Abstract
The SlipChip platform was tested to perform high throughput nanoliter multiplex PCR. The advantages of using the SlipChip platform for multiplex PCR include the ability to preload arrays of dry primers, instrument-free sample manipulation, small sample volume, and high throughput capacity. The SlipChip was designed to preload one primer pair per reaction compartment, and to screen up to 384 different primer pairs with less than 30 nanoliters of sample per reaction compartment. Both a 40-well and a 384-well design of the SlipChip were tested for multiplex PCR. In the geometries used here, the sample fluid was spontaneously compartmentalized into discrete volumes even before slipping of the two plates of the SlipChip, but slipping introduced additional capabilities that made devices more robust and versatile. The wells of this SlipChip were designed to overcome potential problems associated with thermal expansion. By using circular wells filled with oil and overlapping them with square wells filled with the aqueous PCR mixture, a droplet of aqueous PCR mixture was always surrounded by the lubricating fluid. In this design, during heating and thermal expansion, only oil was expelled from the compartment and leaking of the aqueous solution was prevented. Both 40-well and 384-well devices were found to be free from cross-contamination, and end point fluorescence detection provided reliable readout. Multiple samples could also be screened on the same SlipChip simultaneously. Multiplex PCR was validated on the 384-well SlipChip with 20 different primer pairs to identify 16 bacterial and fungal species commonly presented in blood infections. The SlipChip correctly identified five different bacterial or fungal species in separate experiments. In addition, the presence of the resistance gene mecA in methicillin resistant Staphylococcus aureus (MRSA) was identified. The SlipChip will be useful for applications involving PCR arrays, and lays the foundation for new strategies for diagnostics, point-of-care devices, and immobilization based arrays.
Introduction
This paper describes a method of performing nanoliter-scale multiplex PCR on a preloaded SlipChip. Since its introduction, multiplex PCR has been successfully applied in many areas from research to clinical diagnostics, including genetic analysis of cancer cells,1,2 monitoring of genetic variability and clonal evolution,3 and identification of infectious diseases caused by viruses, bacteria, fungi, and parasites.4,5 The conventional method for performing multiplex PCR is to load multiple primers to amplify multiple target templates in one reaction compartment.5 The throughput of this approach is generally limited to less than 10 targets per compartment because of uneven amplification rates of different targets, mutual interference of multiple primers, and a limiting number of existing fluorophors required for detection. Multiplex PCR can also be performed using PCR multi-well plates,6 but this method usually requires a large amount of reagent and sample.
Another strategy is to use many miniaturized compartments, each containing a primer pair for a different target. Implementing this strategy requires approaches for handling small volumes of liquid, such as provided by microfluidics. Microfluidic technology has advantages over traditional PCR platforms, e.g. smaller reaction volume, high-throughput capacity, and portability. Miniaturized high performance PCR components7-22 and systems integrating such PCR components with sample preparation to provide sample-to-answer platforms23-29 have been created. For example, components use microchambers, microchannels, microwell arrays, microdroplets, or valves to compartmentalize PCR reactions. A universally accepted platform has not yet emerged, as it is desirable to simplify microfluidic approaches further, e.g. eliminating the need for complex fabrication or pumping equipment.
Here we evaluate the performance of high-throughput, multiplex PCR on the SlipChip30-34 platform. The SlipChip allows microliter volumes of solutions to be effectively distributed among hundreds of nanoliter compartments without requiring pumps or robotic dispensing. It also allows preloading and storage of multiple reagents without cross contamination. It has been validated for protein crystallization30-32 and nanoliter immunoassays.33 SlipChip relies on encoding a program for manipulating fluids into an array of wells and ducts imprinted in two plates. These plates are then brought in contact “face to face” and the program is executed by slipping the two plates relative to one another. Here, we tested a 40-well and a 384-well design of the SlipChip to perform multiplex PCR. The 384-well SlipChip was designed to perform 384 simultaneous PCR reactions to identify up to 384 different templates on a single 10 μL sample with end-point fluorescence detection. An array of primer pairs was directly deposited in the wells of the SlipChip and allowed to dry at room temperature. The entire SlipChip experiment with preloaded primers can be setup easily by the user with a single pipetting step. PCR reactions were performed on a standard thermal cycler and were initiated by a single slipping step without requiring instruments.
Results and Discussion
We first tested PCR on the SlipChip with the design containing 40 wells and two inlets for two different samples (Figure 1 A, B). The 40-well SlipChip was made from soda-lime glass (See Experimental Section in Supporting Information) with a final dimension of 4 cm by 2 cm by 0.07 cm for each plate. This device can be used to screen two different samples simultaneously, with up to 20 different primer pairs for each sample. The top plate (Figure 1C) contained the fluid inlet, square wells (side length of 640 μm, depth of 70 μm) rectangular wells (length of 570 μm, width of 230 μm, depth of 70 μm), and outlets (not shown). The bottom plate (Figure 1D) contained circular wells (diameter of 560 μm, depth of 30 μm) and the ducts for introduction of the sample (width of 150 μm, depth of 30 μm). Different primer pairs were preloaded into the bottom circular wells and allowed to dry under room temperature (Figure 1D). The top and bottom plates were then submerged under mineral oil and assembled to form a continuous fluidic path (Figure 1E). The PCR master mixture, a solution containing SsoFast EvaGreen Supermix, 1 mg/mL BSA, and template DNA (or water for the control experiments), was introduced into the SlipChip by pipetting (Figure 1F). In this geometry, the sample fluid spontaneously broke up into discrete volumes even before slipping. This breakup of a continuous stream into discrete volumes is valuable for applications where compartmentalization is required, such as stochastic confinement35,36 and digital PCR.37 After injection of the sample, the top plate was slipped down to overlap the square wells with the circular wells on the bottom plate (Figure 1G, H), and the dry primers preloaded in the circular wells dissolved in the sample introduced from square wells. The rectangular wells on the top plate also aligned with the middle of the duct on the bottom plate. The aqueous solution formed a circular droplet in the wells due to the surface tension (Figure 1G, H), and the volume of solution in each compartment was estimated using AutoCAD software to be 26 nL. As our goal in this paper was to characterize intrinsic performance of the SlipChip, we have not attempted to reuse SlipChips after a surface wash, for the fear of potential contamination from experiment to experiment. Such contamination would have made make our results inconclusive as we wished to test cross-contamination among wells. We do know that the SlipChip can be re-used after the previous PCR product is removed by thoroughly cleaning the chip with piranha (H2SO4: H2O2 = 3:1). Then, after plasma cleaning, the SlipChip can be silanized again for re-use (See Experimental Section in Supporting Information).
Figure 1.
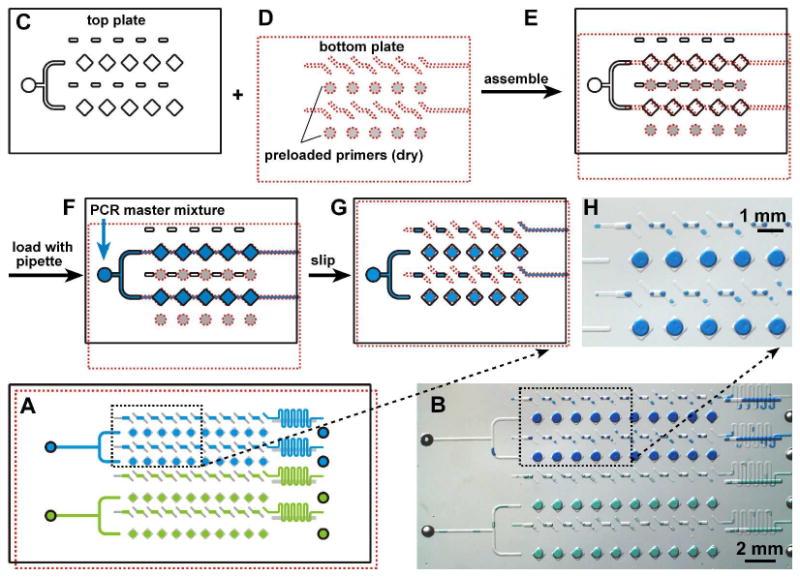
Assembly and operation of the preloaded multiplex PCR SlipChip. A) Schematic drawing shows the PCR SlipChip after introducing two different samples (blue and green) and slipping to combine the sample with the preloaded primers. B) A bright field image of the experiment described in A) using food dyes instead of the samples. C) The top plate of the PCR SlipChip contained square wells, rectangular wells, sample inlets, and outlets (outlets not shown) and D) the bottom plate of the PCR SlipChip contained ducts for the samples and preloaded circular wells with different PCR primers. E) The SlipChip was assembled and the two plates were aligned such that the sample wells and sample ducts lined up to form a continuous fluidic path. F) The sample, the PCR master mixture, was flowed through the fluidic path to load the sample wells. G) The PCR SlipChip was slipped to align the square sample wells with the circular primer wells. When the PCR mixture touched the primer on the bottom of the primer well, the PCR primer dissolved in and mixed with the reaction mixture. After the SlipChip was slipped, thermal cycling was performed. H) Microphotograph shows the formation of droplets within the wells after the aqueous dye (representing the PCR mixture) was slipped to contact wells containing oil.
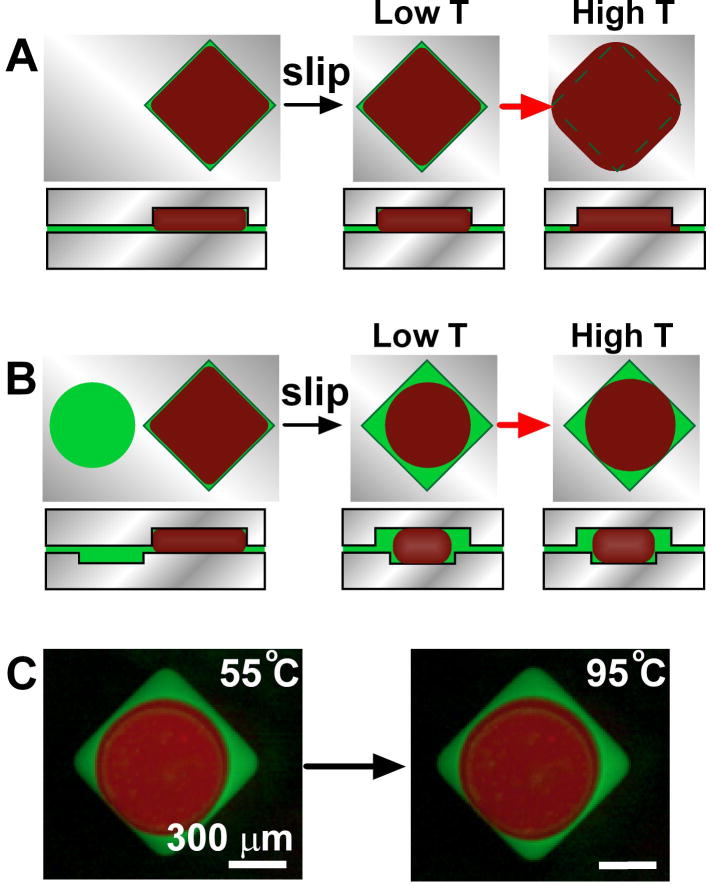
We addressed the issue of thermal expansion during thermal cycling by careful design of the SlipChip. The material of the SlipChip (glass), the lubricating fluid (mineral oil), and the sample (the aqueous PCR mixture) all have different thermal expansion coefficients. The aqueous reaction mixture and the oil should expand more than the glass when the temperature of the SlipChip is increased from room temperature to the annealing temperature (55 °C) and then to the denaturation temperature (95 °C). This thermal expansion could cause the reaction mixture to be expelled from each compartment into the gap between the two plates, potentially leading to cross-contamination (Figure 2A). To confirm that this was a potential problem, and to characterize thermal expansion, we used an aqueous solution containing red quantum dots and mineral oil containing green quantum dots to study the fluid movement during thermal cycling (Experimental Section and Figure S1 in Supporting Information). When using the SlipChip with only square wells, the aqueous solution filled the square well completely, and after an increase in temperature, the aqueous solution entered the gap between the two plates of the SlipChip. To ensure that no fluid enters the gap and no cross-contamination could take place, we changed the design of the SlipChip to use it for PCR experiments (Figure 2B, C). In this design, we found that when a smaller, circular well containing oil in the bottom plate was brought into contact with a square well containing aqueous solution in the top plate, due to the surface tension the aqueous solution would form a droplet surrounded by mineral oil within the hydrophobic well (Figure 2B-C). When the temperature was increased, the aqueous solution expanded inside the reaction compartment and the mineral oil expanded and moved out of the well through the gap between the top and bottom plates in the SlipChip, serving as a buffer material. In this design, the oil was preferentially expelled from the reaction compartment under both “kinetic” and “thermodynamic” control. Because the oil was located closer to the entrance of gap between the plates, we expected it to be expelled preferentially even at very high rates of expansion, when the wetting preferences and capillary forces may matter less. Because the oil preferentially wets the hydrophobic surfaces of the SlipChip, we also expected it to enter the gap preferentially at very slow rates of expansion, when the capillary effects are likely to control the outcome. In addition, we determined that the shape and size of the bottom well influenced formation of a single droplet of consistent size in the center of the two wells. While there are many variations of well shapes and sizes that can provide this effect, we found that the wells used here (see the Experimental Section in Supporting Information) provided consistent results. Finally, the rectangular wells on the top plate overlapped with the ducts on the bottom plate to address the issue of thermal expansion of the solution remaining in the ducts.
Figure 2.
Control of thermal expansion during thermocycling on SlipChip by changing well geometry. A) Top and side view schematic drawings of a square well containing aqueous PCR reaction mixture (red) and mineral oil (green). The square well was completely filled when aqueous solution was introduced (left). After slipping to break the fluidic connections to other wells (not shown here), the aqueous solution filled the square well completely (middle, low T), and after an increase in temperature, the aqueous solution entered the gap between the two plates of the SlipChip (right, high T). B) Top and side view schematic drawings of a shallow circular well(bottom plate) containing oil (green) next to a deep square well (top plate) filled with aqueous PCR reaction mixture (red) (left). After slipping, the aqueous solution would form a droplet surrounded by mineral oil within the hydrophobic well due to the surface tension (middle, low T). When the temperature was increased, the aqueous solution expanded inside the reaction compartment and the mineral oil expanded and moved out of the well through the gap between the top and bottom plates in the SlipChip, serving as a buffer material (right, high T). C) Top view of fluorescent microphotographs from experiments described as in B). Aqueous solution containing red quantum dots (red) in square well was slipped and overlaid with circular well containing mineral oil staining with green quantum dots (green). An aqueous droplet was formed in hydrophobic well due to the surface tension at 55 °C (left). After increase of temperature to 95 °C, the aqueous solution expanded but still stayed inside the reaction compartment (right). No leakage of the aqueous solution outside of the reaction compartment was observed.
To evaluate the performance of the SlipChip, we tested PCR in the SlipChip (the 40-well design, Figure 1) by amplifying the nuc gene in S. aureus genomic DNA (gDNA). Primers for the S. aureus nuc gene38 were preloaded into the circular wells of the bottom plate of the SlipChip and allowed to dry under room temperature. The PCR master mixture (EvaGreen supermix, 5 pg/μL S. aureus gDNA, and 1 mg/mL BSA) was injected into the channels to fill the top two rows of wells. The bottom two rows of wells were filled with the same aqueous PCR mixture but gDNA template was replaced by nuclease-free water. The square wells in the top plate and circular wells in the bottom plate were overlapped by slipping the two plates of the SlipChip relative to one another (Figure 1G). The SlipChip was placed into the thermal cycler on a flat in situ adaptor for PCR amplification. We confirmed that no cross contamination occurred between different rows in the SlipChip as only wells containing template showed amplification (Figure 3). Fluorescent intensity increased significantly (> 5 fold, p < 0.0001, n = 20) only in the wells containing gDNA (Figure 3A-C), and all 20 wells containing template showed amplification after thermal cycling, verifying the robustness of the PCR SlipChip. The fluorescent intensity was uniform across the 20 wells containing template. A real time imaging system can be used to further verify the amplification rate in each well. After thermal cycling, the top plate of the SlipChip was slipped back to re-form the continuous fluidic path. The PCR product from the top two rows (containing template) and the bottom two rows (control) was recovered individually by replacing the aqueous solution in each fluidic path with mineral oil. A gel electrophoresis experiment was performed and indicated successful on-chip amplification and the correct size of the amplification product (∼ 270 bp) (Figure 3D).
Figure 3.
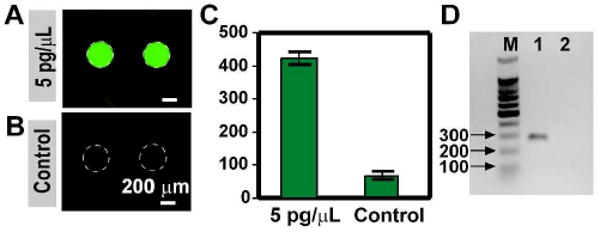
PCR was successful and robust on the SlipChip platform. A) Microphotograph of wells containing S. aureus (MSSA) gDNA showed significant increase of fluorescence intensity after thermal cycling. B) Microphotograph of the control wells showed no increase of fluorescence. C) Analysis of the fluorescence intensity of the wells containing template vs. the control wells. The average fluorescent intensity of the wells containing template was significantly higher than the control wells (p < 0.0001, n = 20). The fluorescent intensity of the wells before thermal cycling was around 80 units. D) Gel electrophoresis of the DNA amplified in the SlipChip. Column M contained the 100 bp DNA ladder, Column 1 contained sample recovered from wells loaded with template, and Column 2 contained sample collected from control wells. Only one molecular weight of DNA (∼ 270 bp) was found in wells containing template, and no DNA was found in control wells without template.
Next, we tested the cross contamination among adjacent wells by preloading the primer pairs for the nuc gene in S. aureus and the mecA gene39 in Methicillin-resistant S. aureus (MRSA) on the chip alternatively in the same row, and injecting PCR master mixture containing 100 pg/μL of Methicillin-sensitive S. aureus (MSSA) genomic DNA into the SlipChip (primer pairs can be found in the Supporting Information). Because the nuc gene is commonly present in S. aureus but the mecA gene is not, all ten wells preloaded with the primers for the nuc gene showed a significant increase in fluorescence intensity after thermal cycling, and none of the wells loaded with primers for the mecA gene increased in fluorescence intensity (p < 0.0001, n = 10) (Figure 4 A-B). Combined with the results above (Figure 3), we concluded that each well was an isolated reaction condition, and there was no communication among wells.
Figure 4.
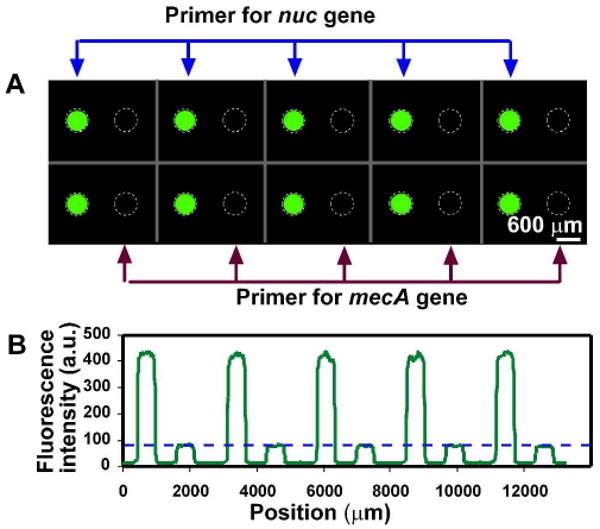
No cross-contamination among adjacent wells was seen after PCR on the SlipChip. Primer pairs for the nuc gene in S. aureus and the mecA gene in Methicillin-resistant S. aureus (MRSA) were preloaded on the chip alternatively in the same row. PCR master mixture, containing 100 pg/μL of S. aureus (MSSA) genomic DNA, was injected into the SlipChip. A) Fluorescent microphotograph shows that only wells preloaded primers for the nuc gene (left in each pair of wells) showed fluorescent signal corresponding to DNA amplification. B) A line scan across the middle of the top row in (A) shows that fluorescence intensity of wells loaded with nuc primers were more than 5 fold higher than that of the wells loaded with mecA primers (p < 0.0001, n = 10) The blue dashed line in (B) represents the fluorescence intensity before thermal cycling in the wells.
Furthermore, we demonstrated that the SlipChip containing 384 wells, which can be preloaded with up to 384 different pairs of primers, can be applied for high-throughput multiplex PCR. This chip was designed analogously to the 40-well SlipChip shown in Figure 1. The top plate contained the fluid inlet, square wells (side length of 420 μm, depth of 70 μm) and rectangular wells (length of 320 μm, width of 200 μm, depth of 70 μm). The bottom plate contained circular wells (diameter of 340 μm, depth of 30 μm) and the ducts for introduction of the sample (width of 200 μm, depth of 30 μm). The dimension of each plate was 7.6 cm by 2.6 cm by 0.07cm. The volume of solution in each compartment was estimated to be 7 nL. In this paper, we tested this platform in the context of 16 different pathogens that are commonly present in blood infections by using 20 different primer pairs preloaded on the SlipChip. Primer sequences were selected from previous publications (Supporting Information Table S1), and the PCR master mixture was combined with cells at a final concentration of approximately 106 cfu/mL. This guaranteed the presence of targeted cells—106 cfu/mL corresponds to ∼7 cells per well. In parallel, we have demonstrated that PCR on SlipChip is effective down to the single molecule level;37 while this sensitivity is important for clinical studies that are likely to contain very few microbes, we did not pursue it here. In this study, the SlipChip was divided into 28 independent regions, and a primer pair for each pathogen was preloaded as 4 by 4 matrices to test the reproducibility of PCR reactions (Figure 5B, regions A3-A7, B3-B7, C3-C7, D3-D7). Primers for pBad template were preloaded in the two columns of wells at the edges of the SlipChip (Figure 5B, regions A1, B1, C1, D1) as a positive internal control. A purified pBad 331 bp template (final concentration 1 pg/uL) was added to the PCR master mixture before loading. This SlipChip design was loaded via dead-end filling.40 Two columns next to the wells containing primers for pBad (Figure 5B, regions A2, B2, C2, D2) were left empty as a negative control for cross-contamination. The position and name of the deposited primer pairs targeting each pathogen are shown in Table 1, and the sequence of the deposited primer pairs are shown in Supporting Information Table S1. A bright-field image of 384-well SlipChip after filling with red dye and slipping was used to show the entire layout of the SlipChip design (Figure 5A).
Figure 5.
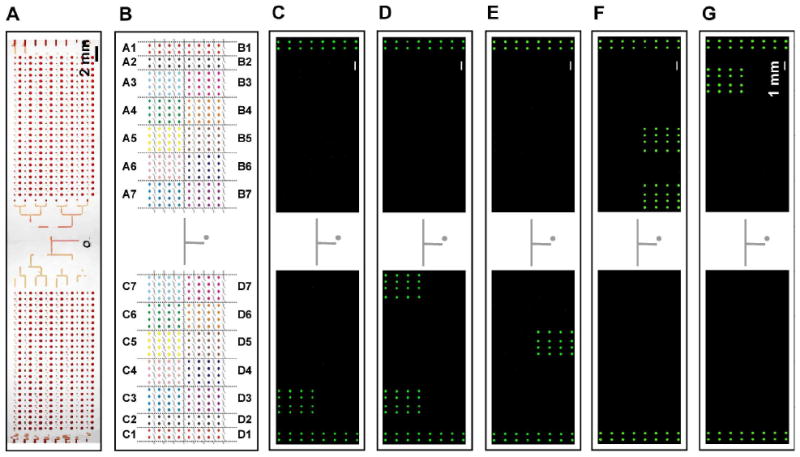
Detection of pathogens by PCR using panels of preloaded primers on the 384-well SlipChip. A panel of 20 different primer pairs was preloaded onto the SlipChip to differentiate and genotype 16 different pathogens, with one additional primer pair for a positive internal control. A) Bright field image of the whole chip filled with red dye and slipped into position. B) A schematic drawing of the 384-well SlipChip designed for high-throughput multiplex PCR. Different colors indicate wells preloaded with different primer pairs (see Table 1). (C-G) Fluorescence microphotographs showed that the SlipChip correctly identified and genotyped different microbes by using PCR. In all experiments, regions for positive control (pBad primer pair, A1, B1, C1, and D1) increased in fluorescence intensity while regions for negative control (no primer pairs, A2, B2, C2, and D2) did not. All samples were loaded with pBad 331 bp template as positive internal controls. Fluorescent images were stitched after acquisition (See Experimental Section in Supporting Information). (C) When loaded with MSSA, only region C3 (S. aureus nuc primer) showed increase of fluorescence intensity; D) When loaded with MRSA, both regions C3 (S. aureus nuc primer) and C7 (MRSA mecA primer) showed increase of fluorescence intensity; E) When loaded with C. albicans, region D5 (C. albicans calb primer) showed increase of fluorescence intensity; F) When loaded with P. aeruginosa, regions B5 (P. aeruginosa vic primer) and B7 (Pseudomonas 16S rRNA primer) showed increase of fluorescence intensity; G) When loaded with E. coli, region A3 (E. coli nlp primer) wells showed increase of fluorescence intensity.
Table 1.
Position and name of pre-deposited primer pairs
| Position | Target gene for control/pathogen |
|---|---|
| A1-D1 | pBad 333 bp ds DNA |
| A2-D2 | empty |
| A3 | Escherichia coli nlp4 |
| A4 | Streptococcus pyogenes fah4 |
| A5 | Streptococcus pyogenes OppA4 |
| A6 | Streptococcus pneumoniae cinASP4 |
| A7 | Streptococcus pneumoniae plySP4 |
| B3 | Enterococcus faecium bglB4 |
| B4 | Enterococcus faecalis ace4 |
| B5 | Pseudomonas aeruginosa vic52 |
| B6 | Streptococcus agalactiae cpsY4 |
| B7 | Pseudomonas 16S rRNA52 |
| C3 | Staphylococcus aureus nuc38 |
| C4 | Staphylococcus epidermidis agrC4 |
| C5 | Streptococcus mutans dltA4 |
| C6 | Proteus mirabilis aad4 |
| C7 | MRSA mecA39 |
| D3 | Candida tropicalis ctr53 |
| D4 | Candida glabrata cgl53 |
| D5 | Candida albicans calb53 |
| D6 | Klebsiella pneumoniae cim4 |
| D7 | Klebsiella pneumoniae acoA4 |
The SlipChip PCR was able to identify cells robustly, as only the regions preloaded with the appropriate primers showed a significant increase in fluorescent signal (Figure 5C-G). In all experiments, the regions for positive controls (A1, B1, C1, D1) showed an increase in fluorescent signal and regions for negative controls (A2, B2, C2, D2) did not. The SlipChip was able to identify correctly MSSA (Figure 5C, nuc gene in region C3), MRSA (Figure 5D, nuc gene in region C3 and mecA gene in region C7), Candida albicans (Figure 5E, calb gene in region D5), P. aeruginosa (Figure 5F, vic gene in region B5 and Pseudomonas 16S rRNA gene in region B7), and E. coli (Figure 5G, nlp gene in region A3).
Conclusion
This paper demonstrated a SlipChip platform for high-throughput multiplex PCR, with robust performance and lack of false negatives, false positives and cross-contamination. We used up to 384 nanoliter-scale reactions for multiplex PCR with a preloaded array of primer pairs, and chips with larger numbers of wells are straightforward to create. The PCR SlipChip can be loaded simply by pipetting, avoiding any requirements for complex injection methods. It is not limited to testing a single sample, illustrated here by testing two different samples simultaneously on a single preloaded SlipChip. The approach shown here to prevent cross-contamination due to thermal expansion will be valuable in other applications of the SlipChip that require changing of temperature- such as testing of protein stability and protein-ligand interactions41-43- and applications that might operate under widely different environmental temperatures, e.g. in field testing.
In addition to distinguishing a large number of different species in one experiment, the SlipChip will also be capable of providing quantitative results, either by integrating real-time imaging techniques for multiplex real-time PCR,44 or by using a large number of wells for each primer pair to enable counting the number of all amplicon copies in one experiment to perform multiplex digital PCR.15,45 The PCR SlipChip design can be modified to use the primer pairs established in current PCR array technology, such as those sold by SABiosciences,6 but with a much higher density, smaller size, and smaller reaction volume. It can also adapt the current technologies of microarray printing, such as inkjet, microjet deposition, and spotting technologies,46 to preload primers on the SlipChip.
The SlipChip encloses sample volumes in a layer of lubricating fluid, just as droplet-based platforms do.47,48 As long as the surfaces of the devices are preferentially wetted by the lubricating fluid, there is no need to incorporate additional control of the surface chemistry of the device. Control of surface chemistry takes place at the interface between the aqueous solution and the immiscible oil. This interfacial control is well-established for PCR, routinely performed under oil, and can be extended to other applications with some advantages provided by the fluorous lubricating fluids and fluorous amphiphiles,49,50 which can be used for difficult tasks such as suppressing interfacial aggregation of amyloid beta peptides.51
SlipChip-based PCR will be applicable to other situations requiring amplification of nucleic acids, for example, high-throughput multiplex DNA amplification performed before sequencing.20 Simple handling of small volumes on the multiplex SlipChip will be especially valuable for the analysis of cellular heterogeneity in a wide range of research and diagnostics contexts.54-56 PCR on SlipChip will also be useful for the detection of medically important bacterial infections,57 genetic diseases and genetic mutations,58,59 and food or water contaminants.60,61 The current platform can also be adapted to perform reverse transcription PCR for RNA amplification for RNA virus detection,62 study gene expression,63 and investigate cellular heterogeneity.64 This SlipChip can also be used in other applications that require preloaded arrays of reagents with multiplex and high throughput capacity, temperature control, or changing temperature, such as protein crystallization,30-32,34 testing of protein stability and protein-ligand interactions,41-43 heterogeneous immunoassays,33 DNA hybridization,65 DNA-protein interaction,66 and chromatin immuno-precipitation (ChIP).67 The SlipChip-based PCR method should enable inexpensive diagnostics, as the SlipChip itself can be fabricated inexpensively and requires no complex equipment or specialized knowledge to operate. When dried reagents are preloaded onto the SlipChip, it is also easy to transport and store. If integrated with isothermal amplification methods such as loop-mediated amplification (LAMP),68,69 recombinase polymerase amplification (RPA),70 nucleic acid sequence based amplification (NASBA),71,72 transcription-mediated amplification (TMA),73,74 helicase-dependent amplification (HAD),75,76 rolling-circle amplification (RCA),77,78 and strand-displacement amplification (SDA),75, 79,80 and with simple readouts, the SlipChip should also make nucleic acid testing available in resource-limited settings.
Supplementary Material
Acknowledgments
This work was supported by the NIH Director's Pioneer Award program, part of the NIH Roadmap for Medical Research (1 DP1 OD003584). Part of this work was performed at the Materials Research Science and Engineering Centers microfluidic facility (funded by the National Science Foundation). We thank Heidi Park for contributions to writing and editing this manuscript
Footnotes
Supporting Information Available. Experimental Section and supporting figures and tables. This material is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Kambouris M, Jackson CE, Feldman GL. Hum Mutat. 1996;8:64–70. doi: 10.1002/(SICI)1098-1004(1996)8:1<64::AID-HUMU9>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 2.Casilli F, Di Rocco ZC, Gad S, Tournier I, Stoppa-Lyonnet D, Frebourg T, Tosi M. Hum Mutat. 2002;20:218–226. doi: 10.1002/humu.10108. [DOI] [PubMed] [Google Scholar]
- 3.Oliveira DC, de Lencastre H. Antimicrob Agents Chemother. 2002;46:2155–2161. doi: 10.1128/AAC.46.7.2155-2161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palka-Santini M, Cleven BE, Eichinger L, Kronke M, Krut O. BMC Microbiol. 2009;9:1. doi: 10.1186/1471-2180-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Westh H, Lisby G, Breysse F, Boddinghaus B, Chomarat M, Gant V, Goglio A, Raglio A, Schuster H, Stuber F, Wissing H, Hoeft A. Clin Microbiol Infect. 2009;15:544–551. doi: 10.1111/j.1469-0691.2009.02736.x. [DOI] [PubMed] [Google Scholar]
- 6.SABiosciences Corporation Homepage. 2010 http://sabiosciences.com/
- 7.Kopp MU, de Mello AJ, Manz A. Science. 1998;280:1046–1048. doi: 10.1126/science.280.5366.1046. [DOI] [PubMed] [Google Scholar]
- 8.Khandurina J, McKnight TE, Jacobson SC, Waters LC, Foote RS, Ramsey JM. Anal Chem. 2000;72:2995–3000. doi: 10.1021/ac991471a. [DOI] [PubMed] [Google Scholar]
- 9.Lagally ET, Medintz I, Mathies RA. Anal Chem. 2001;73:565–570. doi: 10.1021/ac001026b. [DOI] [PubMed] [Google Scholar]
- 10.Koh CG, Tan W, Zhao MQ, Ricco AJ, Fan ZH. Anal Chem. 2003;75:4591–4598. doi: 10.1021/ac0343836. [DOI] [PubMed] [Google Scholar]
- 11.Lagally ET, Scherer JR, Blazej RG, Toriello NM, Diep BA, Ramchandani M, Sensabaugh GF, Riley LW, Mathies RA. Anal Chem. 2004;76:3162–3170. doi: 10.1021/ac035310p. [DOI] [PubMed] [Google Scholar]
- 12.Hashimoto M, Chen PC, Mitchell MW, Nikitopoulos DE, Soper SA, Murphy MC. Lab Chip. 2004;4:638–645. doi: 10.1039/b406860b. [DOI] [PubMed] [Google Scholar]
- 13.Pal R, et al. Lab Chip. 2005;5:1024–1032. doi: 10.1039/b505994a. [DOI] [PubMed] [Google Scholar]
- 14.Xiang Q, Xu B, Fu R, Li D. Biomed Microdevices. 2005;7:273–279. doi: 10.1007/s10544-005-6069-8. [DOI] [PubMed] [Google Scholar]
- 15.Ottesen EA, Hong JW, Quake SR, Leadbetter JR. Science. 2006;314:1464–1467. doi: 10.1126/science.1131370. [DOI] [PubMed] [Google Scholar]
- 16.Marcus JS, Anderson WF, Quake SR. Anal Chem. 2006;78:956–958. doi: 10.1021/ac0513865. [DOI] [PubMed] [Google Scholar]
- 17.Wang J, Chen ZY, Corstjens P, Mauk MG, Bau HH. Lab Chip. 2006;6:46–53. doi: 10.1039/b511494b. [DOI] [PubMed] [Google Scholar]
- 18.Beer NR, Hindson BJ, Wheeler EK, Hall SB, Rose KA, Kennedy IM, Colston BW. Anal Chem. 2007;79:8471–8475. doi: 10.1021/ac701809w. [DOI] [PubMed] [Google Scholar]
- 19.Lindstrom S, Hammond M, Brismar H, Andersson-Svahn H, Ahmadian A. Lab Chip. 2009;9:3465–3471. doi: 10.1039/b912596e. [DOI] [PubMed] [Google Scholar]
- 20.Tewhey R, et al. Nat Biotechnol. 2009;27:1025–1031. doi: 10.1038/nbt.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Doorn R, Klerks MM, van Gent-Pelzer MPE, Speksnijder AGCL, Kowalchuk GA, Schoen CD. Appl Environ Microbiol. 2009;75:7253–7260. doi: 10.1128/AEM.00796-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emanuel PA, Bell R, Dang JL, McClanahan R, David JC, Burgess RJ, Thompson J, Collins L, Hadfield T. J Clin Microbiol. 2003;41:689–693. doi: 10.1128/JCM.41.2.689-693.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu RH, Yang JN, Lenigk R, Bonanno J, Grodzinski P. Anal Chem. 2004;76:1824–1831. doi: 10.1021/ac0353029. [DOI] [PubMed] [Google Scholar]
- 24.Easley CJ, Karlinsey JM, Bienvenue JM, Legendre LA, Roper MG, Feldman SH, Hughes MA, Hewlett EL, Merkel TJ, Ferrance JP, Landers JP. Proc Natl Acad Sci U S A. 2006;103:19272–19277. doi: 10.1073/pnas.0604663103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Legendre LA, Bienvenue JM, Roper MG, Ferrance JP, Landers JP. Anal Chem. 2006;78:1444–1451. doi: 10.1021/ac0516988. [DOI] [PubMed] [Google Scholar]
- 26.Dineva MA, Mahilum-Tapay L, Lee H. Analyst. 2007;132:1193–1199. doi: 10.1039/b705672a. [DOI] [PubMed] [Google Scholar]
- 27.Pipper J, Zhang Y, Neuzil P, Hsieh TM. Angew Chem Int Edit. 2008;47:3900–3904. doi: 10.1002/anie.200705016. [DOI] [PubMed] [Google Scholar]
- 28.Rossney AS, Herra CM, Brennan GI, Morgan PM, O'Connell B. J Clin Microbiol. 2008;46:3285–3290. doi: 10.1128/JCM.02487-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Welch DF, Ginocchio CC. J Clin Microbiol. 2010;48:22–25. doi: 10.1128/JCM.02268-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du WB, Li L, Nichols KP, Ismagilov RF. Lab Chip. 2009;9:2286–2292. doi: 10.1039/b908978k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li L, Du W, Ismagilov R. J Am Chem Soc. 2010;132:106–111. doi: 10.1021/ja908555n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li L, Du W, Ismagilov RF. J Am Chem Soc. 2010;132:112–119. doi: 10.1021/ja908558m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu WS, Chen DL, Du WB, Nichols KP, Ismagilov RF. Anal Chem. 2010;82:3276–3282. doi: 10.1021/ac100044c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li L, Ismagilov RF. Ann Rev Biophys. 2010;39:139–158. doi: 10.1146/annurev.biophys.050708.133630. [DOI] [PubMed] [Google Scholar]
- 35.Boedicker JQ, Vincent ME, Ismagilov RF. Angew Chem Int Ed. 2009;48:5908–5911. doi: 10.1002/anie.200901550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vincent ME, Liu W, Haney EB, Ismagilov RF. Chem Soc Rev. 2010;39:974–984. doi: 10.1039/b917851a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen F, Du W, Luchetta EM, Fok A, Ismagilov RF. Lab Chip. 2010 doi: 10.1039/c004521g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brakstad OG, Aasbakk K, Maeland JA. J Clin Microbiol. 1992;30:1654–1660. doi: 10.1128/jcm.30.7.1654-1660.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shrestha NK, Tuohy MJ, Hall GS, Isada CM, Procop GW. J Clin Microbiol. 2002;40:2659–2661. doi: 10.1128/JCM.40.7.2659-2661.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li L, Karymov M, Nichols KP, Ismagilov RF. Langmuir. 2010 doi: 10.1021/la101460z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matulis D, Kranz JK, Salemme FR, Todd MJ. Biochemistry. 2005;44:5258–5266. doi: 10.1021/bi048135v. [DOI] [PubMed] [Google Scholar]
- 42.Ericsson UB, Hallberg BM, DeTitta GT, Dekker N, Nordlund P. Anal Biochem. 2006;357:289–298. doi: 10.1016/j.ab.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 43.Niesen FH, Berglund H, Vedadi M. Nat Protoc. 2007;2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 44.Mackay IM. Clin Microbiol Infec. 2004;10:190–212. doi: 10.1111/j.1198-743x.2004.00722.x. [DOI] [PubMed] [Google Scholar]
- 45.Vogelstein B, Kinzler KW. P Natl Acad Sci USA. 1999;96:9236–9241. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heller MJ. Annu Rev Biomed Eng. 2002;4:129–153. doi: 10.1146/annurev.bioeng.4.020702.153438. [DOI] [PubMed] [Google Scholar]
- 47.Song H, Tice JD, Ismagilov RF. Angew Chem Int Ed. 2003;42:768–772. doi: 10.1002/anie.200390203. [DOI] [PubMed] [Google Scholar]
- 48.Song H, Chen DL, Ismagilov RF. Angew Chem Int Ed. 2006;45:7336–7356. doi: 10.1002/anie.200601554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roach LS, Song H, Ismagilov RF. Anal Chem. 2005;77:785–796. doi: 10.1021/ac049061w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kreutz JE, Li L, Roach LS, Hatakeyama T, Ismagilov RF. J Am Chem Soc. 2009;131:6042–6043. doi: 10.1021/ja808697e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meier M, Kennedy-Darling J, Choi SH, Norstrom EM, Sisodia SS, Ismagilov RF. Angew Chem Int Edit. 2009;48:1487–1489. doi: 10.1002/anie.200805225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qin X, Emerson J, Stapp J, Stapp L, Abe P, Burns JL. J Clin Microbiol. 2003;41:4312–4317. doi: 10.1128/JCM.41.9.4312-4317.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luo GZ, Mitchell TG. J Clin Microbiol. 2002;40:2860–2865. doi: 10.1128/JCM.40.8.2860-2865.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Howell MD, Diveley JP, Lundeen KA, Esty A, Winters ST, Carlo DJ, Brostoff SW. Proc Natl Acad Sci U S A. 1991;88:10921–10925. doi: 10.1073/pnas.88.23.10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Castilla LH, Couch FJ, Erdos MR, Hoskins KF, Calzone K, Garber JE, Boyd J, Lubin MB, Deshano ML, Brody LC, Collins FS, Weber BL. Nature Genet. 1994;8:387–391. doi: 10.1038/ng1294-387. [DOI] [PubMed] [Google Scholar]
- 56.Trogan E, Choudhury RP, Dansky HM, Rong JX, Breslow JL, Fisher EA. Proc Natl Acad Sci U S A. 2002;99:2234–2239. doi: 10.1073/pnas.042683999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Millar BC, Xu J, Moore JE. Curr Issues Mol Biol. 2007;9:21–39. [PubMed] [Google Scholar]
- 58.Wang YY, et al. Proc Natl Acad Sci U S A. 2005;102:1104–1109. doi: 10.1073/pnas.0408831102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lun FMF, Tsui NBY, Chan KCA, Leung TY, Lau TK, Charoenkwan P, Chow KCK, Lo WYW, Wanapirak C, Sanguansermsri T, Cantor CR, Chiu RWK, Lo YMD. Proc Natl Acad Sci U S A. 2008;105:19920–19925. doi: 10.1073/pnas.0810373105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Norton DM. J AOAC Int. 2002;85:505–515. [PubMed] [Google Scholar]
- 61.Malik S, Beer M, Megharaj M, Naidu R. Environ Int. 2008;34:265–276. doi: 10.1016/j.envint.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 62.Stockton J, Ellis JS, Saville M, Clewley JP, Zambon MC. J Clin Microbiol. 1998;36:2990–2995. doi: 10.1128/jcm.36.10.2990-2995.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bustin SA. J Mol Endocrinol. 2002;29:23–39. doi: 10.1677/jme.0.0290023. [DOI] [PubMed] [Google Scholar]
- 64.Chandran UR, Ma CQ, Dhir R, Bisceglia M, Lyons-Weiler M, Liang WJ, Michalopoulos G, Becich M, Monzon FA. BMC Cancer. 2007;7:64. doi: 10.1186/1471-2407-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dufva M, Petersen J, Poulsen L. Anal Bioanal Chem. 2009;395:669–677. doi: 10.1007/s00216-009-2848-z. [DOI] [PubMed] [Google Scholar]
- 66.Walter G, Bussow K, Cahill D, Lueking A, Lehrach H. Curr Opin Microbiol. 2000;3:298–302. doi: 10.1016/s1369-5274(00)00093-x. [DOI] [PubMed] [Google Scholar]
- 67.Collas P, Dahl JA. Front Biosci. 2008;13:929–943. doi: 10.2741/2733. [DOI] [PubMed] [Google Scholar]
- 68.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Nucleic Acids Res. 2000;28:e63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aryan E, Makvandi M, Farajzadeh A, Huygen K, Bifani P, Mousavi SL, Fateh A, Jelodar A, Gouya MM, Romano M. Microbiol Res. 2009;165:211–220. doi: 10.1016/j.micres.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 70.Piepenburg O, Williams CH, Stemple DL, Armes NA. PLoS Biol. 2006;4:e204. doi: 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Romano JW, Williams KG, Shurtliff RN, Ginocchio C, Kaplan M. Immunol Invest. 1997;26:15–28. doi: 10.3109/08820139709048912. [DOI] [PubMed] [Google Scholar]
- 72.Dimov IK, Garcia-Cordero JL, O'Grady J, Poulsen CR, Viguier C, Kent L, Daly P, Lincoln B, Maher M, O'Kennedy R, Smith TJ, Ricco AJ, Lee LP. Lab Chip. 2008;8:2071–2078. doi: 10.1039/b812515e. [DOI] [PubMed] [Google Scholar]
- 73.Hill C, Bott M, Clark K, Jonas V. Clin Chem. 1995;41:S107–S107. [Google Scholar]
- 74.Chelliserrykattil J, Nelson NC, Lyakhov D, Carlson J, Phelps SS, Kaminsky MB, Gordon P, Hashima S, Ngo T, Blazie S, Brentano S. J Mol Diagn. 2009;11:680–680. [Google Scholar]
- 75.Vincent M, Xu Y, Kong HM. Embo Rep. 2004;5:795–800. doi: 10.1038/sj.embor.7400200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jeong YJ, Park K, Kim DE. Cell Mol Life Sci. 2009;66:3325–3336. doi: 10.1007/s00018-009-0094-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hafner GJ, Yang IC, Wolter LC, Stafford MR, Giffard PM. Biotechniques. 2001;30:852–856. doi: 10.2144/01304rr03. [DOI] [PubMed] [Google Scholar]
- 78.Lizardi PM, Huang X, Zhu Z, Bray-Ward P, Thomas DC, Ward DC. Nat Genet. 1998;19:225–232. doi: 10.1038/898. [DOI] [PubMed] [Google Scholar]
- 79.Walker GT, Fraiser MS, Schram JL, Little MC, Nadeau JG, Malinowski DP. Nucleic Acids Res. 1992;20:1691–1696. doi: 10.1093/nar/20.7.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hellyer TJ, Nadeau JG. Expert Rev Mol Diagn. 2004;4:251–261. doi: 10.1586/14737159.4.2.251. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.