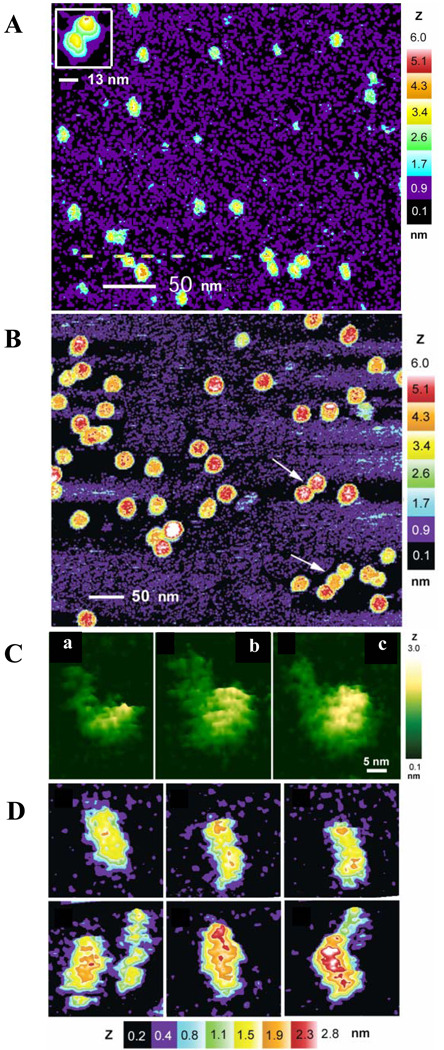
Fig. 2.
Illustrative examples of various self-assembled complexes of amyloidogenic proteins. (A) Diversity of soluble oligomers formed by 16 µm Aβ42. This sample was prepared using the 16 µm Aβ42 solution diluted to 4 µm and immediate addition to the mica surface for AFM. (B) High relative molecular mass oligomers. This sample was prepared after the Aβ42 peptide incubation at a concentration of 80 µm for 3 h followed by dilution to 40 µm before depositing on the mica surface. (C) High-resolution AFM images of monomeric (a), dimeric (b) and tetrameric (c) Aβ42 peptide. Here, the diffuse area to the left of the particle in each image corresponds to the disordered N-terminal region of the Aβ42 monomer. This sample was prepared at an initial Aβ42 peptide concentration of 100 µm, which was immediately diluted to 30 µm and incubated with shaking for 1 h before being deposited on the AFM mica surface. (D) High-resolution AFM images of unit-protofibrils formed in a 16 µm Aβ42 peptide solution. This sample was prepared by the dilution of the starting solution to 4 µm either immediately or after 4 h. Images in this plot are courtesy of Professor Steven O. Smith.
α-synuclein oligomers
Similar to Aβ, α-synuclein (which is believed to be a major player in the pathogenesis of PD and other synucleinopathies) fibrillization in vitro is not a simple two-state transition from monomer to fibrils, representing instead a rather complex process involving the formation of oligomeric intermediates of various sizes and morphologies. The prolonged incubation of α-synuclein at different temperatures resulted in a temperature-dependent, progressive aggregation, with dimers being formed first [68]. This temperature-modulated oligomerization was shown to be accompanied by a small but reproducible increase in the ordered secondary structure content. Interestingly, the trapped oligomeric conformation was structurally similar to the premolten globule-like partially folded monomeric confomer induced by low pH or high temperature [68]. Therefore, it has been concluded that the partially folded premolten globule-like conformation of α-synuclein can be stabilized as the protein undergoes a highly selective self-assembly process during prolonged incubation at elevated temperatures [68]. The formation of oxidative dimers and higher-order oligomers with dityrosine cross-links in α-synuclein under the conditions of oxidative stress was also reported [69].
Various oligomers were separated from fibrillar and monomeric α-synuclein by sedimentation followed by gel-filtration chromatography [70,71]. AFM analysis revealed a great morphological diversity of these oligomers, including various spheres (with heights ranging from 2 to 6 nm), chains of spheres (protofibrils) and rings with heights ranging from 3 to 7 nm [70,71]. In this process, spherical protofibrils were formed rapidly, whereas annular species were produced on prolonged incubation of spheres [72]. In addition to the completed rings, doughnuts, the existence of partially formed rings (crescents) has been observed [72]. The formation of both doughnuts and fibrils was shown to require an initial formation of spherical, β-structure-enriched, α-synuclein oligomers. The morphology of the oligomers was shown to be affected by the solution conditions, including the presence of lipids [72–74], organic solvents [75] or metal ions [76]. In fact, the incubation of α-synuclein with different metals for 1 day at 4 °C produced three different classes of oligomer, where Cu2+, Fe3+ and Ni2+ yielded 0.8–4 nm spherical particles, similar to α-synuclein incubated without metal ions, Mg2+, Cd2+ and Zn2+ gave larger, 5–8 nm spherical oligomers, whereas Co2+ and Cd2+ frequent annular (doughnut-like) oligomers, 70–90 nm in diameter with Ca2+ and 22–30 nm in diameter with Co2+ [76]. Analysis of the different α-synuclein oligomers by Raman microscopy revealed that the spheroidal oligomers contained a significant amount of α-helical structure (~47%) and β-sheet structure (~29%). The formation of protofibrils was accompanied by a decrease in the α-helical content to ~37% and a concomitant increase in the β-sheet content to ~54% [77]. Both spheroidal and annular oligomers were proposed to be cytotoxic. In fact, the spherical protofibrils were shown to bind to brain-derived membrane fractions much more tightly than monomeric or fibrillar α-synuclein did [72]. Annular oligomers, known as amyloid pores, are also able to bind to membrane [72], significantly affecting cell viability by influencing the membrane stability [19,20].
Soluble oligomers are often transient, as they are consumed as fibrillization proceeds [20,71,72]. In situ AFM analysis showed that the formation of globular oligomers precedes the appearance of amyloid fibrils and is systematically observed under conditions for accelerated fibrillation, potentially indicating that the oligomers can act as on-pathway intermediates during amyloidogenesis [78]. However, soluble oligomerization can also be an off-pathway reaction, and under some conditions the productive α-synuclein fibrillation is known to be inhibited in favor of soluble oligomer formation. For example, nitrated α-synuclein remains assembled into the oligomeric spheroids even after incubation for a very prolonged time [79]. Furthermore, the addition of nitrated α-synuclein substantially inhibited the fibrillation of the nonmodified protein in a concentration-dependent manner [79,80]. Preferential oligomerization was also found when α-synuclein was co-incubated with cigarette smoke components, such as nicotine and hydroquinone [81], various flavonoids [82] or substoichiometric concentrations of 3,4-dihydroxyphenylacetic acid [83], or as a result of methionine oxidation [84].
Although in the majority of the analyzed cases soluble α-synuclein oligomers were cytotoxic and dramatically affected membrane permeability [19,20,72], there are several cases when oligomerization of this protein resulted in the formation of nontoxic species. In fact, this finding is rather expected due the highly heterogeneous nature of protein aggregates (including oligomers) caused either by the heterogeneous starting materials or by multiple pathways of assembly, or by both these factors. Therefore, it is difficult to believe that all the soluble oligomers, with their astonishing morphological variability, will be similarly cytotoxic [81]. In agreement with this reasoning, the flavonoid baicalein was shown to inhibit α-synuclein fibrillation via the stabilization of soluble oligomers that possessed very specific structural features: being spherical, having a relatively globular structure with a packing density intermediate between that of premolten globules and typical globular proteins, having a relatively well-developed secondary structure and being characterized by high thermodynamic stability [85]. These oligomers were able to inhibit fibrillation of baicalein-untreated α-synuclein and, most importantly, did not disrupt the integrity of the biological membrane [85]. Similarly, the oxidation of α-synuclein methionines by H2O2 greatly inhibited fibrillation of this protein in vitro, leading to the formation of relatively stable oligomers, which were not toxic to dopaminergic and GABAergic neurons [84]. These observations clearly show that the soluble oligomer formation from the amyloidogenic protein does not always create harm, and can in fact be beneficial.
Tau protein oligomers
The appearance of neurofibrillary tangles (NFTs), insoluble intracellular fibers of paired helical filaments arising from the misfolding and aggregation of the neuronal-specific microtubule-associated protein tau, is closely correlated with AD progression [86–88]. As with Aβ and α-synuclein, the formation of soluble tau oligomers, rather than that of mature fibrils, is the key to cell death associated with tau aggregation [89]. Recently, structures of tau oligomers that appear in response to heparin-induced aggregation were analyzed by multidimensional NMR [90]. This study revealed that the regions VQIINK280 and VQIVYK311 of tau protein were the major sites of intermolecular interaction in the oligomer, and that these intermolecular interactions were triggered in response to heparin addition [90]. It has also been shown that tau assembly involves two distinct dimers (cysteine-dependent and cysteine-independent) that differ in resistance to reduction [91]. Interestingly, the population of cysteine-dependent tau oligomers increased prior to the detection of fibrils, which was accompanied by an increase in the amounts of cysteine-independent dimer [91]. In addition to the small oligomers discussed above, a granular tau oligomer having a prefilamentous structure was also found [92]. Quantification of frontal cortex samples displaying varying degrees of NFT pathology revealed significantly increased levels of granular tau oligomers, even in brains with a very early neuropathology stage at which clinical symptoms of AD and NFTs in frontal cortex are believed to be absent [92]. Based on these observations, it has been concluded that the increase in granular tau oligomer levels occurs prior to the detectable formation of NFTs and before individuals manifest clinical symptoms of AD, suggesting that granular tau oligomer levels may represent a very early sign of NFT formation and AD [92].
Mammalian prion protein oligomers
In the prion diseases, the autocatalytic conversion of the cellular form of the prion protein (PrPC) to an alternative conformation (scrapie form of the prion protein, PrPSc) is assumed to take place [93]. Here, PrPC is monomeric and sensitive to protease digestion, whereas PrPSc is richer in β-sheet content, has a low solubility and is resistant to protease digestion [93]. Furthermore, PrPSc can convert PrPC into its pathogenic PrPSc [93]. By analogy with other conformational diseases, prion protein oligomers and/or prefibrillar aggregates might be cytotoxic [94,95]. In agreement with this hypothesis, the most infectious species were shown to be soluble oligomers of the prion protein, 17–27 nm in diameter and 300–600 kDa in mass, derived from the disaggregation of PrPSc [96]. Furthermore, these oligomers were able to efficiently convert the PrPC into a protease-resistant form in an in vitro assay [96]. Being incubated at low pH, the full-length mouse prion protein was shown to transform into an equilibrium mixture of soluble β-rich oligomers and α-rich monomers [97]. With time, these β-rich oligomers were assembled into the worm-like amyloid fibrils in a step-wise manner, and potentially via multiple routes [97].
Yeast prion protein oligomers
The fact that amyloids in general, and PrPSc in particular, can self-propagate suggests that the structure of the final fibrillar state might be determined by the structural information encoded in the initial nucleus formed in the very early phase of protein fibrillation. A systematic analysis of fibrillation of yeast prion protein Sup35 supported this hypothesis, and showed that the structural variability in the initial nucleus was a crucial determining factor of the diversity of prion strain conformations and resulting strain phenotypes [98]. In fact, an intriguing correlation was found between the reversible formation of soluble oligomers at low temperature and the ability of Sup35 to form the Sc4 prion conformation that leads to the induction of strong [PSI+] phenotypes. The oligomer formation was driven by the non-native aromatic interactions outside the amyloid core, which specifically led to the formation of highly infectious strain conformations with more limited amyloid cores [98]. Based on this interesting study, the authors concluded that transient non-native interactions in the initial oligomers could play a crucial role in the subsequent determination of the diversity of amyloid conformations and resulting prion strain phenotypes [98].
Insulin oligomers
The physiological form of insulin is a zinc-stabilized hexamer. However, in 20% acetic acid, insulin is monomeric [99]. A systematic analysis of insulin fibrillation in 20% acetic acid revealed the existence of noticeable structural changes occurring before the onset of fibril formation [100]. In this study, at least two different types of oligomeric intermediate between the native monomer and fibrils were detected. These intermediates had significantly different underlying structures, being easily distinguishable by FTIR, CD and 8-anilinonaphthalene-1-sulfonate binding, and corresponded to the significantly different association state, as determined by the dynamic light scattering. Both oligomeric intermediates had non-native conformations, indicating that fibrillation occurred from a β-rich structure that is significantly different from the native fold [100]. The existence of significant amounts of oligomeric species of insulin prior to the appearance of mature fibrils and throughout the fibril elongation process was further confirmed by dynamic light scattering [100] and time-lapse AFM [101]. SAXS analysis of oligomeric species accumulated at the early fibrillation stages of insulin revealed the peculiar morphology where oligomer appeared as a bead-on-a-string assembly of six units, each with dimensions comparable with those of insulin monomers [102].
Oligomers of the immunoglobulin light chain
Light chain amyloidosis (or primary amyloidosis) originates from the formation and systemic deposition (especially in the kidneys) of fibrils of monoclonal immunoglobulin light chain variable domains in patients suffering from multiple myeloma [103]. LEN is a κIV immunoglobulin light chain variable domain from a patient suffering from multiple myeloma, the recombinant form of which is a dimer that represents an established system for the in vitro characterization of light chain amyloid proteins [104]. A systematic analysis of the LEN fibrillation process revealed an inverse concentration dependence due to the formation of the off-pathway soluble oligomers (probably octamers) at a high protein concentration [105,106]. In fact, these soluble off-pathway oligomeric species were formed at high protein concentrations prior to the appearance of fibrils, significantly slowing down the kinetics of fibril formation, as compared with the fibrillation rates measured at much lower protein concentrations. However, fibrils were still observed eventually at high protein concentrations, despite the initial trapping of most of the protein as soluble off-pathway oligomers. Because most of the protein was present in these off-pathway intermediates at relatively early times of aggregation, and because all the proteins eventually formed fibrils, a structural rearrangement from the nonfibril-prone off-pathway oligomers to a more fibril-prone species must occur at later aggregation stages. The corresponding structural changes, being monitored by a variety of techniques, revealed a significant increase in the disordered secondary structure content, an increase in the solvent accessibility and a decrease in the intrinsic stability of the soluble oligomers [105]. More specifically, the fibrillation of the dimeric LEN can be described by the following model. First, the transition to a partially folded but relatively native-like conformation, I, takes place. At this stage, LEN preserves its dimeric state. Then, a soluble oligomer (an octamer), I8, is formed from the partially folded dimer. This octamer is comprised of partially folded LEN molecules, the conformations of which correspond to those of the initially formed partially folded intermediate I. With time, these I8 oligomers undergo a transformation into a second class of soluble oligomers (I*8)n and the component molecules undergo a conformational change leading to a less ordered structure, I*. At the final stage, the exponential growth of fibrils occurs. In contrast to I, the conformation of I* is much more disordered, as detected by probes of secondary structure, increased susceptibility to proteolysis, increased H/D exchange and decreased conformational stability [105]. Interestingly enough, this structural reorganization, which leads to a more unfolded conformation in the higher oligomers, accounting for the observed decrease in stability and increase in solvent accessibility, is driven by the self-association of the I8 oligomers. Based on these observations it has been concluded that LEN represents a unique fibrillating system in which soluble off-pathway oligomeric intermediates have been shown to be the major transient species, and in which fibrillation occurs from a relatively unfolded conformation present in these intermediates [105].
Concluding remarks
Mounting evidence indicates that uncontrolled (or unwanted) self-aggregation of amyloidogenic proteins represents a fundamental basis for the development of various types of amyloid-related degenerative disease. Aggregation is a very complex self-assembly process characterized by an astonishing polymorphism of final products. Proteins are known to self-assemble into various aggregated forms, e.g. soluble amyloid oligomers, amyloid fibrils and amorphous aggregates. The choice between the three aggregation pathways, fibrillation, amorphous aggregate formation and oligomerization, is determined by the amino acid sequence and by the peculiarities of the protein environment. However, these forms are often found at the end of the aggregation reaction. In addition, each of these aggregated species can be present in several morphologically and structurally different forms. This polymorphism reflects the wide variation in the association states of amyloidogenic proteins and diversity of intermolecular interactions stabilizing final self-aggregated forms. For example, Aβ oligomers are known to be characterized by a wide range of association degree (from <10 to >100 kDa), and the biochemical properties of Aβ oligomers and their pathogeneity depend on their sizes and structures. Furthermore, even similarly sized Aβ oligomers can be characterized by a dramatic structural (and potentially pathogenic) polymorphism. In all likelihood, this polymorphism reflects the existence of multiple independent and competing assembly pathways leading to aggregation. In relation to the pathogenesis, amyloid oligomers (which are oligomeric but soluble states of amyloidogenic proteins), rather than insoluble amyloid fibrils, are now believed to be responsible for the initiation of neurodegenerative cascades of corresponding diseases. These oligomers are inevitably formed during the aggregation process, both as crucial intermediates in the fibrillogenesis and as independent off-pathway entities. The fact that various amyloidogenic proteins can form toxic soluble oligomers clearly suggests that amyloid oligomers can be considered as the general key factors in the pathogenesis of various degenerative diseases. Recent studies have clearly shown that the formation and toxicity mechanisms of various amyloid oligomers can also be different from one another. Therefore, the therapeutic strategies targeted at the inhibition of fibrillation or at the dissolving of preformed fibrils can be potentially harmful, as the prevention of aggregation may cause the formation or stabilization of toxic oligomer states. On the other hand, because oligomers are formed via various pathways, not all oligomers are equally cytotoxic, and several cases were reported when amyloidogenic proteins were forced to form some nontoxic oligomers. Importantly, under some conditions, the formation of such nontoxic oligomers can very effectively compete with the fibrillation process and the preformed nontoxic oligomers can serve as potent fibrillation inhibitors.
Acknowledgements
I am very thankful to Professor Steven O. Smith for providing high-resolution AFM images of various oligomeric forms of Aβ42 peptide. I express my deepest gratitude to Alexey Uversky for carefully reading and editing the manuscript. This work was supported in part by the Program of the Russian Academy of Sciences for ‘Molecular and cellular biology’, by grants R01 LM007688-01A1 and GM071714-01A2 from the National Institutes of Health and grant EF 0849803 from the National Science Foundation. I gratefully acknowledge the support of the IUPUI Signature Centers Initiative.
Abbreviations
- AD
Alzheimer’s disease
- ADDL
Aβ-derived diffusible ligand
- AFM
atomic force microscopy
- NFT
neurofibrillary tangle
- PD
Parkinson’s disease
- PrPC
cellular form of the prion protein
- PrPSc
scrapie form of the prion protein.
References
- 1.Bellotti V, Mangione P, Stoppini M. Biological activity and pathological implications of misfolded proteins. Cell Mol Life Sci. 1999;55:977–991. doi: 10.1007/s000180050348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dobson CM. Protein misfolding, evolution and disease. Trends Biochem Sci. 1999;24:329–332. doi: 10.1016/s0968-0004(99)01445-0. [DOI] [PubMed] [Google Scholar]
- 3.Kelly JW. The alternative conformations of amyloidogenic proteins and their multi-step assembly pathways. Curr Opin Struct Biol. 1998;8:101–106. doi: 10.1016/s0959-440x(98)80016-x. [DOI] [PubMed] [Google Scholar]
- 4.Rochet JC, Lansbury PT., Jr Amyloid fibrillogenesis: themes and variations. Curr Opin Struct Biol. 2000;10:60–68. doi: 10.1016/s0959-440x(99)00049-4. [DOI] [PubMed] [Google Scholar]
- 5.Uversky VN, Talapatra A, Gillespie JR, Fink AL. Protein deposits as the molecular basis of amyloidosis. I. Systemic amyloidoses. Med Sci Monitor. 1999;5:1001–1012. [Google Scholar]
- 6.Uversky VN, Talapatra A, Gillespie JR, Fink AL. Protein deposits as the molecular basis of amyloidosis. II. Localized amyloidosis and neurodegenerative disorders. Med Sci Monitor. 1999;5:1001–1012. [Google Scholar]
- 7.Westermark P, Benson MD, Buxbaum JN, Cohen AS, Frangione B, Ikeda S, Masters CL, Merlini G, Saraiva MJ, Sipe JD. Amyloid fibril protein nomenclature – 2002. Amyloid. 2002;9:197–200. doi: 10.3109/13506120209114823. [DOI] [PubMed] [Google Scholar]
- 8.Merlini G, Bellotti V. Molecular mechanisms of amyloidosis. N Engl J Med. 2003;349:583–596. doi: 10.1056/NEJMra023144. [DOI] [PubMed] [Google Scholar]
- 9.Uversky VN. Neuropathology, biochemistry, and biophysics of alpha-synuclein aggregation. J Neurochem. 2007;103:17–37. doi: 10.1111/j.1471-4159.2007.04764.x. [DOI] [PubMed] [Google Scholar]
- 10.Saraiva MJ. Transthyretin amyloidosis: a tale of weak interactions. FEBS Lett. 2001;498:201–203. doi: 10.1016/s0014-5793(01)02480-2. [DOI] [PubMed] [Google Scholar]
- 11.Singleton A, Gwinn-Hardy K, Sharabi Y, Li ST, Holmes C, Dendi R, Hardy J, Crawley A, Goldstein DS. Association between cardiac denervation and parkinsonism caused by alpha-synuclein gene triplication. Brain. 2004;127:768–772. doi: 10.1093/brain/awh081. [DOI] [PubMed] [Google Scholar]
- 12.Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, Hulihan M, Peuralinna T, Dutra A, Nussbaum R, et al. alpha-Synuclein locus triplication causes Parkinson's disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 13.Farrer M, Kachergus J, Forno L, Lincoln S, Wang DS, Hulihan M, Maraganore D, Gwinn-Hardy K, Wszolek Z, Dickson D, et al. Comparison of kindreds with parkinsonism and alpha-synuclein genomic multiplications. Ann Neurol. 2004;55:174–179. doi: 10.1002/ana.10846. [DOI] [PubMed] [Google Scholar]
- 14.Verdone G, Corazza A, Viglino P, Pettirossi F, Giorgetti S, Mangione P, Andreola A, Stoppini M, Bellotti V, Esposito G. The solution structure of human beta2-microglobulin reveals the prodromes of its amyloid transition. Protein Sci. 2002;11:487–499. doi: 10.1110/ps.29002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shikama Y, Kitazawa J, Yagihashi N, Uehara O, Murata Y, Yajima N, Wada R, Yagihashi S. Localized amyloidosis at the site of repeated insulin injection in a diabetic patient. Intern Med. 49:397–401. doi: 10.2169/internalmedicine.49.2633. [DOI] [PubMed] [Google Scholar]
- 16.Sacchettini JC, Kelly JW. Therapeutic strategies for human amyloid diseases. Nat Rev Drug Discov. 2002;1:267–275. doi: 10.1038/nrd769. [DOI] [PubMed] [Google Scholar]
- 17.Bhak G, Choe YJ, Paik SR. Mechanism of amyloidogenesis: nucleation-dependent fibrillation versus double-concerted fibrillation. BMB Rep. 2009;42:541–551. doi: 10.5483/bmbrep.2009.42.9.541. [DOI] [PubMed] [Google Scholar]
- 18.Tan SY, Pepys MB. Amyloidosis. Histopathology. 1994;25:403–414. doi: 10.1111/j.1365-2559.1994.tb00001.x. [DOI] [PubMed] [Google Scholar]
- 19.Lashuel HA, Hartley D, Petre BM, Walz T, Lansbury PT., Jr Neurodegenerative disease: amyloid pores from pathogenic mutations. Nature. 2002;418:291. doi: 10.1038/418291a. [DOI] [PubMed] [Google Scholar]
- 20.Caughey B, Lansbury PT. Protofibrils, pores, fibrils, and neurodegeneration: separating the responsible protein aggregates from the innocent bystanders. Annu Rev Neurosci. 2003;26:267–298. doi: 10.1146/annurev.neuro.26.010302.081142. [DOI] [PubMed] [Google Scholar]
- 21.Ferreira ST, Vieira MN, De Felice FG. Soluble protein oligomers as emerging toxins in Alzheimer's and other amyloid diseases. IUBMB Life. 2007;59:332–345. doi: 10.1080/15216540701283882. [DOI] [PubMed] [Google Scholar]
- 22.Nimmrich V, Grimm C, Draguhn A, Barghorn S, Lehmann A, Schoemaker H, Hillen H, Gross G, Ebert U, Bruehl C. Amyloid beta oligomers (A beta(1–42) globulomer) suppress spontaneous synaptic activity by inhibition of P/Q-type calcium currents. J Neurosci. 2008;28:788–797. doi: 10.1523/JNEUROSCI.4771-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nixon RA. Autophagy in neurodegenerative disease: friend, foe or turncoat? Trends Neurosci. 2006;29:528–535. doi: 10.1016/j.tins.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 24.Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem. 2009;78:959–991. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- 25.Huang X, Cuajungco MP, Atwood CS, Hartshorn MA, Tyndall JD, Hanson GR, Stokes KC, Leopold M, Multhaup G, Goldstein LE, et al. Cu(II) potentiation of Alzheimer abeta neurotoxicity. Correlation with cell-free hydrogen peroxide production and metal reduction. J Biol Chem. 1999;274:37111–37116. doi: 10.1074/jbc.274.52.37111. [DOI] [PubMed] [Google Scholar]
- 26.Huang X, Atwood CS, Hartshorn MA, Multhaup G, Goldstein LE, Scarpa RC, Cuajungco MP, Gray DN, Lim J, Moir RD, et al. The A beta peptide of Alzheimer's disease directly produces hydrogen peroxide through metal ion reduction. Biochemistry. 1999;38:7609–7616. doi: 10.1021/bi990438f. [DOI] [PubMed] [Google Scholar]
- 27.Cuajungco MP, Goldstein LE, Nunomura A, Smith MA, Lim JT, Atwood CS, Huang X, Farrag YW, Perry G, Bush AI. Evidence that the beta-amyloid plaques of Alzheimer's disease represent the redox-silencing and entombment of abeta by zinc. J Biol Chem. 2000;275:19439–19442. doi: 10.1074/jbc.C000165200. [DOI] [PubMed] [Google Scholar]
- 28.Opazo C, Huang X, Cherny RA, Moir RD, Roher AE, White AR, Cappai R, Masters CL, Tanzi RE, Inestrosa NC, et al. Metalloenzyme-like activity of Alzheimer's disease beta-amyloid. Cu-dependent catalytic conversion of dopamine, cholesterol, and biological reducing agents to neurotoxic H(2)O(2) J Biol Chem. 2002;277:40302–40308. doi: 10.1074/jbc.M206428200. [DOI] [PubMed] [Google Scholar]
- 29.Barnham KJ, Ciccotosto GD, Tickler AK, Ali FE, Smith DG, Williamson NA, Lam YH, Carrington D, Tew D, Kocak G, et al. Neurotoxic, redox-competent Alzheimer's beta-amyloid is released from lipid membrane by methionine oxidation. J Biol Chem. 2003;278:42959–42965. doi: 10.1074/jbc.M305494200. [DOI] [PubMed] [Google Scholar]
- 30.Macario AJ, Conway de Macario E. Sick chaperones, cellular stress, and disease. N Engl J Med. 2005;353:1489–1501. doi: 10.1056/NEJMra050111. [DOI] [PubMed] [Google Scholar]
- 31.Muchowski PJ, Wacker JL. Modulation of neurodegeneration by molecular chaperones. Nat Rev Neurosci. 2005;6:11–22. doi: 10.1038/nrn1587. [DOI] [PubMed] [Google Scholar]
- 32.Almeida CG, Takahashi RH, Gouras GK. Beta-amyloid accumulation impairs multivesicular body sorting by inhibiting the ubiquitin-proteasome system. J Neurosci. 2006;26:4277–4288. doi: 10.1523/JNEUROSCI.5078-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Serpell LC, Sunde M, Blake CC. The molecular basis of amyloidosis. Cell Mol Life Sci. 1997;53:871–887. doi: 10.1007/s000180050107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kodali R, Wetzel R. Polymorphism in the intermediates and products of amyloid assembly. Curr Opin Struct Biol. 2007;17:48–57. doi: 10.1016/j.sbi.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 35.Petkova AT, Leapman RD, Guo Z, Yau WM, Mattson MP, Tycko R. Self-propagating, molecular-level polymorphism in Alzheimer's beta-amyloid fibrils. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- 36.Wetzel R, Shivaprasad S, Williams AD. Plasticity of amyloid fibrils. Biochemistry. 2007;46:1–10. doi: 10.1021/bi0620959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goldsbury C, Frey P, Olivieri V, Aebi U, Muller SA. Multiple assembly pathways underlie amyloid-beta fibril polymorphisms. J Mol Biol. 2005;352:282–298. doi: 10.1016/j.jmb.2005.07.029. [DOI] [PubMed] [Google Scholar]
- 38.Gosal WS, Morten IJ, Hewitt EW, Smith DA, Thomson NH, Radford SE. Competing pathways determine fibril morphology in the self-assembly of beta2-microglobulin into amyloid. J Mol Biol. 2005;351:850–864. doi: 10.1016/j.jmb.2005.06.040. [DOI] [PubMed] [Google Scholar]
- 39.Uversky VN, Fernandez A, Fink AL. Structural and conformational prerequisites of amyloidogenesis. In: Uversky VN, Fink AL, editors. Protein Misfolding, Aggregation and Conformational Diseases. New York, NY: Springer Science+Business Media, LLC; 2006. pp. 1–20. [Google Scholar]
- 40.Griffith JS. Self-replication and scrapie. Nature. 1967;215:1043–1044. doi: 10.1038/2151043a0. [DOI] [PubMed] [Google Scholar]
- 41.Uversky VN, Eliezer D. Biophysics of Parkinson's disease: structure and aggregation of alpha-synuclein. Curr Protein Pept Sci. 2009;10:483–499. doi: 10.2174/138920309789351921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morris AM, Watzky MA, Finke RG. Protein aggregation kinetics, mechanism, and curve-fitting: a review of the literature. Biochim Biophys Acta. 2009;1794:375–397. doi: 10.1016/j.bbapap.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 43.Ferrone F. Analysis of protein aggregation kinetics. Methods Enzymol. 1999;309:256–274. doi: 10.1016/s0076-6879(99)09019-9. [DOI] [PubMed] [Google Scholar]
- 44.Ferrone FA, Hofrichter J, Sunshine HR, Eaton WA. Kinetic studies on photolysis-induced gelation of sickle cell hemoglobin suggest a new mechanism. Biophys J. 1980;32:361–380. doi: 10.1016/S0006-3495(80)84962-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wegner A, Savko P. Fragmentation of actin filaments. Biochemistry. 1982;21:1909–1913. doi: 10.1021/bi00537a032. [DOI] [PubMed] [Google Scholar]
- 46.Baskakov IV. Branched chain mechanism of polymerization and ultrastructure of prion protein amyloid fibrils. FEBS J. 2007;274:3756–3765. doi: 10.1111/j.1742-4658.2007.05916.x. [DOI] [PubMed] [Google Scholar]
- 47.Lomakin A, Chung DS, Benedek GB, Kirschner DA, Teplow DB. On the nucleation and growth of amyloid beta-protein fibrils: detection of nuclei and quantitation of rate constants. Proc Natl Acad Sci USA. 1996;93:1125–1129. doi: 10.1073/pnas.93.3.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo J, Taddei N, Ramponi G, Dobson CM, Stefani M. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 2002;416:507–511. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- 49.Sakono M, Zako T. Amyloid oligomers: formation and toxicity of Abeta oligomers. FEBS J. 2010;277:1348–1358. doi: 10.1111/j.1742-4658.2010.07568.x. [DOI] [PubMed] [Google Scholar]
- 50.Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer's amyloid beta-peptide. Nat Rev Mol Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 51.LaFerla FM, Green KN, Oddo S. Intracellular amyloid-beta in Alzheimer’s disease. Nat Rev Neurosci. 2007;8:499–509. doi: 10.1038/nrn2168. [DOI] [PubMed] [Google Scholar]
- 52.Klein WL, Krafft GA, Finch CE. Targeting small Abeta oligomers: the solution to an Alzheimer's disease conundrum? Trends Neurosci. 2001;24:219–224. doi: 10.1016/s0166-2236(00)01749-5. [DOI] [PubMed] [Google Scholar]
- 53.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 54.Glabe CG. Structural classification of toxic amyloid oligomers. J Biol Chem. 2008;283:29639–29643. doi: 10.1074/jbc.R800016200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roychaudhuri R, Yang M, Hoshi MM, Teplow DB. Amyloid beta-protein assembly and Alzheimer disease. J Biol Chem. 2009;284:4749–4753. doi: 10.1074/jbc.R800036200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kuo YM, Emmerling MR, Vigo-Pelfrey C, Kasunic TC, Kirkpatrick JB, Murdoch GH, Ball MJ, Roher AE. Water-soluble Abeta (N-40, N-42) oligomers in normal and Alzheimer disease brains. J Biol Chem. 1996;271:4077–4081. doi: 10.1074/jbc.271.8.4077. [DOI] [PubMed] [Google Scholar]
- 57.Podlisny MB, Ostaszewski BL, Squazzo SL, Koo EH, Rydell RE, Teplow DB, Selkoe DJ. Aggregation of secreted amyloid beta-protein into sodium dodecyl sulfate-stable oligomers in cell culture. J Biol Chem. 1995;270:9564–9570. doi: 10.1074/jbc.270.16.9564. [DOI] [PubMed] [Google Scholar]
- 58.Walsh DM, Tseng BP, Rydel RE, Podlisny MB, Selkoe DJ. The oligomerization of amyloid beta-protein begins intracellularly in cells derived from human brain. Biochemistry. 2000;39:10831–10839. doi: 10.1021/bi001048s. [DOI] [PubMed] [Google Scholar]
- 59.Bernstein SL, Dupuis NF, Lazo ND, Wyttenbach T, Condron MM, Bitan G, Teplow DB, Shea J-E, Ruotolo BT, Robinson CV, et al. Amyloid-β protein oligomerization and the importance of tetramers and dodecamers in the aetiology of Alzheimer's disease. Nat Chem. 2009;1:326–331. doi: 10.1038/nchem.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Walsh DM, Klyubin I, Fadeeva JV, Cullen WK, Anwyl R, Wolfe MS, Rowan MJ, Selkoe DJ. Naturally secreted oligomers of amyloid beta protein potently inhibit hippocampal long-term potentiation in vivo. Nature. 2002;416:535–539. doi: 10.1038/416535a. [DOI] [PubMed] [Google Scholar]
- 61.Walsh DM, Klyubin I, Fadeeva JV, Rowan MJ, Selkoe DJ. Amyloid-beta oligomers: their production, toxicity and therapeutic inhibition. Biochem Soc Trans. 2002;30:552–557. doi: 10.1042/bst0300552. [DOI] [PubMed] [Google Scholar]
- 62.Ono K, Condron MM, Teplow DB. Structure-neurotoxicity relationships of amyloid beta-protein oligomers. Proc Natl Acad Sci USA. 2009;106:14745–14750. doi: 10.1073/pnas.0905127106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lesne S, Koh MT, Kotilinek L, Kayed R, Glabe CG, Yang A, Gallagher M, Ashe KH. A specific amyloid-beta protein assembly in the brain impairs memory. Nature. 2006;440:352–357. doi: 10.1038/nature04533. [DOI] [PubMed] [Google Scholar]
- 64.Lambert MP, Barlow AK, Chromy BA, Edwards C, Freed R, Liosatos M, Morgan TE, Rozovsky I, Trommer B, Viola KL, et al. Diffusible, nonfibrillar ligands derived from Abeta 1–42 are potent central nervous system neurotoxins. Proc Natl Acad Sci USA. 1998;95:6448–6453. doi: 10.1073/pnas.95.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hoshi M, Sato M, Matsumoto S, Noguchi A, Yasutake K, Yoshida N, Sato K. Spherical aggregates of beta-amyloid (amylospheroid) show high neurotoxicity and activate tau protein kinase I/glycogen synthase kinase-3beta. Proc Natl Acad Sci USA. 2003;100:6370–6375. doi: 10.1073/pnas.1237107100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lashuel HA, Lansbury PT., Jr Are amyloid diseases caused by protein aggregates that mimic bacterial pore-forming toxins? Q Rev Biophys. 2006;39:167–201. doi: 10.1017/S0033583506004422. [DOI] [PubMed] [Google Scholar]
- 67.Bitan G, Kirkitadze MD, Lomakin A, Vollers SS, Benedek GB, Teplow DB. Amyloid beta-protein (Abeta) assembly: Abeta 40 and Abeta 42 oligomerize through distinct pathways. Proc Natl Acad Sci USA. 2003;100:330–335. doi: 10.1073/pnas.222681699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Uversky VN, Lee HJ, Li J, Fink AL, Lee SJ. Stabilization of partially folded conformation during alpha-synuclein oligomerization in both purified and cytosolic preparations. J Biol Chem. 2001;276:43495–43498. doi: 10.1074/jbc.C100551200. [DOI] [PubMed] [Google Scholar]
- 69.Souza JM, Giasson BI, Chen Q, Lee VM, Ischiropoulos H. Dityrosine cross-linking promotes formation of stable alpha-synuclein polymers. Implication of nitrative and oxidative stress in the pathogenesis of neurodegenerative synucleinopathies. J Biol Chem. 2000;275:18344–18349. doi: 10.1074/jbc.M000206200. [DOI] [PubMed] [Google Scholar]
- 70.Conway KA, Lee SJ, Rochet JC, Ding TT, Harper JD, Williamson RE, Lansbury PT., Jr Accelerated oligomerization by Parkinson's disease linked alpha-synuclein mutants. Ann N Y Acad Sci. 2000;920:42–45. doi: 10.1111/j.1749-6632.2000.tb06903.x. [DOI] [PubMed] [Google Scholar]
- 71.Conway KA, Lee SJ, Rochet JC, Ding TT, Williamson RE, Lansbury PT., Jr Acceleration of oligomerization, not fibrillization, is a shared property of both alpha-synuclein mutations linked to early-onset Parkinson's disease: implications for pathogenesis and therapy. Proc Natl Acad Sci USA. 2000;97:571–576. doi: 10.1073/pnas.97.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ding TT, Lee SJ, Rochet JC, Lansbury PT., Jr Annular alpha-synuclein protofibrils are produced when spherical protofibrils are incubated in solution or bound to brain-derived membranes. Biochemistry. 2002;41:10209–10217. doi: 10.1021/bi020139h. [DOI] [PubMed] [Google Scholar]
- 73.Jo E, McLaurin J, Yip CM, St George-Hyslop P, Fraser PE. alpha-Synuclein membrane interactions and lipid specificity. J Biol Chem. 2000;275:34328–34334. doi: 10.1074/jbc.M004345200. [DOI] [PubMed] [Google Scholar]
- 74.Lee HJ, Choi C, Lee SJ. Membrane-bound alpha-synuclein has a high aggregation propensity and the ability to seed the aggregation of the cytosolic form. J Biol Chem. 2002;277:671–678. doi: 10.1074/jbc.M107045200. [DOI] [PubMed] [Google Scholar]
- 75.Munishkina LA, Phelan C, Uversky VN, Fink AL. Conformational behavior and aggregation of alpha-synuclein in organic solvents: modeling the effects of membranes. Biochemistry. 2003;42:2720–2730. doi: 10.1021/bi027166s. [DOI] [PubMed] [Google Scholar]
- 76.Lowe R, Pountney DL, Jensen PH, Gai WP, Voelcker NH. Calcium(II) selectively induces alpha-synuclein annular oligomers via interaction with the C-terminal domain. Protein Sci. 2004;13:3245–3252. doi: 10.1110/ps.04879704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Apetri MM, Maiti NC, Zagorski MG, Carey PR, Anderson VE. Secondary structure of alpha-synuclein oligomers: characterization by Raman and atomic force microscopy. J Mol Biol. 2006;355:63–71. doi: 10.1016/j.jmb.2005.10.071. [DOI] [PubMed] [Google Scholar]
- 78.Hoyer W, Cherny D, Subramaniam V, Jovin TM. Rapid self-assembly of alpha-synuclein observed by in situ atomic force microscopy. J Mol Biol. 2004;340:127–139. doi: 10.1016/j.jmb.2004.04.051. [DOI] [PubMed] [Google Scholar]
- 79.Yamin G, Uversky VN, Fink AL. Nitration inhibits fibrillation of human alpha-synuclein in vitro by formation of soluble oligomers. FEBS Lett. 2003;542:147–152. doi: 10.1016/s0014-5793(03)00367-3. [DOI] [PubMed] [Google Scholar]
- 80.Uversky VN, Yamin G, Munishkina LA, Karymov MA, Millett IS, Doniach S, Lyubchenko YL, Fink AL. Effects of nitration on the structure and aggregation of alpha-synuclein. Brain Res Mol Brain Res. 2005;134:84–102. doi: 10.1016/j.molbrainres.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 81.Hong DP, Fink AL, Uversky VN. Smoking and Parkinson's disease: does nicotine affect alpha-synuclein fibrillation? Biochim Biophys Acta. 2009;1794:282–290. doi: 10.1016/j.bbapap.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Meng X, Munishkina LA, Fink AL, Uversky VN. Molecular mechanisms underlying the flavonoid-induced inhibition of alpha-synuclein fibrillation. Biochemistry. 2009;48:8206–8224. doi: 10.1021/bi900506b. [DOI] [PubMed] [Google Scholar]
- 83.Zhou W, Gallagher A, Hong DP, Long C, Fink AL, Uversky VN. At low concentrations, 3,4-dihydroxyphenylacetic acid (DOPAC) binds non-covalently to alpha-synuclein and prevents its fibrillation. J Mol Biol. 2009;388:597–610. doi: 10.1016/j.jmb.2009.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhou W, Long C, Reaney SH, Di Monte DA, Fink AL, Uversky VN. Methionine oxidation stabilizes non-toxic oligomers of alpha-synuclein through strengthening the auto-inhibitory intra-molecular long-range interactions. Biochim Biophys Acta. 2010;1802:322–330. doi: 10.1016/j.bbadis.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hong DP, Fink AL, Uversky VN. Structural characteristics of alpha-synuclein oligomers stabilized by the flavonoid baicalein. J Mol Biol. 2008;383:214–223. doi: 10.1016/j.jmb.2008.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Selkoe DJ. Altered structural proteins in plaques and tangles: what do they tell us about the biology of Alzheimer’s disease? Neurobiol Aging. 1986;7:425–432. doi: 10.1016/0197-4580(86)90055-2. [DOI] [PubMed] [Google Scholar]
- 87.DeMager PP, Penke B, Walter R, Harkany T, Hartignny W. Pathological peptide folding in Alzheimer's disease and other conformational disorders. Curr Med Chem. 2002;9:1763–1780. doi: 10.2174/0929867023369169. [DOI] [PubMed] [Google Scholar]
- 88.LaFerla FM, Oddo S. Alzheimer's disease: Abeta, tau and synaptic dysfunction. Trends Mol Med. 2005;11:170–176. doi: 10.1016/j.molmed.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 89.Santacruz K, Lewis J, Spires T, Paulson J, Kotilinek L, Ingelsson M, Guimaraes A, DeTure M, Ramsden M, McGowan E, et al. Tau suppression in a neurodegenerative mouse model improves memory function. Science. 2005;309:476–481. doi: 10.1126/science.1113694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Peterson DW, Zhou H, Dahlquist FW, Lew J. A soluble oligomer of tau associated with fiber formation analyzed by NMR. Biochemistry. 2008;47:7393–7404. doi: 10.1021/bi702466a. [DOI] [PubMed] [Google Scholar]
- 91.Sahara N, Maeda S, Murayama M, Suzuki T, Dohmae N, Yen SH, Takashima A. Assembly of two distinct dimers and higher-order oligomers from full-length tau. Eur J Neurosci. 2007;25:3020–3029. doi: 10.1111/j.1460-9568.2007.05555.x. [DOI] [PubMed] [Google Scholar]
- 92.Maeda S, Sahara N, Saito Y, Murayama S, Ikai A, Takashima A. Increased levels of granular tau oligomers: an early sign of brain aging and Alzheimer's disease. Neurosci Res. 2006;54:197–201. doi: 10.1016/j.neures.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 93.Prusiner SB. Prions. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, Glabe CG. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 95.Eghiaian F. Structuring the puzzle of prion propagation. Curr Opin Struct Biol. 2005;15:724–730. doi: 10.1016/j.sbi.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 96.Silveira JR, Raymond GJ, Hughson AG, Race RE, Sim VL, Hayes SF, Caughey B. The most infectious prion protein particles. Nature. 2005;437:257–261. doi: 10.1038/nature03989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jain S, Udgaonkar JB. Evidence for stepwise formation of amyloid fibrils by the mouse prion protein. J Mol Biol. 2008;382:1228–1241. doi: 10.1016/j.jmb.2008.07.052. [DOI] [PubMed] [Google Scholar]
- 98.Ohhashi Y, Ito K, Toyama BH, Weissman JS, Tanaka M. Differences in prion strain conformations result from non-native interactions in a nucleus. Nat Chem Biol. 2010;6:225–230. doi: 10.1038/nchembio.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nielsen L, Khurana R, Coats A, Frokjaer S, Brange J, Vyas S, Uversky VN, Fink AL. Effect of environmental factors on the kinetics of insulin fibril formation: elucidation of the molecular mechanism. Biochemistry. 2001;40:6036–6046. doi: 10.1021/bi002555c. [DOI] [PubMed] [Google Scholar]
- 100.Ahmad A, Uversky VN, Hong D, Fink AL. Early events in the fibrillation of monomeric insulin. J Biol Chem. 2005;280:42669–42675. doi: 10.1074/jbc.M504298200. [DOI] [PubMed] [Google Scholar]
- 101.Podesta A, Tiana G, Milani P, Manno M. Early events in insulin fibrillization studied by time-lapse atomic force microscopy. Biophys J. 2006;90:589–597. doi: 10.1529/biophysj.105.068833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vestergaard B, Groenning M, Roessle M, Kastrup JS, van de Weert M, Flink JM, Frokjaer S, Gajhede M, Svergun DI. A helical structural nucleus is the primary elongating unit of insulin amyloid fibrils. PLoS Biol. 2007;5:e134. doi: 10.1371/journal.pbio.0050134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Solomon A. Clinical implications of monoclonal light chains. Semin Oncol. 1986;13:341–349. [PubMed] [Google Scholar]
- 104.Stevens PW, Raffen R, Hanson DK, Deng YL, Berrios-Hammond M, Westholm FA, Murphy C, Eulitz M, Wetzel R, Solomon A, et al. Recombinant immunoglobulin variable domains generated from synthetic genes provide a system for in vitro characterization of light-chain amyloid proteins. Protein Sci. 1995;4:421–432. doi: 10.1002/pro.5560040309. doi: 10.1002/pro.5560040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Souillac PO, Uversky VN, Fink AL. Structural transformations of oligomeric intermediates in the fibrillation of the immunoglobulin light chain LEN. Biochemistry. 2003;42:8094–8104. doi: 10.1021/bi034652m. doi: 10.1021/bi034652m. [DOI] [PubMed] [Google Scholar]
- 106.Souillac PO, Uversky VN, Millett IS, Khurana R, Doniach S, Fink AL. Elucidation of the molecular mechanism during the early events in immunoglobulin light chain amyloid fibrillation. Evidence for an off-pathway oligomer at acidic pH. J Biol Chem. 2002;277:12666–12679. doi: 10.1074/jbc.M109229200. doi: 10.1074/jbc.M109229200. [DOI] [PubMed] [Google Scholar]