Abstract
The genome-wide association study by Herbert and colleagues identified the INSIG2 single nucleotide polymorphism (SNP) rs7566605 as contributing to increased BMI in ethnically distinct cohorts. The present study sought to further clarify by testing whether SNPs of INSIG2 influenced quantitative adiposity or glucose homeostasis traits in Hispanics of the Insulin Resistance Atherosclerosis Family Study (IRASFS). Using a tagging SNP approach, rs7566605 and 31 additional SNPs were genotyped in 1425 IRASFS Hispanics. SNPs were tested for association with six adiposity measures: BMI, waist circumference (WAIST), waist to hip ratio (WHR), subcutaneous adipose tissue (SAT), visceral adipose tissue (VAT), and VAT to SAT ratio (VSR). SNPs were also tested for association with fasting glucose (GFAST), fasting insulin (FINS), and three measures obtained from the frequently sampled intravenous glucose tolerance test: insulin sensitivity (SI), acute insulin response (AIR), and disposition index (DI). Most prominent association was observed with direct CT-measured adiposity phenotypes, including VAT, SAT, and VSR (P-values range from 0.007 to 0.044 for rs17586756, rs17047718, rs17047731, rs9308762, rs12623648, and rs11673900). Multiple SNP associations were observed with all glucose homeostasis traits (P-values range from 0.001 to 0.031 for rs17047718, rs17047731, rs2161829, rs10490625, rs889904, and rs12623648). Using BMI as a covariate in evaluation of glucose homeostasis traits slightly reduced their association. However, association with adiposity and glucose homeostasis phenotypes is not significant following multiple comparisons adjustment. Trending association after multiple comparisons adjustment remains suggestive of a role for genetic variation of INSIG2 in obesity, but these results require validation.
Keywords: Insulin-induced gene 2, single nucleotide polymorphism, genetic association, adiposity, glucose homeostasis
Introduction
INSIG2 is a ~21.5 Kb gene located on chromosome 2q14 that has been functionally linked to obesity, most notably due to its role in cholesterol and fatty acid synthesis feedback inhibition (1, 2). After conducting a genome-wide association study evaluating the contributions of single-nucleotide polymorphisms (SNPs) to obesity, Herbert et al. reported compelling evidence that SNP rs7566605 was associated with increased Body Mass Index (BMI) in a cohort of the Framingham Heart Study (FHS). Their finding of association of rs7566605 with BMI was replicated in ethnically distinct populations, including several European-derived and an African American cohort (3). The rs7566605 SNP is located ~10 Kb upstream of the transcription start site for Insulin-induced gene 2 (INSIG2).
Subsequent to the report by Herbert et al., association studies of INSIG2 have been reported in several populations. A majority of these replication studies have focused efforts solely on the initial rs7566605 and BMI. Several of these replication studies did not corroborate the INSIG2 findings in European American, Indian, Chinese, and Afro-Caribbean samples (4–11). However, rs7566605 association with BMI was reported in other European-derived cohorts (12). In addition, a recent study reported that a SNP in the INSIG1 promoter was associated with higher plasma glucose and post-load plasma glucose in middle-aged men (13). This finding implies additional consideration of a role for INSIG2 in glucose homeostasis maintenance. The diverse results from these studies suggest that INSIG2 (and rs7566605) could evince its genetic effect through other adiposity phenotypes, such as those affecting adiposity at specific depots rather than whole-body BMI. Furthermore, the variance in association results could be explained if other SNPs near or within INSIG2, and in linkage disequilibrium (LD) with rs7566605, have true biological effects that vary between populations and/or also require differing levels of power for detection.
In light of these observations, we evaluated INSIG2 using a systematic SNP tagging approach in a Hispanic study sample from the Insulin Resistance and Atherosclerosis Family Study (IRASFS). The IRASFS possesses more sophisticated measures of adipose tissue distribution that are assessed using computed tomography (CT), as well as standard anthropometric adiposity measures, and detailed measures of glucose homeostasis (14). We hypothesized that SNPs of INSIG2 will be associated with multiple measures of adiposity and/or glucose homeostasis, further clarifying the role of this well-studied gene.
Methods and Procedures
Subjects
The IRASFS is a multi-center family-based study designed to discern genetic components of quantitative measures of adiposity and glucose homeostasis in Hispanic and African Americans. IRASFS subjects were recruited from three sites: San Luis Valley, CO, a rural Hispanic population; San Antonio, TX, an urban Hispanic population; and Los Angeles, CA, an African American population. The study design, recruitment, and phenotyping are described elsewhere in more detail (14, 15). We have chosen to focus this report on the 90 multigenerational Hispanic families (1425 individuals). Study of INSIG2 polymorphisms in our African American cohort is underway. Individuals were recruited over a 2.5 year period at the clinical centers on the basis of large family size, rather than disease phenotype. Proband recruitment required at least four living siblings and five living offspring among the four siblings. Family size ranged from 4 to 39 individuals. All subjects provided informed consent. Though the subjects were recruited on the basis of family size and not disease status, approximately 13.7% were diagnosed with type 2 diabetes (by self report). The actual number of diabetics in the total Hispanic cohort is n=181 (14.2%), confirmed during clinical exams. Table 1 summarizes the primary phenotypes measured on study participants.
Table 1.
Demographics of the IRASFS total Hispanic cohort.
| Hispanic individuals in IRASFS | ||
|---|---|---|
| Phenotype | Mean (SD) | Median |
| Demographic | ||
| Age (yrs) | 42.8 (14.6) | 41.3 |
| Female gender (%) | 58 | |
| Adiposity | ||
| Body Mass Index (kg/m2) (BMI) | 28.9 (6.1) | 28.1 |
| Waist Circumference (cm) | 89.8 (14.3) | 89.1 |
| Weight (kg) | 77.6 (18.2) | 75.5 |
| Waist/Hip Ratio (WHR) | 0.86 (0.08) | 0.85 |
| Subcutaneous Fat Area L4/L5 (cm2) (SAT) | 338.7 (154.7) | 313.7 |
| Visceral Fat Area L4/L5 (cm2) (VAT) | 113.8 (61.2) | 105.9 |
| Visceral: Subcutaneous Ratio (VSR) | 0.38 (0.21) | 0.33 |
| Glucose Homeostasis | ||
| Diabetics in Cohort (n; %) | 181; 14.2% | |
| Insulin Sensitivity (SI, ×10−5 min−1/[pmol/L]) | 2.0 (1.9) | 1.5 |
| Acute Insulin Response (AIR, pmol/L) | 695 (655) | 534 |
| Disposition index (DI; 10−5 min−1) | 1205.3 (1238.2) | 918.7 |
| glucose effectiveness (SG; min−1) | 0.02 (.009) | 0.02 |
| fasting insulin (FINS) | 16.1 (12.9) | 13 |
| fasting glucose (FGLU; mg/dL) | 103.2 (34.8) | 93.5 |
Insulin Resistance Atherosclerosis Family Study (IRASFS)
Phenotypes
The IRASFS subjects have been extensively phenotyped with adiposity and glucose homeostasis quantitative traits. Body Mass Index (BMI, kg/m2) was obtained from height and weight measurements taken for each subject at the study exam. Waist circumference (WAIST, cm) was taken with a standard metric tape measurer and consisted, at minimum, of the circumference between the tenth rib and the iliac crest. Hip circumference was also obtained using a standard metric tape measure and consisted of the maximum circumference of the buttocks. The waist-to-hip ratio (WHR) was calculated as the ratio of the WAIST and hip circumference measurements. In addition to standard anthropometric measures of adiposity, the IRASFS incorporates direct measures of adiposity at specific abdominal depots with the use of computed tomography (CT) scanning. Visceral and subcutaneous adipose tissue (VAT; SAT; respectively, cm2) were acquired in a standard protocol consisting of a single scout of the abdomen that was followed by two 10mm thick axial images (obtained at L2–L3 and L4–L5 spinal discs). The CT images were transferred to magnetic tape and sent to a reading center at the University of Colorado Health Sciences for analysis (15). The visceral to subcutaneous ratio (VSR) was obtained by calculating the ratio of the visceral to subcutaneous adipose tissue estimates.
Measures of glucose homoeostasis were assessed using the frequently sampled intravenous glucose tolerance test (FSIGT), and minimal model analyses (MINMOD software program), in order to calculate insulin sensitivity (SI) and glucose effectiveness (SG; 16, 17). Acute insulin response (AIR) was calculated as the mean insulin increment in plasma insulin concentration above basal during the first 8 minutes following glucose infusion. Disposition Index (DI) was calculated as DI=AIR × SI. Plasma glucose (GFAST) and insulin (FINS) values were obtained using standard methods. Diabetes-affected individuals were excluded from SNP association analysis with glucose homeostasis traits.
DNA Preparation
Genomic DNA was purified from whole blood PUREGENE DNA isolation kits (Gentra Inc., Minneapolis, MN, USA). Quantification of the purified DNA was performed by fluorometric assay (Hoefer DyNA Quant 200 fluorometer; Hoefer Pharmacia Biotech Inc., San Francisco, CA, USA). All of the samples were diluted to a final concentration of 5 ng/μl.
SNP Genotyping and Selection
Genotyping was performed using the iPlex MassARRAY SNP genotyping system (Sequenom Inc., San Diego, CA, USA), which utilizes mass tagging to differentiate between alleles (18). SNPs were selected to cover a 52.46 Kb genomic region, which included the INSIG2 gene, 26 Kb 5′ to the gene, and 8 Kb 3′ to the gene. At the time of SNP selection for this study, only the Phase 2 HapMap Project data was available, and thus did not include a Hispanic population. However, existing Phase 2 HapMap Project population tagging SNPs had been shown to be largely transferable as tagging SNPs for other ethnic cohorts, such as the IRASFS Hispanics (19). Thus, the Hapmap pairwise tagging algorithm was run to select CEU HapMap tagSNPs with an r2 threshold of 0.8 at the MAF cut-off of 5%. The tagging SNPs were supplemented with additional HapMap (www.hapmap.org) genotyped SNPs, as well as those of dbSNP. According to the tagging function of HapMap, over 90% of the CEU haplotypic variation in the genomic region is captured by the 31 selected SNPs at an r2 threshold of 0.8. This same set of SNPs also captures over 75% of the haplotypic variation in the INSIG2 genomic region at an r2 threshold of 0.8 in the YRI HapMap population.
Each pedigree has previously been examined for consistency of stated family structure and is described in detail elsewhere (20). Maximum likelihood estimates of allele frequencies were computed using the largest set of unrelated individuals (n=228) and tested for departures from Hardy-Weinberg equilibrium proportions (HWE) using a chi squared goodness of fit test. The largest set of unrelated individuals was also used to calculate the D′ and r2 inter-SNP linkage disequilibrium (LD) statistics. Each of the SNPs evaluated in this work were examined for Mendelian inconsistencies in their genotypes using the program PEDCHECK (21). Any genotypes inconsistent with Mendelian inheritance that could not be resolved by examination of the genotyping data were converted to missing.
Statistical Analysis
Single-SNP analysis was performed using the variance components method as implemented in the software package, SOLAR (http://www.sfbr.org/solar/). Briefly, analysis consisted of the two degree of freedom overall test of genotypic association and the three individual models defined by the a priori genetic models (e.g., dominant, additive, recessive). To minimize type 1 error, we reported the individual genetic models only when the genotypic association test (2df test) suggested an association, or after adjusting the individual genetic model P-values by a principal component analysis method. Tests were computed by adjusting measures of adiposity and glucose homeostasis for age, gender and recruitment center (San Antonio, TX and San Luis Valley, CO) and, in parallel, adjusting for age, gender, BMI, and recruitment center. When necessary, quantitative traits were transformed to best approximate the distributional assumptions of the test (i.e., conditional normality and homogeneity of variance).
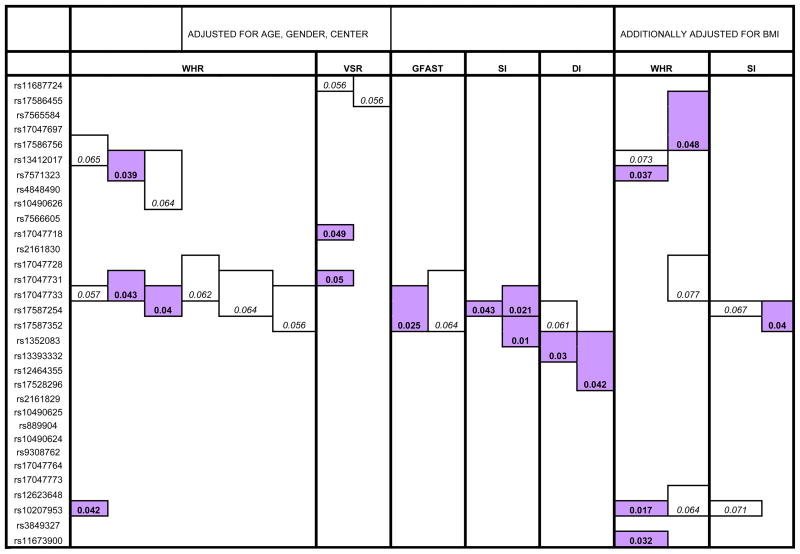
The data were also analyzed using the quantitative pedigree disequilibrium test (QPDT), using one, two, three, and four marker moving windows, which assess single SNP and haplotype association. The moving window analysis considers progressive combinations of physically adjacent SNPs as part of multi-SNP haplotypes. The method determines whether those haplotypes are under- or over- transmitted to offspring in family units whose quantitative trait variance departs from that expected based upon the quantitative trait variance and mean of the population (22). A global p-value is generated by the QPDT, which represents the overall significance based on the association tests of all the individual allele combinations of that haplotype, and these are what are initially reported (in Figure 2). Additionally reported are the most associated combinations of alleles for each globally associated haplotype (Supplementary Table 2). The QPDT uses the expectation-maximization algorithm to estimate the haplotype frequencies and is generally robust to potential population stratification.
Figure 2.
Global P-values from QPDT association analysis of single and multi-marker SNP haplotypes with adiposity and glucose homeostasis phenotypes. Italicized numbers represent trending association of a single-SNP or haplotype with a trait (P<.08) and bold numbers in shaded boxes represent association (P<.05). Only those phenotypes with two or more trending/associated elements are shown here, others are excluded. SAT is not included under BMI adjusted results due to high correlation with BMI. Results adjusted for age, gender, and center are on the left and those adjusted for age, gender, center, and BMI are on the right. These results have not been adjusted for multiple comparisons or inter-SNP correlation.
Waist to Hip Ratio (WHR); Visceral Adipose Tissue (VAT); Subcutaneous Adipose Tissue (SAT); Visceral to Subcutaneous Ratio (VSR); Fasting Glucose (GFAST); Fasting Insulin (FINS); Insulin Sensitivity (SI); Acute Insulin Response (AIR); Disposition Index (DI)
In order to adjust for multiple comparisons, we computed a principal component analysis to estimate the number of independent dimensions in the dataset (23). The number of independent dimensions in the phenotypic data was determined using the estimated genetic correlations between adiposity traits (Supplementary Table 3). It was found that 3 dimensions accounted for approximately 96% of the variation in the adiposity traits (data not shown). The number of independent dimensions in the genotypic data was determined using the pairwise D′ and r2 statistics, calculated and represented in Supplementary Figure 1. It was found that 9 dimensions accounted for approximately 95% of the variation in the genotyped SNPs (data not shown). Subsequently, the estimated number of independent dimensions across the dataset (9 × 3 = 27) is used to determine a standard Bonferroni threshold of statistical significance (.05/27 = .002).
Results
The present study genotyped rs7566605 and 31 additional Phase 2 HapMap CEU tagSNPs in 1425 Hispanic Americans of the IRASFS. In total, 17 genotyped SNPs are in the region 5′ to INSIG2, 9 are in introns of the genic region, and 6 are within the 3′ UTR (locations represented to scale in Supplementary Figure 1). As such, the present study depicts one of the first higher density haplotype block structures of the INSIG2 genomic region. Furthermore, now that the Phase 3 Hapmap Project data is available, we were able to compare the IRASFS-genotyped MAFs for INSIG2 tagging SNPs to those represented for the Mexican Ancestry in Los Angeles, California cohort (MEX). As expected, we found that the INSIG2 SNP MAFs are more reflective of those of the MEX cohort than those of the CEU cohort.
Only one SNP, rs3849327, was found to depart significantly from HWE (Supplementary Table 1). Inter-SNP D′ calculation revealed that linkage disequilibrium (LD) was generally high (mean D′ > .80), and inspection of results shows three major haplotype blocks (Gabriel method; Supplementary Figure 1). The largest haplotype block is ~28Kb and encompasses a large portion of the promoter and 5′ end of the genic region (Haplotype Block 1), the second haplotype block is ~3Kb and encompasses portions of the second and third introns (Haplotype Block 2), and the third haplotype block is less than 1Kb within the 3′-UTR (Haplotype Block 3). Additionally, r2 calculations show an overall low degree of inter-SNP correlation (grand mean r2 < .13), consistent with the selection of tagging SNPs for genotyping. However, inter-SNP correlation was significantly higher within and near Gabriel-defined Haplotype Blocks 1 and 2, where the mean r2 is ~.90 across 7 distinct intra-correlated SNP elements, consisting of 20 variants among them (mean calculated using variants with pairwise r2 statistics > .70; Supplementary Figure 1).
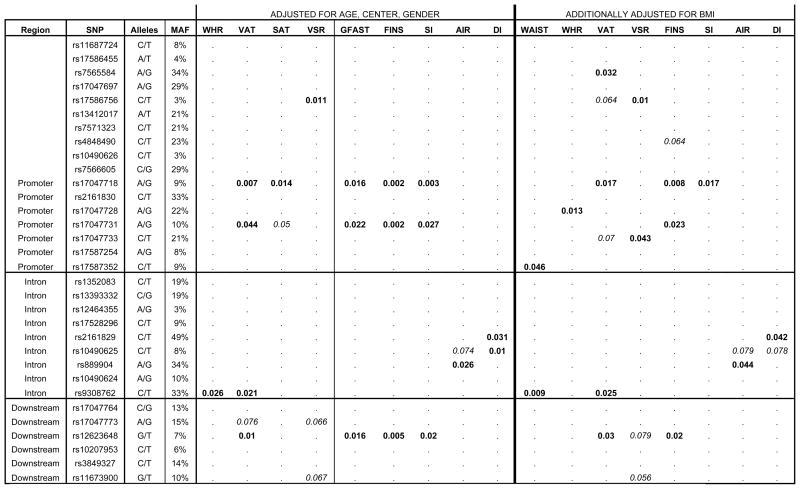
The results of single-SNP association analyses using the 2df test of SOLAR are depicted in Figure 1, while significant a priori genetic model test results for 2df-associated SNPs are depicted in Table 2. The SNP rs7566605 did not show evidence of association with any measure of adiposity or glucose homeostasis. The minor allele frequency (MAF) of rs7566605 was similar to that observed in previous studies (~30%), and it was in LD with virtually every other genotyped SNP (mean D′ with other variants = .936; D′ range =.05–1.0). Moreover, rs7566605 shows low inter-SNP correlation with all but two variants, rs17047697 (r2=.96) and rs2161830 (r2=.75), which also were not associated with any measures of adiposity or glucose homeostasis. Overall, associated SNPs under SOLAR were spread throughout the surveyed genomic region, with a higher concentration in the promoter and 5′ region of the gene. Standard anthropometric measures of adiposity (e.g. BMI, WAIST, and WHR) had little evidence of association with INSIG2 SNPs. However, one low frequency SNP showed evidence of association with BMI and WAIST (MAF ~3%, rs10490626, P=.016–.053), and another with WHR (MAF ~33%, rs9308762, P=.026). The SNP rs10490626 has a genotyped MAF < 5%, which is reflective of a bias in tag SNP selection towards the CEU HapMap population, as a Mexican American cohort in the HapMap Phase 3 release also has a lower rs10490626 MAF. More striking was evidence of association with CT-measured adiposity phenotypes, including VAT, SAT, and VSR. Most prominent are 3 moderately correlated SNPs that were associated with VAT (rs17047718 in promoter, rs17047731 in promoter, rs12623648 in 3′-UTR MAF=~7%–10%; P=.007–.044; mean r2 is ~.65). The intronic SNP rs9308762 was also associated with VAT (MAF=33%; P=.021).
Figure 1.
Single SNP Genotypic Association (2df test) Results in the IRASFS Hispanics. The minor allele frequency (MAF) and SNP designations are in the far left columns. Results adjusted by standard parameters (age, gender, center) and those adjusted by standard parameters and BMI are presented. SAT is not included in the BMI adjusted results due to high correlation with BMI (r2≥.90). Associated P-values are in bold (P<.05) and those trending towards association are italicized (P<.10). A dot (“.”) represents no association. These results have not been adjusted for multiple comparisons or inter-SNP correlation. BMI, WAIST, and BMI-ADJUSTED GFAST not included below due to scarce association.
Body Mass Index (BMI); Waist Circumference (WAIST); Waist to Hip Ratio (WHR); Visceral Adipose Tissue (VAT); Subcutaneous Adipose Tissue (SAT); Visceral to Subcutaneous Ratio (VSR); Fasting Glucose (GFAST); Fasting Insulin (FINS); Insulin Sensitivity (SI); Acute Insulin Response (AIR); Disposition Index (DI)
Table 2.
Genotypic means and significant a priori genetic model association results for prominently associated SNPs under the SOLAR 2DF test from Figure 1. These results have not been adjusted for multiple comparisons or inter-marker correlation.
Each of the genotypic mean values have accompanying standard deviations in parentheses. Second from left column lists the genomic region the SNP was located within, the third from left notes the genetic model under which the SNP was associated (as well as the p-value by which it was associated in parenthesis), the fourth from left notes the traits the SNP was associated with, the fifth from left notes the MAF, and the final 3 columns hold the genotypic means. A majority of the SNPs were associated under a recessive model. The additive model represents an effect that is dosage dependent with the minor allele, the dominant model represents an effect that is maximized when only one copy of the minor allele is inherited, and the recessive mode represents an effect that is significant only when two copies of the minor allele are inherited. 1/1 represents individuals homozygous for the major allele, ½represents heterozygous individuals, and 2/2 represents minor allele homozygotes.
| Genotypic Means (SD) |
|||||||
|---|---|---|---|---|---|---|---|
| SNP | Region | Genetic Model Association | Trait | MAF | 1,1 | 1,2 | 2,2 |
| rs17047718 | Promoter | Recessive (P=.002) | VAT | 9% | 107 (54) | 102 (64) | 57 (31) |
| (P=.012) | SAT | 320 (156) | 310 (153) | 219 (115) | |||
| (P=.007) | GFAST | 93 (10) | 94 (8) | 85 (9) | |||
| (P=.004) | FINS | 15 (10) | 14 (8) | 8 (5) | |||
| (P=.002) | SI | 2 (2) | 2 (2) | 4 (2) | |||
| rs17047731 | Promoter | Recessive (P=.019) | VAT | 10% | 114 (60) | 118 (70) | 67 (51) |
| (P=.026) | SAT | 349 (155) | 345 (157) | 221 (144) | |||
| (P=.006) | GFAST | 94 (10) | 94 (9) | 86 (9) | |||
| (P=.002) | FINS | 15 (10) | 17 (14) | 7 (5) | |||
| (P=.008) | SI | 2 (2) | 2 (2) | 4 (2) | |||
| rs2161829 | Intron 2 | Additive/Recessive (P=.013/.012) | DI | 49% | 1235 (1166) | 1296 (1203) | 1441 (1297) |
| rs889904 | Intron 2 | Additive/Dominant (P=.045/.008) | AIR | 8% | 831 (695) | 696 (577) | 765 (694) |
| rs9308762 | Intron 3 | Recessive (P=.010) | VAT | 33% | 115 (61) | 116 (61) | 94 (94) |
| rs12623648 | 3′-UTR | Recessive (P=.003) | VAT | 7% | 114 (56) | 115 (55) | 61 (35) |
| (P=.004) | GFAST | 94 (10) | 94 (9) | 84 (6) | |||
| (P=.039) | FINS | 14 (10) | 15 (8) | 7 (5) | |||
| (P=.016) | SI | 2 (2) | 3 (3) | 5 (2) | |||
Visceral Adipose Tissue (VAT); Subcutaneous Adipose Tissue (SAT); Fasting Glucose (GFAST); Fasting Insulin (FINS); Insulin Sensitivity (SI); Minor Allele Frequency (MAF)
When the single-SNP adiposity phenotype results incorporated adjustment for BMI, in addition to age, gender, and center, there were additional associations with WAIST and WHR (rs17587352, rs9308762, MAF=9–33% P=.009–.046). Adjustment for BMI also had varying effects on associations with CT-measured adiposity phenotypes. SNP associations with VAT largely persisted after BMI adjustment, save for the loss of one SNP (rs17047731, P=.196), and the appearance of an additional association (rs7565584, MAF=34%, P=.032). The SNP rs17586756 remains associated with VSR (P=.011), while rs17047733 shows newly modest association with VSR (MAF=21%; P=.043). The BMI adjusted single-SNP association results are also depicted in Figure 1.
Interestingly, in addition to evidence of association with obesity, correlated SNPs rs17047718, rs17047731, and rs12623648 accounted for the association with glucose homeostasis phenotypes gauging insulin resistance (Figure 1). These SNPs shared strong associations with GFAST, FINS, and SI, and appear to follow a recessive model with minor allele homozygotes exhibiting a protective effect (MAF=7%–9%; P=.001–.027). 3 SNPs in intron 2 were associated with glucose homeostasis phenotypes that measure β-cell function, including AIR and DI. The SNPs rs2161829 (MAF=49%) and rs10490625 (MAF=7%) were associated with DI (P=.010–.031), and rs889904 was associated with AIR (MAF=34%, P=.026). The effects of prominently associated SNPs rs17047718, rs17047731, and rs12623648 on glucose homeostasis and adiposity phenotypes are represented in Table 2.
When the single-SNP association analysis with glucose homeostasis traits was performed adjusting for BMI, evidence of association with GFAST was no longer observed. Significant association with SI was also reduced (P=.017–.149). Meanwhile, all association with FINS persisted. The SNP rs2161829 remained associated with DI, while rs10490625 did not (P=.078). Lastly, rs889904’s association with AIR persisted.
Single and multi-marker SNP association with adiposity and glucose homeostasis phenotypes was also assessed by the QPDT method. Generally, QPDT associations were more modest than those observed with SOLAR, but were consistent in both the localization of and the traits involved in genetic associations. However, there are exceptions, as exemplified in the lack of single-marker association of rs17047718, rs17047731, and rs12623648 (Figure 2). Though “1 marker haplotype” testing using QPDT is roughly equivalent in premise to the single-SNP testing of SOLAR reported in Figure 1, it has considerably reduced power due to loss of data in implementing the analysis method (see Methods and Procedures). The most associated individual allele combinations underlying the significant global P-values reported here can be found in Supplementary Table 2. 1, 2, 3, and 4 adjacent SNP haplotypes, as formed by QPDT’s moving window analysis, were modestly associated with adiposity phenotypes, including WHR (P=.039–.043) and VSR (P=.049–.050). 1, 2, 3, and 4 marker haplotypes were modestly associated with glucose homeostasis phenotypes, including GFAST (P=.025), SI (P=.010–.043), and DI (P=.030–.042). Similar results were seen following QPDT analysis adjusted for BMI, as well as age, gender, and center. Single and multi-marker SNP haplotypes were associated with BMI-adjusted adiposity measures, including WHR (P=.017–.048) and VAT (P=.042). Single and multi-marker SNP haplotypes were also associated with BMI-adjusted glucose homeostasis measures, including GFAST (P=.046) and SI (P=.040).
Given the prominence and linkage disequilibrium of the moderately correlated SNPs rs17047718, rs17047731, and rs12623648 in the presented single-SNP associations (Figure 1), we conducted an additional QPDT test involving only these three SNPs. The test revealed moderate association with VSR. Single and multi-marker SNP haplotypes including SNPs rs17047718 and rs17047731 were associated with VSR (p=.005–.049). Following the additional adjustment of the dataset for BMI, single and multi-marker SNP haplotypes including rs17047718 and rs17047731 were again associated with VSR (p=.009–.087). These results are consistent with the inter-SNP correlation pattern of these three variants. That is, rs17047718 and rs17047731 have a pairwise r2 of .84, but each of the preceding variants share a pairwise r2 of ~.55 with rs12623648. These results are more clearly depicted in Supplementary Figure 2.
The potential for type I error as a result of multiple testing raises concerns about these results. The principal component analysis described previously estimated that a majority of the genotypic variation in the dataset could be explained in 9 independent dimensions, while the phenotypic variation in the dataset could be explained in 3 independent dimensions. Therefore, 27 total dimensions are sufficient to explain a majority of the variance in the dataset, and are then used to establish a new threshold of significance to account for type I error. The standard Bonferroni correction yields a new threshold of p=.0018 for significance (p=.05/27). Though some of the associated SNPs approach significance by this estimated threshold, none of the more notable SNPs actually survive the correction (ex. rs17047718’s noted association with VAT, p=.007, and FINS, p=.002, prior to adjustment for BMI; Figure 1).
Discussion
The insulin-induced genes INSIG1 and INSIG2 inhibit Sterol Response Element Binding Protein (SREBP) activation in response to high cholesterol and are partially regulated by insulin. When uninhibited by INSIGs or other genes, SREBPs act as transcription factors promoting expression of endogenous cholesterol and fatty acid synthesis genes. As their namesake suggests, INSIG function is affected by insulin, which is aberrantly secreted in obesity and diabetes (1, 2). Clearly, there are many avenues by which genetically-driven alterations in the transcription or function of INSIG2 could contribute to excess circulation of cholesterol and fatty acids, leading to increased adiposity. The findings of Herbert et al. and some subsequent replication studies have suggested a role for inherited genetic variation of INSIG2 in human obesity. The mixed nature of all INSIG2 genetic studies (especially those targeting BMI) suggests that INSIG2 polymorphisms might affect obesity through additional modalities. Association of an INSIG1 SNP with plasma glucose, the insulin-responsiveness of the INSIG proteins, and findings of SREBP binding sites near metabolic genes, imply an additional role for INSIG2 in the regulation of glucose homeostasis (1, 2, 13).
Our findings suggest an association of INSIG2 with human obesity, but also its potential involvement in physiological control of glucose homeostasis. Relevant to prior reports, we observed no evidence for association between rs7566605 and any phenotypic measures of adiposity (or glucose homeostasis) in the IRASFS. The strongest observed SNP associations with obesity measures was with CT-measured fat deposits at specific abdominal depots (VAT, SAT, VSR; Figure 1). Many of the same SNPs associated with adiposity levels also accounted for a majority of the association with glucose homeostasis measures, which included phenotypes gauging insulin response/resistance (FINS, GFAST, and SI). Significant evidence of association with AIR and DI was also observed, most of which was within intron 2 of INSIG2. The most prominent of the adiposity- or glucose homeostasis-associated INSIG2 SNPs were rs17047731, rs17047718, and rs12623648. The preceding three SNPs exhibited moderate inter-SNP correlation, as well as some haplotypic association with VSR, and so they are not completely independent elements (Supp Figure 2). Likewise, a similar amount of high inter-SNP correlation exists between many other INSIG2 SNPs of the promoter and the first and second introns (e.g. 20 SNPs can be approximately grouped into 7 intra-correlated elements with a mean r2 ≥ .90 in this region). A visual representation of the data regarding inter-SNP correlations is presented in Supplementary igure 1 (Results; Figure 1; Supp Figure 1).
Generally, a mechanism by which an INSIG2 SNP may affect adiposity or glucose homeostasis is by altering the gene’s transcription or response to insulin/cholesterol, resulting in aberrant SREBP activation and thus gene transcription (13). Given INSIG2’s regulatory role in the amount of circulating fatty acids and cholesterol, as well as the inability of BMI to distinguish between lean and fat mass, abdominal fat depot size (VAT, SAT, VSR) may be a more sensitive measure of the gene’s function. Adjustment for BMI caused the appearance of some additional single SNP associations with adiposity phenotypes, but did not alter a majority of the statistical significance of previous associations (Figure 1). These effects of BMI adjustment further confirm that BMI is not as consistent a gauge of INSIG2 function as direct adiposity measures (Figure 1). Furthermore, this could be an indication that downstream SREBPs initiate transcription of genes involved in bodily fat patterning, which may explain the persistent association of INSIG2 SNPs with VAT after BMI adjustment.
These results have not been adjusted for type I error as presented, and should thus be considered with caution. One perspective on accounting for type I error would utilize a standard Bonferroni correction for all SNP and trait comparisons, but this may be too conservative (23, 24). Such a correction may be too stringent because there is a strong prior hypothesis that this gene harbors obesity-associated variants, which is based on a body of genetic and functional evidence (25–27). Secondly, a predetermined set of adiposity and glucose homeostasis traits were used to test each variant for genetic association, rather than “fishing” for evidence (24). Thirdly, the glucose homeostasis traits could be considered secondary and/or exploratory analyses for the purposes of determining the number of conducted tests. Lastly, there is a nontrivial amount of inter-trait and inter-SNP correlation present in this data set, which makes the number of conducted tests difficult to determine (23, 24). For instance, the estimated genetic correlation between BMI and WAIST, WHR, SAT, or VAT ranges from an r2 of .61 to .94 (Supplementary Table 3). Furthermore, as discussed, the inter-SNP D′ and r2 statistics have shown that most INSIG2 SNPs are within a region of high, but not complete, overall LD (Results, Supplementary Figure 1).
Therefore, we utilized a principal components method to estimate the number of independent dimensions in the dataset given the genotypic and phenotypic correlation (Methods, Results). The number of independent dimensions (n=27) was then used in a standard Bonferroni adjustment to obtain a new threshold of significance (p=.05/27). The newly estimated threshold of p=.0018 technically excludes all observed association with adiposity measures. Thus, multiple comparisons adjustment does not strictly allow an interpretation of statistically significant difference in INSIG2 SNP genotypic means and variances. Nevertheless, we believe that we present multiple comparisons-adjusted suggestive evidence of association, which is both consistent with and augmentative to prior genetic studies of INSIG2. Furthermore, the correlations between SNPs and adiposity phenotypes have made the appropriate method of adjustment for type I error, and thus the proper α, somewhat speculative (23, 24).
We feel the findings following adjustment for multiple comparisons do not meet the criteria for firm acceptance of a null hypothesis. That is, the null hypothesis that genetic variation within and near INSIG2 has no bearing on adiposity or glucose homeostasis traits. Despite the inferences based on our suggestive results that follow, it remains possible that this and other reports of INSIG2 genetic association are type I errors, thus requiring cautious validation. It also remains a possibility that the inconsistent association of INSIG2 SNPs is due to inter-study differences in statistical power, levels of genetic heterogeneity, and gene-environment interaction, such as in populations of different ethnicity as studied here (24, 28).
It has been hypothesized that rs7566605 associations with BMI are observed only in cases of pre-existing or extreme obesity, implying a prerequisite genetic or environmental predisposition (6, 9, 11, 12). Replicated associations with rs7566605 have most consistently appeared with varying forms of qualitative obesity or overweight/obese subgroup analyses (9, 12, 29–31). Meanwhile, evaluation of rs7566605’s contribution to BMI as a continuous trait and/or in cohorts not selected for obesity most often report more modest or negative results (6, 8, 11, 12, 32–37). The results of testing rs7566605 for association with BMI as a continuous trait in the IRASFS Hispanics, recruited on the basis of family-size and are on average of the overweight classification, are consistent with the latter trend. However, the above trend is imperfect and some qualitative obesity analyses have reported no rs7566605 association with BMI (7, 11, 32–34, 36).
One possibility is that selecting and/or sub-grouping subjects for higher BMI may indirectly select from the higher ranges in alternative obesity phenotypes (such as VAT). The extent of indirect augmentation of an alternative adiposity phenotype would be subject to individual and inter-population differences, which have been most apparent in comparisons of race, gender, or age (38–40). Subsequently, the continuing mixed association between rs7566605 and BMI could be because INSIG2 contribution to BMI is secondary to its contribution to an alternative adiposity phenotype. Of relevance to the current study are the results of Boes et al., which has tested rs7566605’s association with CT-measured abdominal fat area using linear regression, and report no association of rs7566605 with VAT (or lipoprotein metabolism) in accordance with our results (34). From this, it seems likely that rs7566605 is not directly associated with CT-derived abdominal adiposity measures.
Rather, the present association of rs17047718, rs17047731, and rs12623648 with VAT and measures of glucose homeostasis implies that rs7566605’s prior associations with BMI were an indirect result of physiologically meaningful SNPs in LD. We found that rs7566605 is in LD with all but one of the SNPs that showed association in our study (rs12623648), and lies within a large ~28Kb haplotype block. SNPs of true genetic effect in LD with rs7566605 would exert biological influences that some studies may not have the statistical power, or the phenotypic capacity, to detect.
One study has putatively identified a physiologically functional SNP (−102G/A) just upstream of the transcription start site of INSIG2. The novel SNP was modestly associated with BMI in two cohorts of European descent. Moreover, the A allele of SNP −102G/A was related to decreased nuclear protein DNA binding, decreased DNA competiveness for nuclear proteins, and a decreased trend of mRNA expression in primary subcutaneous fat biopsies (P-value was .07) compared to the G allele (41). The SNP lies within the demonstrated large LD block that encompasses the INSIG2 promoter, the first intron, as well as rs7566605, rs17047718, and rs17047731. Furthermore, INSIG2 −102G/A possessed a MAF similar to the primarily associated SNPs in our study (e.g. 10%), a high D′ (D′ is .96) but low r2 (r2 is .04) with rs7566605, and a reduced BMI accompanied the minor allele in two European cohorts (Supplementary Figure 1, Table 2). These findings are consistent with our data and primary inferences, suggesting that the SNPs we present may also have functional relevance. Further study of −102G/A in other cohorts is needed, but it is possible that a haplotype including such a functional SNP, rs17047718, and rs17047731 is the genetic mechanism for increased adiposity involving INSIG2 in our population.
The study also detected association between INSIG2 SNPs and measures of glucose homeostasis. Association between INSIG2 SNPs and glucose homeostasis phenotypes could reflect a direct influence of INSIG on glucose homeostasis or, alternatively, could reflect the indirect influence of adipose tissue (42, 43) on these measures. Evidence of association with glucose homeostasis measures is inconclusively reduced when BMI is added as a covariate in the analysis (Figure 1, Figure 2). In the single-SNP results, BMI adjustment caused the moderately inter-correlated and prominently associated SNPs rs17047731, rs17047718, and rs12623648, to have a reduction in association with GFAST (P=.05–.10), SI (P=.02–.11), and FINS (P=.01–.02). We further explored this possibility by adjusting for VAT, but not BMI (data not shown). VAT adjustment caused the three prominently associated SNPs to have a similar reduction in significance of association with GFAST (P=.14–.17), FINS (P=.03–.08), and SI (P=.07–.28). Finally, adjustment of the three SNPs by BMI and VAT was found to have a reduction in significance of the association with GFAST (P=.15–.20), FINS (P=.04–.09), and SI (P=.06–.31), which closely resembled the effects of VAT adjustment alone. These explorations imply that the mechanism by which these three SNPs affect glucose homeostasis is their effect on adiposity levels, especially fat area of abdominal depots. It should be emphasized, though; that adjustment for measures of adiposity reduces, but does not eliminate, evidence of association with glucose homeostasis traits. Therefore, we cannot exclude an adiposity-independent role for INSIG2 in regulation of glucose homeostasis.
This study has investigated INSIG2 genetic variants and tested these for association with several adiposity and glucose homeostasis phenotypes in the IRASFS Hispanic population. Though we do not observe replication of rs7566605’s association with adiposity measures in our study, we do observe novel association of SNPs (most notably rs17047731, rs17047718, and rs12623648) with more direct measures of adiposity and measures of glucose homeostasis. The SNP rs7566605 was not correlated with the novel associated SNPs via high r2, but was in LD with practically every SNP by high D′. This underscores the need for more extensive haplotype block tagging approaches in addressing histories of mixed association in candidate genes, due to the potential link between regional LD and said mixed association. The novel SNP associations with adiposity and glucose homeostasis traits reported above should be validated through analysis in other cohorts, especially given the present results of multiple comparisons adjustment. In conclusion, more extensive study of the genomic interval suggests that polymorphism of the INSIG2 genic region plays a role in human obesity and diabetes. Future study of INSIG2 should consider genetic variants near rs7566605, alternative adiposity phenotypes, and functional evaluation of novel or associated SNPs.
Supplementary Material
Acknowledgments
This research was supported in part by NIH grants HL060894, HL060931, HL060944, and HL061019. We would like to acknowledge also the helpful comments of the reviewers of Obesity.
Footnotes
Supplementary Material is available at www.nature.com/obesity
References
- 1.Engelking LJ, Liang G, Hammer RE, Takaishi K, Kuriyama H, Evers BM, et al. Schoenheimer effect explained-feedback regulation of cholesterol synthesis in mice mediated by INSIG proteins. The Journal of Clinical Investigation. 2005;115:2489–2498. doi: 10.1172/JCI25614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 3.Herbert A, Gerry NP, McQueen MB, Heid IM, Pfeufer A, Illig T, et al. A Common Genetic Variant Is Associated with Adult and Childhood Obesity. Science. 2006;312:279–283. doi: 10.1126/science.1124779. [DOI] [PubMed] [Google Scholar]
- 4.Dina C, Meyre D, Samson C, Tichet J, Marre M, Jouret B, et al. Comment on “A Common Genetic Variant is Associated with Adult and Childhood Obesity. Science. 2007;315:187b. doi: 10.1126/science.1129402. [DOI] [PubMed] [Google Scholar]
- 5.Feng Y, Dong H, Xiang Q, Hong X, Wilker E, Zhang Y, et al. Lack of association between rs7566605 and obesity in a Chinese population. Human Genetics. 2006;20:743–745. doi: 10.1007/s00439-006-0258-2. [DOI] [PubMed] [Google Scholar]
- 6.Hall DH, Rahman T, Avery PJ, Keavney B. INSIG2 promoter polymorphism and obesity related phenotypes: association study in 1428 members of 248 families. BMC Medical Genetics. 2006;7:83–89. doi: 10.1186/1471-2350-7-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar J, Sunkishala RR, Karthikeyan G, Sengupta S. The common genetic variant upstream of INSIG2 gene is not associated with obesity in Indian population. Clinical Genetics. 2007;71:415–418. doi: 10.1111/j.1399-0004.2007.00795.x. [DOI] [PubMed] [Google Scholar]
- 8.Loos RJF, Barroso I, O’Rahilly S, Wareham NJ. Comment on “A Common Genetic Variant Is Associated with Adult and Childhood Obesity. Science. 2007;315:187c. doi: 10.1126/science.1130012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosskopf D, Bornhorst A, Rimmbach C, Schwann C, Kayser A, Kruger A, et al. Comment on “A Common Genetic Variant is Associated with Adult and Childhood Obesity. Science. 2007;315:187d. doi: 10.1126/science.1130571. [DOI] [PubMed] [Google Scholar]
- 10.Skelly T, Pinheiro AP, Lange LA, Sullivan PF. Is rs7566605, a SNP near INSIG2, associated with body mass in a randomized clinical trial of antipsychotics in schizophrenia? Molecular Psychiatry. 2007;12:321–322. doi: 10.1038/sj.mp.4001956. [DOI] [PubMed] [Google Scholar]
- 11.Smith AJP, Cooper JA, Li LK, Humphries SE. INSIG2 gene polymorphism is not associated with obesity in Caucasian, Afro-Caribbean and Indian subjects. International Journal of Obesity. 2007:1–3. doi: 10.1038/sj.ijo.0803645. [DOI] [PubMed] [Google Scholar]
- 12.Lyon HN, Emilsson V, Hinney A, Heid IM, Lasky-Su J, Zhu X, et al. The Association of a SNP Upstream of INSIG2 with Body Mass Index is Reproduced in Several but Not All Cohorts. PLoS Genetics. 2007;3:627–633. doi: 10.1371/journal.pgen.0030061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krapivner S, Chernogubova E, Ericsson M, Ahlbeck-Glader C, Hamsten A, van’t Hooft FM. Human evidence for the involvement of insulin-induced gene 1 in the regulation of plasma glucose concentration. Diabetologia. 2007;50:94–102. doi: 10.1007/s00125-006-0479-x. [DOI] [PubMed] [Google Scholar]
- 14.Henkin L, Bergman RN, Bowden DW, Ellsworth DL, Haffner SM, Langefeld CD, et al. Genetic epidemiology of insulin resistance and visceral adiposity. The IRAS Family Study design and methods. Ann Epidemiol. 2003;13:211–217. doi: 10.1016/s1047-2797(02)00412-x. [DOI] [PubMed] [Google Scholar]
- 15.Norris JM, Langefeld CD, Scherzinger AL, Rich SS, Bookman E, Beck SR, et al. Quantitative trait loci for abdominal fat and BMI in Hispanic-Americans and African-Americans: the IRAS Family Study. International Journal of Obesity. 2005;29:67–77. doi: 10.1038/sj.ijo.0802793. [DOI] [PubMed] [Google Scholar]
- 16.Boston RC, Stefanovski D, Moate PJ, Sumner AE, Watanabe RM, Bergman RN. MINMOD Millennium: a computer program to calculate glucose effectiveness and insulin sensitivity from the frequently sampled intravenous glucose tolerance test. Diabetes Technol Ther. 2003;5(6):1003–1015. doi: 10.1089/152091503322641060. [DOI] [PubMed] [Google Scholar]
- 17.Pacini G, Bergman RN. MINMOD: a computer program to calculate insulin sensitivity and pancreatic responsivity from the frequently sampled intravenous glucose tolerance test. Comput Methods Programs Biomed. 1986;23(2):113–122. doi: 10.1016/0169-2607(86)90106-9. [DOI] [PubMed] [Google Scholar]
- 18.Buetow KH, Edmonson M, MacDonald R, Clifford R, Yip P, Kelley J, et al. High-throughput development and characterization of a genomewide collection of gene-based single nucleotide polymorphism markers by chip-based matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Proc Natl Acad Sci U S A. 2001;98:581–584. doi: 10.1073/pnas.021506298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Bakker PIW, Burtt NP, Graham RR, Guiducci C, Yelensky R, Drake JA, et al. Transferability of tag SNPs in genetic association studies in multiple populations. Nature Genetics. 2006;38:1298–1303. doi: 10.1038/ng1899. [DOI] [PubMed] [Google Scholar]
- 20.O’Connell JR. Zero-recombinant haplotyping: applications to fine mapping using SNPs. Genet Epidemiol. 2000;19S1:S64–70. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI10>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 21.O’Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang S, Zhang K, Li J, Sun F, Zhao H. Test for association for quantitative traits in general pedigrees: the quantitative pedigree disequilibrium test. Genet Epidemiol. 2001;S1:S370–S375. doi: 10.1002/gepi.2001.21.s1.s370. [DOI] [PubMed] [Google Scholar]
- 23.Gao X, Starmer J, Martin ER. A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet Epidemiol. 2008;32:361–369. doi: 10.1002/gepi.20310. [DOI] [PubMed] [Google Scholar]
- 24.van den Oord EJCG. Controlling false discoveries in genetic studies. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:637–644. doi: 10.1002/ajmg.b.30650. [DOI] [PubMed] [Google Scholar]
- 25.Roeder K, Devlin B, Wasserman L. Improving power in genome-wide association studies: weights tip the scale. Genet Epidemiol. 2007;31:741–747. doi: 10.1002/gepi.20237. [DOI] [PubMed] [Google Scholar]
- 26.Genovese CR, Roeder K, Wasserman L. False discovery control with P-value weighting. Biometrika. 2006;93:509–524. [Google Scholar]
- 27.Roeder K, Bacanu SA, Wasserman L, Devlin B. Using linkage genome scans to improve power of association in genome scans. Am J Hum Genet. 2006;78:243–252. doi: 10.1086/500026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Colhoun HM, McKeigue PM, Davey SG. Problems of reporting genetic associations with complex outcomes. Lancet. 2003;361:865–872. doi: 10.1016/s0140-6736(03)12715-8. [DOI] [PubMed] [Google Scholar]
- 29.Chu X, Erdman R, Susek M, Gerst H, Derr K, Al-agha M, et al. Association of morbid obesity with FTO and INSIG2 allelic variants. Arch Surg. 2008;143(3):235–241. doi: 10.1001/archsurg.2007.77. [DOI] [PubMed] [Google Scholar]
- 30.Zhang J, Lin R, Wang F, Lu M, Lin RY, Wang SZ, et al. A common polymorphism is associated with body mass index in Uyghur population. Diabetes Res Clin Pract. 2008;81(2):e11–3. doi: 10.1016/j.diabres.2008.03.022. [DOI] [PubMed] [Google Scholar]
- 31.Hotta K, Nakamura M, Nakata Y, Matsuo T, Kamohara S, Kotani K, et al. INSIG2 gene rs7566605 polymorphism is associated with severe obesity in Japanese. J Hum Genet. 2008;53(9):857–862. doi: 10.1007/s10038-008-0317-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuzuya M, Ando F, Iguchi A, Shimokata H. No association between rs7566605 variant and being overweight in Japanese. Obesity. 2007;15(11):2531–4. doi: 10.1038/oby.2007.301. [DOI] [PubMed] [Google Scholar]
- 33.Tabara Y, Kawamoto R, Osawa H, Nakura J, Makino H, Miki T, et al. No association between rs7566605 polymorphism and being overweight in Japanese population. Obesity. 2008;16(1):211–5. doi: 10.1038/oby.2007.25. [DOI] [PubMed] [Google Scholar]
- 34.Boes E, Kollerits B, Heid IM, Hunt SC, Pichler M, Paulweber B, et al. INSIG2 polymorphism is neither associated with BMI nor with phenotypes of lipoprotein metabolism. Obesity. 2008;16(4):827–33. doi: 10.1038/oby.2007.132. [DOI] [PubMed] [Google Scholar]
- 35.Oki K, Yamane K, Kamei N, Asao T, Awaya T, Kohno N. The single nucleotide polymorphism upstream of insulin-induced gene 2 (INSIG2) is associated with the prevalence of hypercholesterolaemia, but not with obesity, in Japanese American women. Br J Nutr. 2008;23:1–6. doi: 10.1017/S0007114508006557. [DOI] [PubMed] [Google Scholar]
- 36.Andreasen CH, Mogensen MS, Borch-Johnsen K, Sandbaek A, Lauritzen T, Sorensen TI, et al. Non-replication of genome-wide based associations between common variants in INSIG2 and PFKP and obesity in studies of 18,014 Danes. PLoS ONE. 2008;3(8):e2872. doi: 10.1371/journal.pone.0002872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang L, Wu Y, Li H, Yu Z, Li X, Liu Y, et al. Potential association of INSIG2 rs7566605 polymorphism with body weight in a Chinese subpopulation. Eur J Hum Genet. 2008;16(6):759–61. doi: 10.1038/ejhg.2008.8. [DOI] [PubMed] [Google Scholar]
- 38.Santosa S, Jensen MD. Why are we shaped differently, and why does it matter? Am J Phsiol Endrocrinol Metab. 2008;295(3):E531–5. doi: 10.1152/ajpendo.90357.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bouchard C, Despres JP, Mauriege P. Genetic and nongenetic determinants of regional fat distribution. Endocr Rev. 1993;14(1):72–93. doi: 10.1210/edrv-14-1-72. [DOI] [PubMed] [Google Scholar]
- 40.Deurenberg P, Deurenberg-Yap M. Validity of body composition methods across ethnic population groups. Forum Nutr. 2003;56:299–301. [PubMed] [Google Scholar]
- 41.Krapivner S, Popov S, Chemogubova E, Hellenius M, Fisher RM, Hamsten A, van’t Hooft FM. Insulin-induced gene 2 involvement in human adipocyte metabolism and body weight regulation. J Clin Endocrinol Metab. 2008;93(5):1995–2001. doi: 10.1210/jc.2007-1850. [DOI] [PubMed] [Google Scholar]
- 42.Dahlman I, Arner P. Obesity and polymorphisms in genes regulating human adipose tissue. International Journal of Obesity. 2007;31(11):1629–1641. doi: 10.1038/sj.ijo.0803657. [DOI] [PubMed] [Google Scholar]
- 43.Lundgren M, Buren J, Ruge T, Myrnas T, Eriksson JW. Glucocorticoids down-regulate glucose uptake capacity and insulin-signaling proteins in omental but not subcutaneous human adipocytes. J Clin Endocrinol Metab. 2004;89:2989–2997. doi: 10.1210/jc.2003-031157. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.