Abstract
Many neurodegenerative diseases exhibit protein accumulation and increased oxidative stress. Therapeutic strategies include clearing aggregate-prone proteins by enhancing autophagy or decreasing oxidative stress with antioxidants. Many autophagy-inducing stimuli increase reactive oxygen species (ROS), raising concerns that the benefits of autophagy up-regulation may be counterbalanced by ROS toxicity. Here we show that not all autophagy inducers significantly increase ROS. However, many antioxidants inhibit both basal and induced autophagy. By blocking autophagy, antioxidant drugs can increase the levels of aggregate-prone proteins associated with neurodegenerative disease. In fly and zebrafish models of Huntington's disease, antioxidants exacerbate the disease phenotype and abrogate the rescue seen with autophagy-inducing agents. Thus, the potential benefits in neurodegenerative diseases of some classes of antioxidants may be compromised by their autophagy-blocking properties.
INTRODUCTION
Neurodegenerative diseases, such as Parkinson's disease (PD), and polyglutamine expansion diseases, such as Huntington's disease (HD), represent a significant burden in terms of individual morbidity, mortality and wider social and economic costs (1). Treatment remains only symptomatic as unfortunately there are currently no therapies that modify the progression of the disease (2). Considerable research activity has been directed towards understanding common pathological mechanisms that might be amenable to therapeutic intervention. Oxidative stress is one common feature of these diseases that has been extensively investigated (3). Reactive oxygen species (ROS) are formed by the incomplete reduction of oxygen and are produced at low levels under normal physiological conditions as a result of mitochondrial respiration and a number of other processes. The cell is able to absorb some increase in ROS via a variety of antioxidant defences. When these defences are overwhelmed, ‘oxidative stress’ is said to occur and this ultimately leads to cell death. There is a large amount of in vitro and in vivo evidence suggesting that oxidative stress occurs during neurodegeneration, and, as a result, ameliorating oxidative stress with antioxidant drugs has been a major focus of therapeutic research (4).
A second hallmark of these diseases is the accumulation and aggregation of misfolded pathogenic proteins. Clearance of disease-associated proteins, such as mutant alpha-synuclein, ataxin-3, tau and huntingtin, can be achieved by up-regulating the bulk protein degradative process of macroautophagy (5). Furthermore, autophagy seems integral to neuronal health as inhibition of autophagy results in neurodegeneration even in the absence of disease (6). During autophagy, mutant protein is sequestered along with cytoplasm in a double membrane to form an autophagosome. Subsequent to fusion of the autophagosome and lysosome, autophagosomal substrates are degraded. Autophagy is tightly regulated by a number of pathways, of which the most extensively studied involves the protein kinase mTOR (mammalian target of rapamycin), which negatively regulates autophagy. Inhibition of this pathway with the drug rapamycin enhances autophagy (7). More recently, drugs that do not act via mTOR (‘mTOR-independent’ agents) have been identified (8), and new fundamental mechanisms regulating autophagy have been described (9). Drugs which up-regulate autophagy have been shown to alleviate toxicity in mouse models of HD and spinocerebellar ataxia type 3, and, as a result, autophagy-enhancing drugs represent an alternative potential disease-modifying therapy (10,11).
Recently, the themes of oxidative stress and autophagy have been brought together. Starvation, a classic autophagy-inducing stimulus, has been shown to increase both cellular levels of ROS and autophagy (12). One mechanism by which autophagy appears to be regulated by ROS is the redox regulation of the essential autophagy protein Atg4 (12). This protein is required for the conjugation of Atg8 to phosphatidylethanolamine, a reaction that is in turn essential for the induction of autophagy. It seems unlikely that this is the only ROS-dependent mechanism involved, as Atg4 not only lipidates Atg8 when oxidized but is also responsible for delipidating and recycling Atg8 when reduced. Thus, in strong oxidizing conditions, increased lipidation of Atg8 will initially occur and induce autophagy, but autophagy may ultimately be inhibited if Atg8 is not recycled.
The importance of autophagy and oxidative stress to both pathology and potential therapies for neurodegeneration has led us to investigate their relationship further. A diverse range of stimuli that induce both ROS and autophagy have been described and autophagy induction by these agents is antagonized by antioxidants (13–15). If all autophagy-inducing drugs increase ROS then this may be a potential barrier to their use in human sufferers of neurodegenerative diseases, because the consequent increases in the already high levels of oxidative stress may outweigh any benefit from autophagy up-regulation. For this reason, we tested whether all autophagy-inducing agents also increase oxidative stress, and found this not to be the case. However, our data suggest that many ROS scavengers block both basal autophagy and autophagy induced by ROS-independent mechanisms. Furthermore, these ROS scavengers can enhance the toxicity of polyglutamine expansions in flies and zebrafish, suggesting that the use of such drugs may be limited in the context of neurodegenerative diseases caused by aggregate-prone proteins which are also autophagy substrates.
RESULTS
Not all autophagy-inducing drugs are associated with an increase in ROS
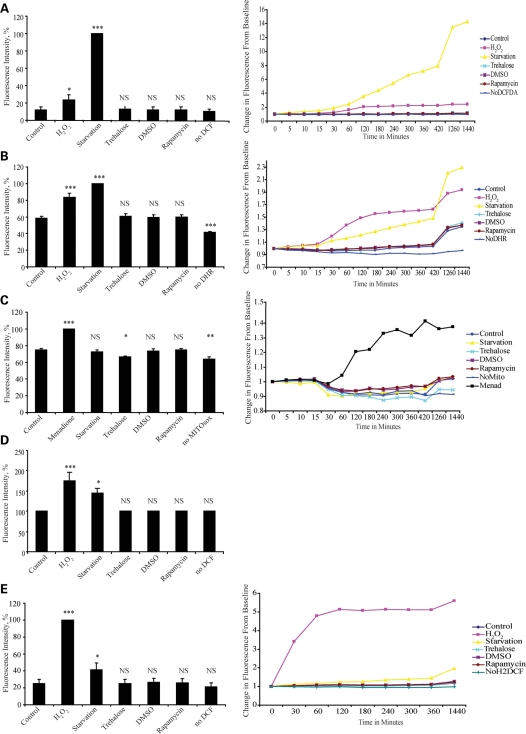
ROS levels are commonly measured using redox sensitive fluorescent probes and fluorescence-activated cell sorting (FACS). This technique may be limited, however, as suspension of cells in phosphate buffered saline (PBS) prior to analysis by FACS will result in starvation and rapid and large increases in ROS (12). In order to avoid this potential artefact, we loaded HeLa cells with one of the three different ROS-sensitive fluorogenic probes [dichlorofluorescin diacetate (DCFDA), dihydrorhodamine 123 and the mitochondrially localized superoxide detector MitoSOX red™] before treating them with autophagy inducers or controls and measured changes in fluorescence over 24 h. Neither rapamycin (an autophagy inducer acting by inhibiting the mTOR pathway), nor trehalose (an autophagy inducer acting via non-mTOR pathways) increased ROS (Fig. 1A–C). In fact, trehalose (a disaccharide with putative antioxidant properties) (16) significantly decreased mitochondrial superoxide production (Fig. 1C). In order to be sure there was no late increase in ROS following treatment with autophagy-inducing agents, we treated cells for 24 h with rapamycin or trehalose, before loading with a ROS sensitive probe and measuring change in signal over 1 h. Again, no increase in ROS following treatment with rapamycin or trehalose was seen (Fig. 1D). We also investigated the effects of autophagy induction on levels of ROS in a second cell line (COS-7) in which we have previously characterized autophagic activity (17). This cell line is of renal origin and therefore may be more sensitive to changes in ROS than HeLa cells. In order to increase the sensitivity of our assay, we used a derivative of DCFDA which increased cellular retention of the probe. Though we saw a much greater increase with positive controls, once again we saw no increase in ROS either with rapamycin or trehalose (Fig. 1E). While we cannot absolutely rule out small increases in ROS induced by rapamycin or trehalose, our data suggest that these agents do not induce the significant increase seen with some other autophagy-inducing stimuli, such as starvation.
Figure 1.
Not all autophagy-inducing agents cause an increase in ROS. Neither trehalose or rapamycin cause an increase in levels of ROS. HeLa cells were loaded with DCFDA (A), dihydrorhodamine 123 (B) or MitoSOX™ red (C), before being treated with H2O2, menadione, rapamycin, trehalose, DMSO or starved in Hanks Balanced Salt Solution (HBSS). Fluorescent signal was serially measured as described in Materials and Methods. The histograms show signal at 24 h as percentage of positive control while adjacent line graphs show change from baseline against time over 24 h. (D) Cells were treated with trehalose, dimethyl sulfoxide (DMSO), rapamycin or neither for 24 h before being loaded with DCFDA and signal read 1 h later. Treatment with hydrogen peroxide or starvation for 1 h after loading provided positive controls. Rapamycin and trehalose cause no increase in signal. (E) COS-7 cells were loaded with Carboxy-H2DCFDA before being treated in a similar way to (A). Only H2O2 and starvation caused significant increases in signal. All experiments were performed with no probe as a negative control (no DCFDA, NoDCFDA; no dihydrorhodamine, NoDHR; no Mito, No MitoSOX™ red; No Carboxy-H2DCFDA, NoH2DCF). Error bars are present for all data points (but are too small to be seen on some) and show standard error of the mean (SEM) for three independent experiments in triplicate (NS, not significant, *P < 0.05, **P < 0.01, ***P < 0.001).
Up-regulation of autophagy by rapamycin and trehalose is impaired by thiol antioxidants
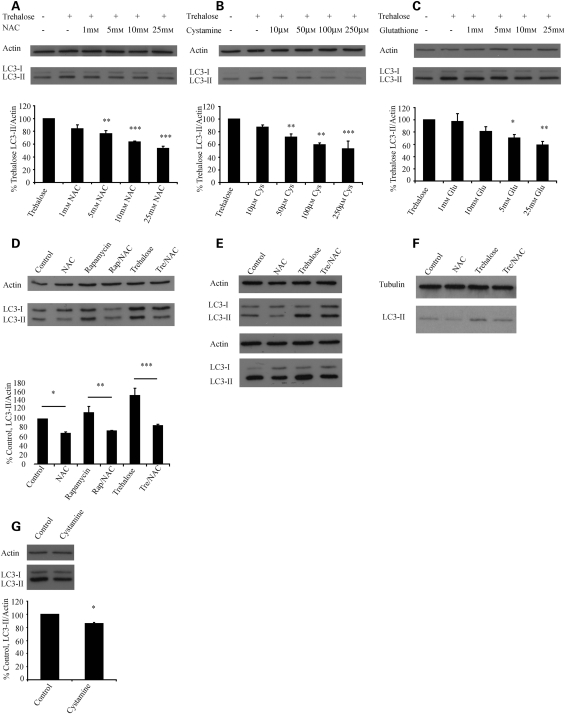
When autophagy induction is mediated by an increase in ROS, increases in autophagy can be inhibited with ROS scavengers (12,14). We next tested whether the impairment of autophagy by antioxidants was limited to situations where the autophagy-inducing agent also significantly increased ROS. We tested the ability of a variety of thiol antioxidants (including those proposed for treatment of HD) (18) to ameliorate the induction of autophagy by trehalose (which does not increase levels of ROS) in COS-7 cells. We found that N-acetyl cysteine (NAC), cystamine [a drug proposed for use in HD, with pleiotropic effects including transglutaminase inhibition, but which is also metabolized to the antioxidant l-cysteine (19)] and glutathione were all able to significantly ameliorate the induction of autophagy by trehalose in a dose-dependent fashion, as measured by levels of the autophagy marker, microtubule-associated protein 1 light chain 3 II (LC3-II) (Fig. 2A–C). LC3-II is formed from LC3-I after it is conjugated to phosphatidylethanolamine, an essential step in autophagy induction. Decreases in levels of LC3-II can be caused by a decrease in autophagosome formation or by increase in autophagosome lysosome fusion. As we are interested in autophagic flux, these experiments were performed in the presence of a saturating dose of bafilomycin A1, a macrolide antibiotic that blocks autophagosome/lysosome fusion (20). Thus, decreases in levels of LC3-II under these conditions represent decreased synthesis, rather than increased autophagosome/lysosome fusion (21). Interestingly, while co-treatment with trehalose and cystamine resulted in no change in LC3-II levels (data not shown), pre-treatment with cystamine did impair induction of autophagy, consistent with the requirement for cystamine to be metabolized to the glutathione precursor l-cysteine in order to influence autophagy (Fig. 2B). Note that while glutathione cannot directly penetrate cells, extracellular glutathione is metabolized by γ-glutamyl-transferase to ultimately form cysteine, which is readily taken up.
Figure 2.
Antioxidant drugs inhibit basal and induced autophagy. NAC, cystamine and glutathione all inhibit trehalose-induced autophagy. COS-7 cells were treated with trehalose and co-treated with the indicated concentrations of NAC (A), pre-treated for 24 h with cystamine (Cys) (B) or co-treated with glutathione (Glu) (C). After 24 h, a saturating dose of bafilomycin A1 was added for the last 4 h prior to harvest. The graphs below each blot show densitometric analysis of LC3-II/actin from three experiments performed in triplicate, with the control condition (trehalose alone) set to 100%. (D) NAC impairs rapamycin induced and basal autophagy. COS-7 cells were treated with 25 mm NAC, rapamycin, rapamycin and NAC, trehalose (Tre) or trehalose and NAC for 24 h. All conditions were treated with bafilomycin for 4 h prior to harvesting. The graph shows densitometric analysis with no treatment (control) set to 100%. (E) The effects of NAC on autophagy are also seen in neurons. Differentiated human primary cortical neurons were treated as indicated (NAC 10 mm) and blotted at 24 h with no further treatment (upper panel) or following 4 h of treatment with bafilomycin A1 (lower panel). Representative data from an experiment performed in triplicate is shown. (F) The effects of NAC are seen in HeLa cells. Hela cells were treated under control conditions or with NAC (15 mm), trehalose, or trehalose and NAC for 24 h before analysis by western blot. All experiments were carried out in the presence of a saturating dose of bafilomycin for the last 4 h. Note that HeLa cells have very low levels of LC3-1. (G) Cystamine can inhibit basal autophagy. COS-7 cells were treated with 250 µm cystamine for 48 h, the last 4 h also being in the presence of bafilomycin before being analysed by western blot. Error bars in all panels represent SEM from at least three independent experiments performed in triplicate (*P < 0.05, **P < 0.01, ***P < 0.001).
Thiol antioxidants can impair basal autophagy
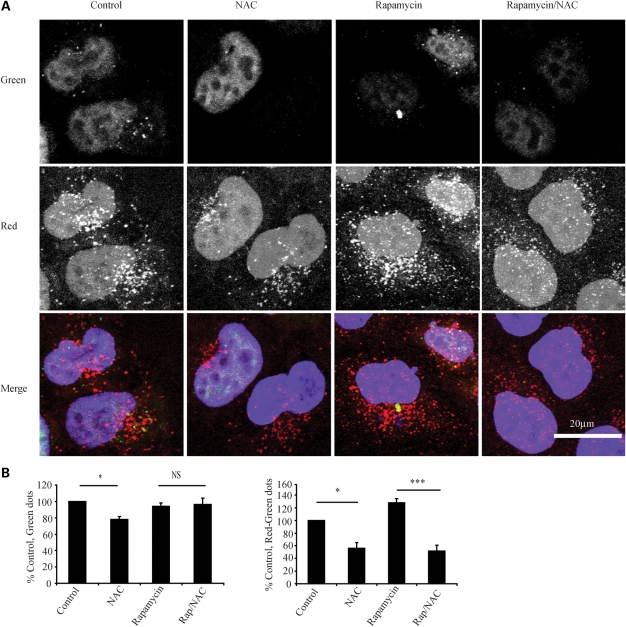
As thiol antioxidants seem to be able to profoundly impair induced autophagy, we examined whether they could impair autophagy induced by mTOR inhibition and basal autophagy. We found that NAC impaired the increase in LC3-II associated with rapamycin treatment and also decreased LC3-II compared with the basal state (Fig. 2D). We confirmed the effect of NAC on basal and inducible autophagy in human primary cortical neurons, with and without bafilomycin (Fig. 2E) and in HeLa cells (Fig. 2F). Pre-treatment with cystamine had similar effects on basal autophagy (Fig. 2G). We next employed a second autophagy assay to confirm our findings. We used a HeLa cell line constitutively expressing LC3 tagged to both green fluorescent protein (GFP) and red fluorescent protein (RFP). Due to the different pKa of these two fluorophores when present on the autophagosome, both red and green are seen, but when autophagosomes fuse with the lysosome, the green signal is quenched in the low pH environment, and only red is seen. Using this technique, it is therefore possible to differentiate and count red dots (autophagolysosomes and autophagosomes), green dots (autophagosomes) and red dots that are not also green (autophagolysosomes). This second autophagy measure was consistent with NAC impairing both basal and inducible autophagy by inhibiting autophagosome synthesis rather than enhancing fusion (Fig. 3A and B).
Figure 3.
NAC impairs autophagosome synthesis in a second assay. (A) NAC significantly decreases both autophagosome and autophagolysosome number. HeLa cells expressing RFP and GFP tagged LC3-II as described in Materials and Methods were treated as shown (NAC 10 mm), before being fixed and subjected to automated counting of red and green dots. Representative confocal microscopy images are shown. (B) Results of automated cell counting. The left panel shows the number of green dots (autophagosomes) and the right panel the number of red minus green dots (autophagolysosomes) relative to control. The graphs shown represent the mean results from five experiments performed in triplicate. Error bars represent SEM (*P < 0.05, ***P < 0.001, NS, non significant).
NAC inhibits the clearance of disease-related proteins
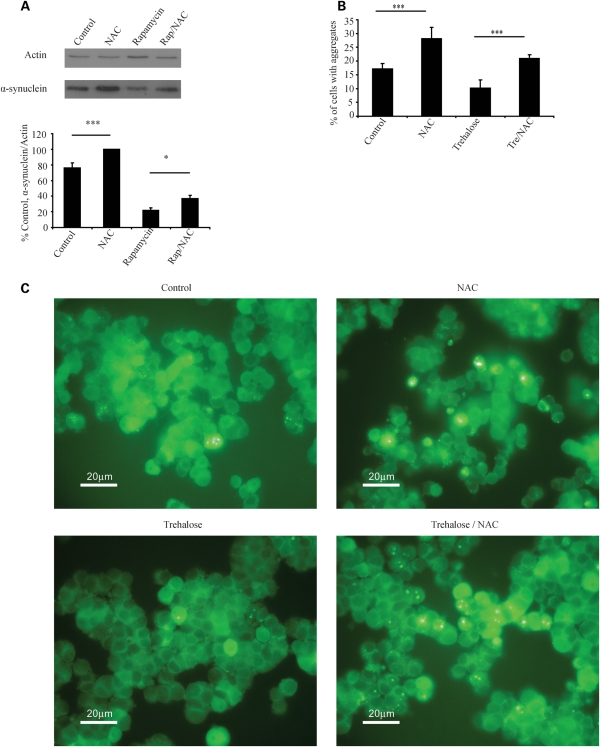
As high doses of NAC impair autophagy, we tested whether it also impaired clearance of disease-related autophagy substrates in neuronal cell models. We found that NAC was able to impair the clearance of an autophagy substrate, haemagglutinin (HA)-tagged mutant A53T α-synuclein, a soluble protein that causes a form of familial PD (22). NAC also significantly impaired the ability of rapamycin to enhance clearance of this substrate (Fig. 4A). Similarly, we found that NAC increased the percentage of cells with aggregates of enhanced green fluorescent protein (EGFP)-tagged mutant huntingtin exon 1 and impaired the ability of trehalose to decrease the percentage of cells with aggregates, a phenomenon that is autophagy dependent (8) (Fig. 4B and C).
Figure 4.
NAC impairs clearance of autophagy substrates. (A) Expression of HA-tagged A53T α-synuclein [a mutant form of the protein that does not form visible aggregates in these cells (22)] was induced for 48 h before transgene expression was switched off. During the switch-off period, cells were treated as shown (NAC 10 mm, Rap, rapamycin) for 24 h before being harvested and analysed by western blotting. The graph shows densitometric analysis with results for NAC set to 100%. (B) NAC increases aggregation of mutant huntingtin. Expression of EGFP tagged huntingtin exon 1 with an expanded (74Q) polyglutamine repeat was induced for 8 h (47). The transgene was then switched off and the cells treated as shown (NAC 10 mm) for 72 h before fixing and counting. The graph shows percentage of cells with aggregates for each condition for a representative experiment, error bars represent standard deviation. For all other panels, error bars represent SEM (*P < 0.05, ***P < 0.001, NS, non significant).
Inhibition of autophagy by antioxidant drugs is not restricted to thiol antioxidants
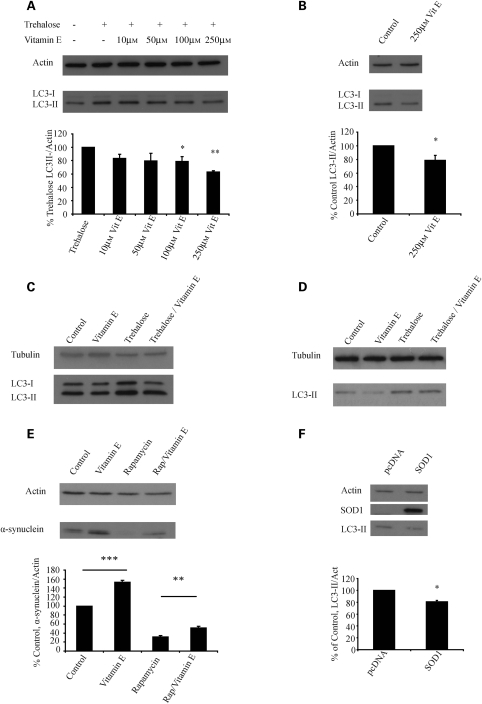
In order to test whether autophagy inhibition by antioxidants is thiol specific, we tested the non-thiol antioxidant vitamin E (DL-alpha tocopherol). Vitamin E has been proposed as a treatment for neurodegenerative disease and has been used in major trials in PD and HD (23,24). We found that vitamin E was similarly able to impair trehalose-induced and basal levels of LC3-II in COS-7 cells, mouse primary cortical neurons and HeLa cells (Fig. 5A–D). Like NAC, it was able to strongly inhibit the degradation of A53T α-synuclein and prevent the enhanced degradation of this substrate in the presence of rapamycin (Fig. 5E).
Figure 5.
Vitamin E and over-expression of SOD can also inhibit autophagy. (A) Vitamin E can inhibit trehalose-induced autophagy. COS-7 cells were treated with trehalose and increasing doses of vitamin E (DL-alpha tocopherol, VitE) for 24 h, before treatment for 4 h with bafilomycin, in a similar manner to the experiments presented in Figure 2A–C. The densitometric analysis shows error bars and statistics from three experiments performed in triplicate as in Figure 2A–C. (B) Vitamin E can inhibit basal autophagy. COS-7 cells were treated with 250 µm vitamin E before being analysed by western blot in a similar manner to Figure 2G. (C) The effects of vitamin E on autophagy are also seen in primary neuronal culture. Differentiated mouse primary cortical neurones were treated as indicated with bafilomycin added for the last 4 h. (D) Vitamin E has similar effects on basal and induced autophagy in HeLa cells. HeLa cells were treated under control conditions or with vitamin E (250 µm), trehalose or trehalose and vitamin E for 24 h before analysis by western blot. All experiments were carried out in the presence of a saturating dose of bafilomycin for the last 4 h. (E) Vitamin E can inhibit the clearance of autophagy substrates associated with neurodegenerative disease. Expression of HA-tagged A53T α-synuclein was induced for 48 h before the cells were treated as shown (vitamin E, 250 µm) for 24 h before being harvested and analysed by western blotting. The graph shows densitometric analysis with results for control set to 100%. (F) COS-7 cells were transfected with human SOD1 and allowed to express for 48 h, the last 4 h with bafilomycin. A representative western blot is shown. The graph shows densitometric analysis from five independent experiments performed in triplicate with pcDNA (control) set to 100%. Error bars represent SEM (*P < 0.05, **P < 0.01, ***P < 0.001, NS, non-significant) for all panels.
As chemical antioxidants often have pleiotropic effects (and therefore could have an influence on autophagic activity by mechanisms other than decreasing oxidative stress), we investigated the effect on autophagy of genetic up-regulation of defences against oxidative stress. Previously, over-expression of the superoxide dismutase gene (SOD2), which scavenges superoxide, has been shown to inhibit induced autophagy associated with increased ROS (25). SOD2 is localized to the mitochondrial matrix, while SOD1 is mainly cytosolic with a smaller fraction in the mitochondrial intermembrane space. In order to test whether SOD1 over-expression can also inhibit autophagy, we transiently over-expressed SOD1 and observed the effect on levels on LC3-II in the presence of bafilomycin. As expected, over-expression of SOD1 decreased basal LC3-II levels (Fig. 5F).
Antioxidants affect multiple canonical mechanisms regulating autophagy
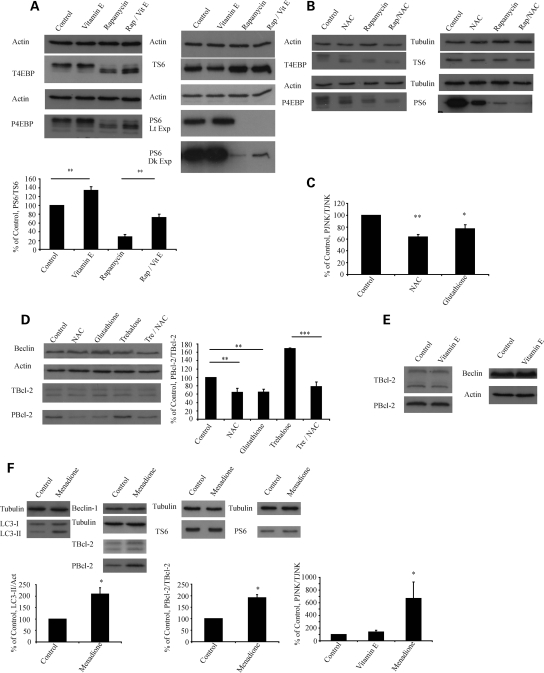
In order to examine potential mechanisms by which antioxidants may influence autophagy, we examined their effect on the mTOR pathway, a classical negative regulator of autophagy, by looking for changes in the phosphorylation status of an mTOR kinase substrate 4E binding protein 1 (4E-BP1) and a protein dependent on mTOR substrate kinase activity, ribosomal protein S6 (S6). Vitamin E enhanced the activity of mTOR but, surprisingly, NAC appeared to be inhibiting mTOR activity—an effect that would be expected to increase, rather than decrease, autophagy (Fig. 6A and B). Thus, while vitamin E influences the mTOR pathway in a way which is consistent with autophagy inhibition, this was not the case for NAC.
Figure 6.
Antioxidants affect different canonical mechanisms regulating autophagy. (A) Vitamin E increases phosphorylation of mTOR substrates. COS-7 cells were treated with 250 µm vitamin E, rapamycin or rapamycin and vitamin E for 24 h before harvest. Lysates were run simultaneously on two gels and probed for mTOR substrates and loading controls. 4E-BP-1 (an mTOR substrate) has multiple phosphorylation sites and runs as four bands with the upper bands representing predominantly phosphorylated forms. Vitamin E increases phosphorylation compared with control and impairs the ability of rapamycin to inhibit the phosphorylation of 4E-BP1. These results were confirmed using a second substrate, ribosomal protein S6. T4E-BP denotes total 4E-BP, P4E-BP phosphorylated 4EBP, TS6 total S6 and PS6 phosphorylated S6. Dk exp denotes dark exposure and Lt exp light exposure. (B) NAC decreases phosphorylation of mTOR substrates. This experiment was performed in a similar manner to (A) but using 25 mm NAC instead of vitamin E. Representative western blots are shown. (C) Thiol antioxidants decrease JNK phosphorylation. COS-7 cells were treated for 24 h with 25 mm NAC or glutathione and the phosphorylation of JNK measured using a FACE ELISA kit. The graph shows phosphorylated (PJNK)/total JNK (TJNK) with control set to 100%. (D) Thiol antioxidants decrease Bcl-2 phosphorylation. COS-7 cells were treated as shown for 24 h and blotted for phosphorylated Bcl-2 (PBcl-2) before stripping and reprobing for total Bcl-2 (TBcl-2). The graph represents densitometric analysis. (E) Vitamin E has no effect on Bcl-2 phosphorylation. COS-7 cells were treated for 24 h with vitamin E or DMSO control and analysed by western blot for levels of phosphorylated Bcl-2 and Beclin 1. No significant differences were seen. (F) The superoxide generating agent menadione increases LC3-II and phosphorylation of JNK and Bcl-2, but has no effect on mTOR substrates or Beclin-1 expression. COS-7 cells were treated with 100 µm menadione for 1 h prior to harvest and analysed by western blot for LC3-II, phosphorylated Bcl-2 and total Bcl-2. The graphs show densitometric analysis with control set to 100%. The effect of menadione and vitamin E on JNK phosphorylation was analysed in a similar way to (C). All panels represent results from three independent triplicate experiments, error bars represent SEM (*P < 0.05, **P < 0.01, ***P < 0.001, NS, non-significant).
Starvation-induced autophagy is associated with increased levels of ROS and has been shown to be mediated by the activation of c-Jun N-terminal protein kinase 1 (JNK1) which, in turn, phosphorylates Bcl-2 and causes it to dissociate from Beclin 1 (26). NAC is able to inhibit both ROS accumulation and autophagy in starved cells (12). Given this, and the unexpected effect of NAC on mTOR, we next tested the effect of thiol antioxidants on JNK activation. NAC and glutathione inhibited the activation of JNK and decreased the phosphorylation of Bcl-2 (Fig. 6C and D). Previous work has shown that oxidative stress in the form of hydrogen peroxide can increase the expression of Beclin-1 and thus induce autophagy (27). We did not find that thiol antioxidants or vitamin E had any significant effect on levels of Beclin-1 (Fig. 6D and E).
Superoxide has recently been described as a specific ROS regulating autophagy, though the exact mechanism remains unknown (25). We found that the superoxide-generating agent, menadione, strongly increased levels of LC3-II at time points as short as 45 min (though, as superoxide is very rapidly converted to hydrogen peroxide, the actual ROS involved in this process is unclear) (Fig. 6F). Menadione had the opposite effect of thiol antioxidants by enhancing JNK and Bcl-2 phosphorylation, but had no effect on mTOR activity or Beclin-1 expression (Fig. 6F). In summary, these results suggest that vitamin E enhances the activity of mTOR, an effect which would be consistent with autophagy inhibition. Thiol ROS scavengers such as NAC inhibit mTOR (which would be expected to induce autophagy) but decrease the phosphorylation of JNK and Bcl-2 which will inhibit autophagy, while the superoxide-generating agent menadione increases levels of LC3-II and the phosphorylation of JNK and Bcl-2.
Thiol antioxidants and over-expression of SOD exacerbate the phenotype seen in Drosophila models of polyglutamine disease
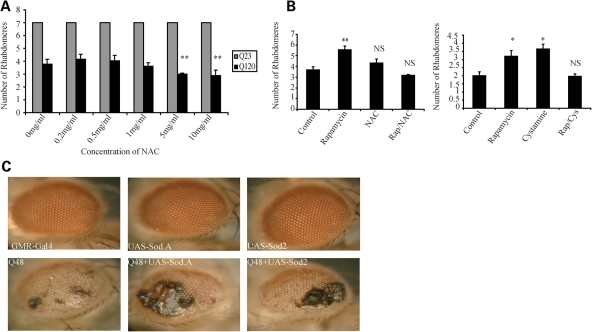
To investigate the potential pathophysiological significance of our findings, we used a Drosophila model of HD that expresses exon 1 of huntingtin with an expanded polyglutamine tract (Q120) in the eye (28). Each eye is made up of many ommatidia which are, in turn, made of eight rhabdomeres, seven of which are visible. The severity of the disease phenotype was measured by counting the number of rhabdomeres per ommatidia—the more severe the degeneration, the fewer rhabdomeres are seen. We treated these flies with increasing doses of NAC and found that low doses of NAC had no significant effect, while high doses exacerbated the neurodegenerative phenotype (Fig. 7A). We used a dose of NAC that did not, on its own, exacerbate the phenotype in the HD flies and tested it alone and in combination with the autophagy-enhancing agent rapamycin. We found that both NAC and cystamine significantly impaired the ability of rapamycin to rescue the HD phenotype (Fig. 7B). Note that rapamycin has been shown to suppress polyglutamine toxicity in flies in an autophagy-dependent manner (29). Though it is possible that rapamycin is inhibiting the rescue seen with cystamine, rather than the other way around, the idea that cystamine is inhibiting autophagy induction is consistent with our cell biology findings. Furthermore, the fact that NAC was able to ameliorate the rescue seen with rapamycin, while exhibiting no significant rescue by itself, suggests that thiol antioxidants are able to inhibit the therapeutic effect of rapamycin.
Figure 7.
Thiol antioxidants and over-expression of SOD can enhance the phonotype in Drosophila models of polyglutamine disease. (A) High-dose NAC exacerbates mutant huntingtin phenotypes. Flies expressing mutant Q120 huntingtin exon-1 in the eye show an increase in neurodegeneration when treated with high dosage of NAC (5–10 mg/ml), while no such phenomena is seen in Q23 control flies. (B) The rescue in Q120 flies seen with rapamycin is abolished when rapamycin is combined with NAC or cystamine. Flies expressing mutant huntingtin exon-1 in eyes showed a significant decrease in neurodegeneration when treated with 100 µm cystamine or 2 µm rapamycin. Neurodegeneration was reversed to control level when the Q120 flies were treated with 500 mg/ml NAC or 100 µm cystamine combined with 2 µm rapamycin (*P < 0.05, **P < 0.01, NS, non-significant). (C) Over-expression of SOD can exacerbate neurodegeneration. Male flies expressing a naked polyglutamine tract (Q48) in the eye show a dramatic increase in neurodegeneration when crossed with flies over-expressing SOD1 or SOD2, while over-expression of SOD1 (Sod.A) or SOD2 has no such effect in control flies.
In order to more specifically examine the role of oxidative stress, we used a second fly model of neurodegeneration which expresses a naked expanded (Q48) polyglutamine stretch which results in a clearly visible eye phenotype. We crossed these flies with flies over-expressing cytosolic and mitochondrial isoforms of SOD, using w118 isogenic males as controls (30–33). We found that over-expression of either SOD1 or SOD2 had no obvious effect on the eye in control flies, but dramatically enhanced the neurodegenerative phenotype seen in Q48 flies (Fig. 7C).
NAC and vitamin E can inhibit autophagic flux in zebrafish and increase aggregation of mutant huntingtin in zebrafish models of polyglutamine disease
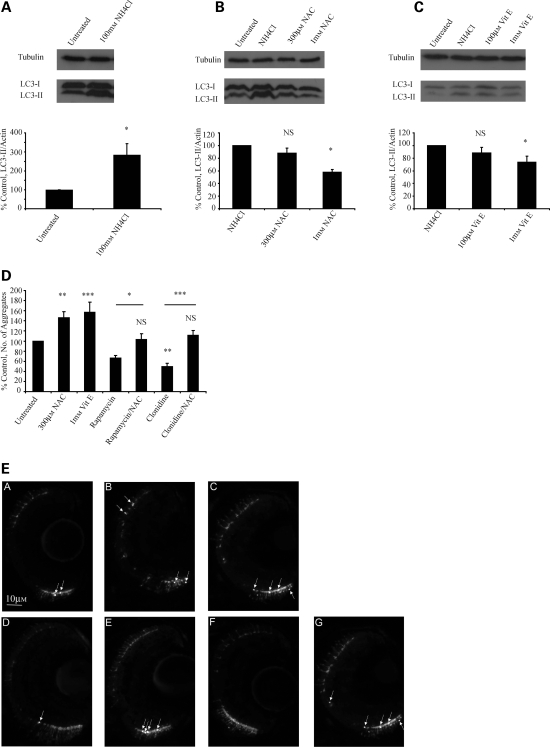
Studies in larval zebrafish have previously demonstrated that LC3-I to LC3-II conversion occurs from 32 h post-fertilization onwards and that changes in LC3-II levels could be measured in the presence of lysosomal protease inhibitors (34). In agreement with this, we observed a concentration-dependant increase in LC3-II levels in larval zebrafish treated with ammonium chloride, with 100 mm ammonium chloride providing reliable and dramatic increases in levels of LC3-II (Fig. 8A). Ammonium chloride blocks autophagosome/lysosome fusion (35) and this assay therefore allows us to investigate in vivo changes in autophagic flux (as opposed to changes in steady-state levels of LC3-II) in much the same way as treatment with bafilomycin in cell cultures.
Figure 8.
NAC and vitamin E can inhibit autophagic flux and increase aggregation in a zebrafish model of Huntington's disease. (A) Ammonium chloride causes significant increases in levels of LC3-II in zebrafish larvae, consistent with an ability to block autophagosome/lysosome fusion. (B and C) In zebrafish, co-treatment with either NAC or vitamin E significantly decreases the levels of LC3-II seen in the presence of ammonium chloride, consistent with their ability to decrease autophagic flux. The panels show western blots for LC3-II against tubulin loading controls. The graphs represent densitometric analysis of three independent experiments. (D and E) NAC and vitamin E increase aggregation, while the autophagy-inducing drugs rapamycin and clonidine decrease aggregation, of EGFP mutant huntingtin in the retina of transgenic zebrafish. (D) shows the average number of aggregates per retina in each condition relative to control. Representative images taken from the retina are shown in (E) with huntingtin aggregates indicated by arrows. (A, control condition; B, NAC treatment; C, vitamin E; D, rapamycin treatment; E, rapamycin and NAC; F, clonidine treatment; G, clonidine and NAC). Note that the rescue seen with rapamycin or clonidine is abolished by co-treatment with NAC (*P < 0.05, **P < 0.01, ***P < 0.001, NS, non-significant).
We found that co-treatment with NAC or vitamin E and ammonium chloride caused significant decreases in LC3-II levels compared with ammonium chloride alone, suggesting that these compounds are able to inhibit autophagic flux (Fig. 8B and C). We tested the pathological relevance of this effect by treating transgenic EGFP-HDQ71 zebrafish and counting huntingtin aggregates in the retina. We found that NAC and vitamin E significantly increased the number of aggregates of mutant huntingtin compared with control (Fig. 8D and E). As predicted, treatment with the autophagy-inducing agents rapamycin and clonidine significantly decreased huntingtin aggregation, but this effect was lost when treatment with these compounds was combined with NAC (Fig. 8D and E). To our knowledge, this is the first demonstration of changes in both autophagic flux and aggregation of autophagy substrates in a vertebrate model of polyglutamine disease.
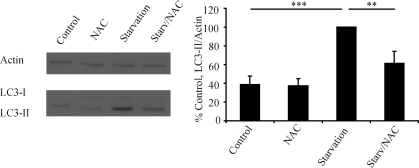
NAC inhibits starvation-induced autophagy in mice
In order to investigate the physiological relevance of our findings, we examined the effect of NAC on starvation-induced autophagy in mice. As expected, starvation strongly increased hepatic LC3-II levels, but this increase was significantly smaller in mice that had been pre-treated with NAC (Fig. 9). We chose the liver as a readout as it is the best-characterized organ for changes in autophagic activity and also has a significant physiological role in adaptation to starvation (36). While no significant changes in LC3-II levels were seen in brain (data not shown), this result was expected as the brain is an organ that is protected from decreases in nutrient availability and therefore would not be expected to show a strong up-regulation of autophagy in response to relatively short periods of starvation. Furthermore, since these experiments could not be performed in the presence of a blocker of autophagosomal/lysosomal fusion, the changes in autophagic flux at the level of synthesis are much more difficult to detect outside of organs which show a strong physiological response to brief periods of starvation.
Figure 9.
NAC inhibits the increase in hepatic LC3-II in starved mice. Mice underwent two periods of starvation of 24 h separated by 90 min of access to food. Following sacrifice, the liver was analysed by western blotting for levels of LC3-II. The graph represents the mean results from five mice per group with starvation (positive control, Starv) set to 100%. Error bars represent SEM (**P < 0.01, ***P < 0.001, NS, non significant).
DISCUSSION
The study presented here contains a number of novel findings. First, we show that drugs that up-regulate autophagy and have been proposed for use in human neurodegenerative disease do not cause detectable increases in ROS production. However, the up-regulation of autophagy by these drugs, and, indeed basal autophagy, are still inhibited by antioxidant drugs, suggesting that some level of ROS production or redox signalling is required for the normal regulation of autophagy. We show that antioxidants have effects on different classic autophagy regulatory pathways, consistent with their ability to inhibit autophagy. Vitamin E has an effect on the mTOR pathway, while thiol-containing antioxidants, such as NAC, decrease the phosphorylation of JNK and Bcl-2. Given the central role of superoxide in autophagy and our observation that menadione has opposite effects on the phosphorylation of JNK and Bcl-2 to NAC, it is possible that NAC exerts its effects by acting downstream of superoxide. However, as the rate constant of thiol groups for the reaction with superoxide is significantly less than that of the abundant SOD, it seems unlikely that superoxide is being directly scavenged. Alternatively, the increased cell thiol content may be blocking or reversing redox changes that occur downstream of superoxide production. We also show in vivo pathological and physiological relevance for these findings including, what is to our knowledge, the first ever vertebrate model of polyglutamine disease which allows quantification of changes in autophagic flux in conjunction with levels of pathological autophagic substrates.
We believe that the data presented here may have significant application to the design of treatment for human neurodegenerative disease. The idea that oxidative stress is pathological in the context of these diseases, and that decreasing oxidative stress is therefore a likely therapeutic strategy, has become a paradigm in the field. Our data suggest that low levels of some forms of ROS or redox signals are essential for autophagy. At least in diseases caused by aggregate-prone proteins which are autophagy substrates, the potential benefits of ROS scavenging may therefore be counterbalanced by an increased load of toxic protein. This is of significance, as supplements with putative antioxidant properties are commonly taken in the general population and even more frequently in those with neurodegenerative disease. In a previously published study from our group, we found that over two-thirds of HD patients were routinely taking supplements with putative antioxidant activity, a similar figure to other published series of PD patients (37,38). Furthermore, previously published series also suggest that many of these patients are unaware of any possible side effects of these supplements and do not tell their treating physician they are taking them (38). This could be a potentially significant confounder in any trial of autophagy-enhancing drugs and indeed it is conceivable that long-term inhibition of autophagy with these agents may even exacerbate the disease course. The only clinical trial of vitamin E in HD published to date showed non-significant worsening of all primary outcome measures, while trials of vitamin E in PD have similarly failed to show striking benefit or harm (23,24).
We do not suggest that treatments aimed at ameliorating oxidative stress may not have their place as therapy, but rather that better understanding of their interaction with other cellular processes may allow for more appropriately designed (and therefore more effective) treatment. For example, since the effects of antioxidants on autophagy appear dose-dependent, it is possible that dose may be crucial in balancing the effects of these drugs, and that higher doses may not necessarily mean greater efficacy. This may be of particular relevance as higher doses of some antioxidants have been proposed for use in HD (3). Additionally, the mechanisms of autophagy regulation by NAC and vitamin E appear distinct (as one might expect from such different antioxidant compounds), indicating that the interaction with autophagy may vary depending on the nature of the antioxidant used. Similarly, it is possible that antioxidant treatment at a time point in the disease when autophagic defence responses have become overwhelmed, or targeting specific cellular compartments, may be more efficacious. Understanding the interaction between various agents that may modify the progression of neurodegenerative diseases has significant current relevance, as clinical trials of both autophagy-enhancing drugs (39) and antioxidant therapies (40) have been performed and combination treatments may appear an attractive therapeutic strategy. Our data suggest that some combinations that appear attractive on the basis of current knowledge (such as rapamycin and cystamine) may not be beneficial.
The demonstration that NAC inhibits the up-regulation of autophagy in vivo may have clinical utility outside of neurodegeneration. NAC has a long safety record in the treatment of acetaminophen poisoning in humans where it is used intravenously at high doses, resulting in serum concentrations comparable to those used in cell and mouse models described here (41). Some malignant tumours up-regulate autophagy as a way of surviving chemotherapy/radiotherapy and to provide nutrient recycling in the avascular tumour core. Autophagy inhibitors may be potential cancer therapies in this context (42). NAC may be particularly attractive in this regard as our data suggest it has the unusual combination of properties of inhibiting both mTOR (which may inhibit tumour growth) and autophagy. Thus, while long-term inhibition of autophagy is likely to be deleterious, it may well be possible to use potent, reversible autophagy inhibitors safely for short periods. For these applications, NAC may be an attractive candidate therapy.
The apparently intimate relationship between ROS and autophagy is just one example of how our appreciation of ROS has moved beyond regarding them simply as a dangerous and unwanted by-product of metabolism. Enhanced understanding of the interaction between these cellular processes is likely to have application to the treatment of human diseases.
MATERIALS AND METHODS
All reagents were from Sigma-Aldrich unless otherwise specified.
Cell culture
COS-7 (African green monkey kidney cells) and HeLa (from human cervical carcinoma origin) were cultured in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% fetal bovine serum, 100 U/ml streptomycin/penicillin and 2 mm l-glutamine. They were maintained at 37°C and 5% CO2. Cells were seeded in six well plates (with cover slips for counting experiments) at densities to allow 60–80% confluence by the time lysates were collected for western blotting. Counting experiments were seeded at higher density to allow 80% confluence prior to transfection. Stable inducible A53T alpha synuclein and HD exon 1 Q74 PC12 cells were cultured in DMEM supplemented with 10% horse serum, 5% fetal bovine serum, 2 mm l-glutamine, 100 U/ml penicillin/streptomycin, 50 µg/ml G418 and 70 µg/ml Hygromycin B. They were kept at 37°C and 10% CO2 and expression was induced by treating them with 1 µg/ml of doxycycline for 48 h (alpha synuclein) or 8 h (HD exon 1), before washing to remove doxycycline and therefore turn off inducible transgene expression. ReNcell CX cells, an immortalized human neuronal progenitor cell line with the ability to readily differentiate into cortical neurons, were cultured according to the manufacturer's instructions (Chemicon, Millpore, MA, USA). Cells were grown in ReNcell NSC maintenance medium supplemented with 20 ng/ml bFGF and 20 ng/ml EGF. Cells were seeded on laminin coated plates and differentiation was initiated with media containing no growth factors for 1 week prior to starting the experiment. Mouse primary cortical neuronal cultures were prepared as described previously (43). Briefly, cortices were dissected from 15- to 16-day-old C57BL/6 mouse embryos and mechanically dissociated in PBS (Ca2+- and Mg2+-free) supplemented with glucose (33 mm). Neurons were plated into multi-well plates coated with 20 µg/ml poly-D-lysine and were maintained in DMEM, supplemented with B-27 (Gibco), 2 mm glutamine, 100 µg/ml streptomycin and 60 µg/ml penicillin, at 37°C and 5% CO2. Cultures were used after 6–7 days in vitro (DIV) when the majority of cells were neuronal and there were no detectable glial elements.
Reagents
Autophagy modifiers were used at the following concentrations. Rapamycin (LC Laboratories) 200 nm, bafilomycin A1 400 nm, trehalose, 100 mm, Menadione 100 µm. Rapamycin was dissolved in DMSO and controls for rapamycin were treated with an equivalent volume of DMSO. Menadione was dissolved in ethanol and menadione controls treated with an equivalent volume of ethanol.
Measurement of ROS
HeLa cells were seeded in a 24-well plate and left overnight. They were incubated with 30 µm DCFDA or Carboxy-H2DCFDA for 10 min, 5 µm MitoSOX™ red for 10 min or 15 µm dihydrorhodamine 123 (all Molecular probes, Invitrogen) for 30 min, before being washed twice and left in rich media, or media containing the relevant treatment. One millimolar hydrogen peroxide (H2O2) (Fisher Scientific) and 100 µM menadione and starvation in HBSS were used as positive controls. Fluorescence was then measured using a CytoFluor Series 4000 multi well plate reader (PerSeptive Biosystems). The plates were returned to an incubator between readings. DCFDA and dihydrorhodamine were measured at excitation 485, emission 530 and MitoSOX™ red at 530/620.
JNK phosphorylation assay
JNK phosphorylation was measured using a colorimetric fast-activated cell-based enzyme-linked immunosorbant assay (FACE ELISA) kit (Active Motif). COS-7 cells were seeded in a 96-well plate and left overnight before being treated for 24 h. They were then fixed, blocked and probed with antibody to phosphorylated or total JNK as per manufacturer's instructions. Absorbance was measured using an OPTImax microplate reader at 450 nm.
Western blot analysis
Prior to analysis, cells were washed with PBS. They were then lysed in radioimmunoprecipitation assay buffer [150 mm NaCl, 1% NP40, 0.5% NaDoC, 0.1% SDS, 50 mm Tris, protease inhibitors (Roche diagnostics)]. A Bio-rad protein assay was then carried out to allow equal loading. The lysates were subjected to a sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS–PAGE) (12%) and proteins were transferred to PVDF. Blots were probed with horseradish peroxidase-conjugated anti-mouse or anti-rabbit IgG at 1:2000. Bands were visualized using ECL™ detection reagent (Amersham). Densitometry was carried out using Image J software and statistically analysed using factorial analysis of variance (ANOVA) using STATVIEW v4.53 (Abacus Concepts) where the control value is set to 100%. The densitometry represents the means of at least three experiments performed in duplicate or triplicate.
Quantification of aggregation
Aggregates were visualized and counted by fluorescence microscopy as previously described (17). Cells were washed once with PBS before being fixed for 20 min in 4% paraformaldehyde in PBS. The cells were washed again in PBS before being mounted in an antifade reagent containing 3 µg/ml 4′,6-diamidino-2-phenylindole (DAPI) to facilitate nuclear staining. Five hundred EGFP positive cells per cover slip were counted by an observer blind to the identity of the slides. The results of the counting experiment were analysed by unconditional logistic regression using SPSS 6.1 (SPSS, Chicago, IL, USA). Images were obtained using a Nikon Eclipse E600 microscope attached to a Nikon DXM1200 digital camera using Nikon ACT-1 acquisition software (Nikon Inc.).
Automated cell counting
HeLa cells expressing RFP/GFP tagged LC3 were seeded on cover slips and treated overnight in the appropriate media. They were then washed in PBS, fixed with 4% paraformaldehyde and mounted in Prolong Gold antifade reagent with DAPI (Invitrogen). The number of red dots and green dots per cell was counted using a Cellomics ArrayScan VTI high content screening reader using the Spot detector V3 Cellomics bioapplication. One thousand cells were counted per cover slip and each experiment was done in triplicate at least four times.
Drosophila experiments
Pseudopupil analysis was performed as previously described (44) crossing virgins of the genotype y w; P,2.4 (Q120) with w1118 isogenic males. The progeny of the above cross was reared on instant fly food containing 500 mg/ml NAC, 100 µm cystamine or 2 µm rapamycin. Combination treatments were done by adding 500 mg/ml NAC or 100 µm cystamine to 2 µm rapamycin. Control flies grown in absence of drug were fed with instant fly food with 0.02% DMSO. Flies were treated with drugs during both the larval and adult stage. To assess the effect of different treatments on Q120 flies, t-test analysis was performed on four independent experiments, each involving the counting of 15 ommatidia was scored from each of 10 individuals, 3 days post-eclosion. Q23 control flies (Q23, P{GMR-HD.Q23) were used to compare the effect of increasing doses of NAC on eye phenotype. To study the effect of SOD on flies expressing an expanded polyglutamine stretch (P{UAS-Q48.myc/flag}31(30), virgins of the genotype w; GMR-GAL4; UAS.Q48myc/flag (Q48) were crossed with males transgenic for SOD, either P{UAS-Sod2.M}UM83(31), or P{UAS-Sod.A}B36(32) or with w1118 isogenic males as control. Overexpression of SOD in the eye does not affect eye phenotype. Male flies expressing either SOD1 (UAS-Sod.A, w; GMR-Gal4/UAS-Sod.A) or SOD2 (UAS-Sod2, w; GMR-Gal4/UAS-Sod2.M) were comparable to control flies (GMR-Gal4, P{GAL4-ninaE.GMR}12) to investigate the effect of SOD over-expression with no polyQ construct. Fly crosses and experiments were performed at 25°C. All crosses for individual experiments were performed at the same time and under the same conditions.
Zebrafish experiments
Maintenance of stocks and collection of embryos
All zebrafish husbandry and experiments were performed in accordance with UK legislation under a license granted by the Home Office and with local ethical approval. Zebrafish were reared under standard conditions on a 14 h light:10 h dark cycle. Embryos were collected from natural spawnings, staged according to established criteria (45) and reared in embryo medium (5 mm NaCl, 0.17 mm KCl, 0.33 mm CaCl2, 0.33 mm Mg2SO4). Experiments measuring endogenous LC3-II levels were performed on wild-type zebrafish of the AB strain. Aggregate counting was performed using the heterozygous larvae from the transgenic rhodopsin::EGFP-HDQ71 zebrafish line (46) (hereafter referred to as transgenic HD zebrafish). Compound exposure experiments were performed in the dark at 28.5°C.
Determination of the maximum tolerated concentration of compounds in larval zebrafish
Compound exposure experiments were performed on wild-type larvae from 2 days post-fertilization (d.p.f.) for 24 h. Concentration response assays were performed over half-log intervals, from 10 µm to 1 mm for ammonium chloride, 1 µm to 1 mm for clonidine, 1 µm to 10 mm for NAC, 1–100 µm for rapamycin, 1 µm to 10 mm for DL-alpha tocopherol, to determine the maximum non-toxic concentration for subsequent autophagy assays (n = 10 larvae per concentration).
Measuring endogenous LC3-II in larval zebrafish
LC3-II assays were performed at the following concentrations: ammonium chloride at 100 mm, NAC at 300 µm and 1 mm, DL-alpha tocopherol at 100 µm and 1 mm for 24 h. Wild-type larvae (n = 30 per treatment group) at 2 d.p.f. were exposed to NAC, DL-alpha tocopherol or embryo medium (untreated control) for 20 h prior to compound replenishment and the addition of ammonium chloride. Larvae were incubated in compounds for a further 4 h then transferred to chilled tubes, homogenized in lysis buffer then processed for western blotting as described above.
Aggregate analysis in the transgenic HD zebrafish
Embryos from out-crossed transgenic HD zebrafish were raised in 0.2 mm 1-phenyl-2-thiourea (PTU) from 1 to 3 d.p.f. to inhibit pigment formation, screened for transgene expression using EGFP fluorescence then washed twice in embryo medium to remove PTU. From 3 to 7 d.p.f., transgenic HD zebrafish larvae were dark-reared in embryo medium alone or embryo medium containing either 300 µm NAC, 1 mm DL-alpha tocopherol, 30 µm rapamycin, 30 µm clonidine (compounds administered alone or in combination). Embryo medium and compounds were replenished daily. At 7 d.p.f., larvae were anesthetized by immersion in 0.2 mg/ml 3-amino benzoic acid ethylester (MS222) then fixed using 4% PFA in PBS at 4°C. Larvae were washed briefly in PBS, allowed to equilibrate in 30% sucrose in PBS then embedded in OCT medium (Tissue-Tek) and frozen on dry ice for subsequent cryosectioning. Sections were cut at 10 µm thickness using a Leica CM3050 cryostat and mounted in 80% glycerol in PBS. The total number of GFP-positive aggregates was counted over 100 µm of the central retina, either side of the optic nerve using a Zeiss Axiophot2 microscope and mean values were calculated using four fish (eight eyes) for each treatment group. Data were compared using ANOVA Representative images were taken using a Sony CCD digital camera.
Mouse experiments
The mice experiments all complied with Home Office project and personal animal licenses. Twenty male C57BL/6 mice were used aged 35–42 days at the start of the study. The mice were divided into four groups, each containing five mice. After 3 weeks of acclimatization, two groups received no treatment while the other two groups were treated with NAC at 1% in their drinking water for 2 weeks. Following this, one of the control groups of mice and one of the groups treated with NAC were starved by for 22.5 h before allowing them 90 min unrestricted access to food. The following day, the same two groups were again starved for 22.5 h before being sacrificed. The liver was dissected and homogenized in lysis buffer [0.25 m Tris, 2.5% Triton, 150 mm NaCl and protease inhibitor cocktail (Roche Scientific)] before being analysed by western blot.
Statistical analyses
P-values for densitometric analysis using multiple conditions were performed using a factorial ANOVA using STATVIEW v4.53 (Abacus Concepts). For densitometry involving two conditions, a two-tailed paired student's t-test was performed on the untransformed data. P-values for counting aggregates were calculated using odds ratio using SPSS Version 6.1 software (SPSS). Experiments were performed at least three times in triplicate unless otherwise indicated.
FUNDING
This work was supported by Action Medical Research and the Rosetrees Trust (Research Training Fellowship to B.U.), the Sackler Foundation, the NIHR Biomedical Research Centre at Addenbrooke's Hospital and MRC Skills Gap Award to A.F., a Medical Research Council Programme Grant (D.C.R. and C.J.O.K.), Wellcome Trust Senior Fellowship (D.C.R.) and an NIHR Biomedical Research Grant at Addenbrooke's Hospital. Funding to pay the Open Access Charge was provided by the Wellcome Trust.
ACKNOWLEDGEMENTS
We thank J. Lawrence Marsh (University of California, Irvine, CA, USA), G. R. Jackson (UCLA School of Medicine, Los Angeles, CA, USA) and Bloomington Drosophila Stock Center for Drosophila stocks and Professor Chris Miller (Institute of Psychiatry, King's College London) for SOD1 constructs. We thank Adrian McNabb, Gay Chalklin, Cheryl Kennerson and Tomasz Dyl for technical support. We would also like to thank Miss Pauline Lansiaux for her help with fly scoring and Drs J. Davies, F. Menzies and B. Ravikumar for helpful comments on the manuscript.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Weintraub D., Comella C.L., Horn S. Parkinson's disease—Part 1: pathophysiology, symptoms, burden, diagnosis, and assessment. Am. J. Manag. Care. 2008;14:S40–S48. [PubMed] [Google Scholar]
- 2.Phillips W., Shannon K.M., Barker R.A. The current clinical management of Huntington's disease. Mov. Disord. 2008;23:1491–1504. doi: 10.1002/mds.21971. doi:10.1002/mds.21971. [DOI] [PubMed] [Google Scholar]
- 3.Stack E.C., Matson W.R., Ferrante R.J. Evidence of oxidant damage in Huntington's disease: translational strategies using antioxidants. Ann. N. Y. Acad. Sci. 2008;1147:79–92. doi: 10.1196/annals.1427.008. [DOI] [PubMed] [Google Scholar]
- 4.Kamat C.D., Gadal S., Mhatre M., Williamson K.S., Pye Q.N., Hensley K. Antioxidants in central nervous system diseases: preclinical promise and translational challenges. J. Alzheimers. Dis. 2008;15:473–493. doi: 10.3233/jad-2008-15314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger Z., Ravikumar B., Menzies F.M., Oroz L.G., Underwood B.R., Pangalos M.N., Schmitt I., Wullner U., Evert B.O., O'Kane C.J., et al. Rapamycin alleviates toxicity of different aggregate-prone proteins. Hum. Mol. Genet. 2006;15:433–442. doi: 10.1093/hmg/ddi458. doi:10.1093/hmg/ddi458. [DOI] [PubMed] [Google Scholar]
- 6.Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. doi:10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 7.Ravikumar B., Futter M., Jahreiss L., Korolchuk V.I., Lichtenberg M., Luo S., Massey D.C., Menzies F.M., Narayanan U., Renna M., et al. Mammalian macroautophagy at a glance. J. Cell Sci. 2009;122:1707–1711. doi: 10.1242/jcs.031773. doi:10.1242/jcs.031773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sarkar S., Davies J.E., Huang Z., Tunnacliffe A., Rubinsztein D.C. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J. Biol. Chem. 2007;282:5641–5652. doi: 10.1074/jbc.M609532200. doi:10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- 9.Pattingre S., Tassa A., Qu X., Garuti R., Liang X.H., Mizushima N., Packer M., Schneider M.D., Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. doi:10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 10.Ravikumar B., Vacher C., Berger Z., Davies J.E., Luo S., Oroz L.G., Scaravilli F., Easton D.F., Duden R., O'Kane C.J., et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet. 2004;36:585–595. doi: 10.1038/ng1362. doi:10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 11.Menzies F.M., Huebener J., Renna M., Bonin M., Riess O., Rubinsztein D.C. Autophagy induction reduces mutant ataxin-3 levels and toxicity in a mouse model of spinocerebellar ataxia type 3. Brain. 2009;133:93–104. doi: 10.1093/brain/awp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scherz-Shouval R., Shvets E., Fass E., Shorer H., Gil L., Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26:1749–1760. doi: 10.1038/sj.emboj.7601623. doi:10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao L., Xu J., Lin Y., Zhao X., Liu X., Chi Z. Autophagy is upregulated in rats with status epilepticus and partly inhibited by Vitamin E. Biochem. Biophys. Res. Commun. 2009;379:949–953. doi: 10.1016/j.bbrc.2008.12.178. doi:10.1016/j.bbrc.2008.12.178. [DOI] [PubMed] [Google Scholar]
- 14.Gao M., Yeh P.Y., Lu Y.S., Hsu C.H., Chen K.F., Lee W.C., Feng W.C., Chen C.S., Kuo M.L., Cheng A.L. OSU-03012, a novel celecoxib derivative, induces reactive oxygen species-related autophagy in hepatocellular carcinoma. Cancer Res. 2008;68:9348–9357. doi: 10.1158/0008-5472.CAN-08-1642. doi:10.1158/0008-5472.CAN-08-1642. [DOI] [PubMed] [Google Scholar]
- 15.Kongsuphol P., Mukda S., Nopparat C., Villarroel A., Govitrapong P. Melatonin attenuates methamphetamine-induced deactivation of the mammalian target of rapamycin signaling to induce autophagy in SK-N-SH cells. J. Pineal. Res. 2009;46:199–206. doi: 10.1111/j.1600-079X.2008.00648.x. [DOI] [PubMed] [Google Scholar]
- 16.Herdeiro R.S., Pereira M.D., Panek A.D., Eleutherio E.C. Trehalose protects Saccharomyces cerevisiae from lipid peroxidation during oxidative stress. Biochim. Biophys. Acta. 2006;1760:340–346. doi: 10.1016/j.bbagen.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 17.Ravikumar B., Duden R., Rubinsztein D.C. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum. Mol. Genet. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. doi:10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- 18.Dubinsky R., Gray C. CYTE-I-HD: phase I dose finding and tolerability study of cysteamine (Cystagon) in Huntington's disease. Mov. Disord. 2006;21:530–533. doi: 10.1002/mds.20756. doi:10.1002/mds.20756. [DOI] [PubMed] [Google Scholar]
- 19.Fox J.H., Barber D.S., Singh B., Zucker B., Swindell M.K., Norflus F., Buzescu R., Chopra R., Ferrante R.J., Kazantsev A., et al. Cystamine increases l-cysteine levels in Huntington's disease transgenic mouse brain and in a PC12 model of polyglutamine aggregation. J. Neurochem. 2004;91:413–422. doi: 10.1111/j.1471-4159.2004.02726.x. doi:10.1111/j.1471-4159.2004.02726.x. [DOI] [PubMed] [Google Scholar]
- 20.Klionsky D.J., Elazar Z., Seglen P.O., Rubinsztein D.C. Does bafilomycin A1 block the fusion of autophagosomes with lysosomes? Autophagy. 2008;4:849–950. doi: 10.4161/auto.6845. [DOI] [PubMed] [Google Scholar]
- 21.Rubinsztein D.C., Cuervo A.M., Ravikumar B., Sarkar S., Korolchuk V., Kaushik S., Klionsky D.J. In search of an ‘autophagomometer. Autophagy. 2009;5:585–589. doi: 10.4161/auto.5.5.8823. doi:10.4161/auto.5.5.8823. [DOI] [PubMed] [Google Scholar]
- 22.Webb J.L., Ravikumar B., Atkins J., Skepper J.N., Rubinsztein D.C. Alpha-Synuclein is degraded by both autophagy and the proteasome. J. Biol. Chem. 2003;278:25009–25013. doi: 10.1074/jbc.M300227200. doi:10.1074/jbc.M300227200. [DOI] [PubMed] [Google Scholar]
- 23.Shoulson I. DATATOP: a decade of neuroprotective inquiry. Parkinson Study Group. Deprenyl and Tocopherol Antioxidative Therapy of Parkinsonism. Ann. Neurol. 1998;44:S160–S166. [PubMed] [Google Scholar]
- 24.Peyser C.E., Folstein M., Chase G.A., Starkstein S., Brandt J., Cockrell J.R., Bylsma F., Coyle J.T., McHugh P.R., Folstein S.E. Trial of d-alpha-tocopherol in Huntington's disease. Am. J. Psychiatry. 1995;152:1771–1775. doi: 10.1176/ajp.152.12.1771. [DOI] [PubMed] [Google Scholar]
- 25.Chen Y., Azad M.B., Gibson S.B. Superoxide is the major reactive oxygen species regulating autophagy. Cell Death Differ. 2009;16:1040–1052. doi: 10.1038/cdd.2009.49. doi:10.1038/cdd.2009.49. [DOI] [PubMed] [Google Scholar]
- 26.Wei Y., Pattingre S., Sinha S., Bassik M., Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol. Cell. 2008;30:678–688. doi: 10.1016/j.molcel.2008.06.001. doi:10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y., McMillan-Ward E., Kong J., Israels S.J., Gibson S.B. Oxidative stress induces autophagic cell death independent of apoptosis in transformed and cancer cells. Cell Death Differ. 2008;15:171–182. doi: 10.1038/sj.cdd.4402233. doi:10.1038/sj.cdd.4402233. [DOI] [PubMed] [Google Scholar]
- 28.Jackson G.R., Salecker I., Dong X., Yao X., Arnheim N., Faber P.W., MacDonald M.E., Zipursky S.L. Polyglutamine-expanded human huntingtin transgenes induce degeneration of Drosophila photoreceptor neurons. Neuron. 1998;21:633–642. doi: 10.1016/s0896-6273(00)80573-5. doi:10.1016/S0896-6273(00)80573-5. [DOI] [PubMed] [Google Scholar]
- 29.Wang T., Lao U., Edgar B.A. TOR-mediated autophagy regulates cell death in Drosophila neurodegenerative disease. J. Cell Biol. 2009;186:703–711. doi: 10.1083/jcb.200904090. doi:10.1083/jcb.200904090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marsh J.L., Walker H., Theisen H., Zhu Y.Z., Fielder T., Purcell J., Thompson L.M. Expanded polyglutamine peptides alone are intrinsically cytotoxic and cause neurodegeneration in Drosophila. Hum. Mol. Genet. 2000;9:13–25. doi: 10.1093/hmg/9.1.13. doi:10.1093/hmg/9.1.13. [DOI] [PubMed] [Google Scholar]
- 31.Missirlis F., Hu J., Kirby K., Hilliker A.J., Rouault T.A., Phillips J.P. Compartment-specific protection of iron-sulfur proteins by superoxide dismutase. J. Biol. Chem. 2003;278:47365–47369. doi: 10.1074/jbc.M307700200. doi:10.1074/jbc.M307700200. [DOI] [PubMed] [Google Scholar]
- 32.Anderson P.R., Kirby K., Hilliker A.J., Phillips J.P. RNAi-mediated suppression of the mitochondrial iron chaperone, frataxin, in Drosophila. Hum. Mol. Genet. 2005;14:3397–3405. doi: 10.1093/hmg/ddi367. doi:10.1093/hmg/ddi367. [DOI] [PubMed] [Google Scholar]
- 33.Ryder E., Blows F., Ashburner M., Bautista-Llacer R., Coulson D., Drummond J., Webster J., Gubb D., Gunton N., Johnson G., et al. The DrosDel collection: a set of P-element insertions for generating custom chromosomal aberrations in Drosophila melanogaster. Genetics. 2004;167:797–813. doi: 10.1534/genetics.104.026658. doi:10.1534/genetics.104.026658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He C., Bartholomew C.R., Zhou W., Klionsky D.J. Assaying autophagic activity in transgenic GFP-Lc3 and GFP-Gabarap zebrafish embryos. Autophagy. 2009;5:520–526. doi: 10.4161/auto.5.4.7768. doi:10.4161/auto.5.4.7768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hart P.D., Young M.R. Ammonium chloride, an inhibitor of phagosome–lysosome fusion in macrophages, concurrently induces phagosome–endosome fusion, and opens a novel pathway: studies of a pathogenic mycobacterium and a nonpathogenic yeast. J. Exp. Med. 1991;174:881–889. doi: 10.1084/jem.174.4.881. doi:10.1084/jem.174.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinet W., De Meyer G.R., Andries L., Herman A.G., Kockx M.M. In situ detection of starvation-induced autophagy. J. Histochem. Cytochem. 2006;54:85–96. doi: 10.1369/jhc.5A6743.2005. doi:10.1369/jhc.5A6743.2005. [DOI] [PubMed] [Google Scholar]
- 37.Underwood B.R., Broadhurst D., Dunn W.B., Ellis D.I., Michell A.W., Vacher C., Mosedale D.E., Kell D.B., Barker R.A., Grainger D.J., et al. Huntington disease patients and transgenic mice have similar pro-catabolic serum metabolite profiles. Brain. 2006;129:877–886. doi: 10.1093/brain/awl027. doi:10.1093/brain/awl027. [DOI] [PubMed] [Google Scholar]
- 38.Wolfrath S.C., Borenstein A.R., Schwartz S., Hauser R.A., Sullivan K.L., Zesiewicz T.A. Use of nutritional supplements in Parkinson's disease patients. Mov. Disord. 2006;21:1098–1101. doi: 10.1002/mds.20902. doi:10.1002/mds.20902. [DOI] [PubMed] [Google Scholar]
- 39.Fornai F., Longone P., Cafaro L., Kastsiuchenka O., Ferrucci M., Manca M.L., Lazzeri G., Spalloni A., Bellio N., Lenzi P., et al. Lithium delays progression of amyotrophic lateral sclerosis. Proc. Natl Acad. Sci. USA. 2008;105:2052–2057. doi: 10.1073/pnas.0708022105. doi:10.1073/pnas.0708022105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Graf M., Ecker D., Horowski R., Kramer B., Riederer P., Gerlach M., Hager C., Ludolph A.C., Becker G., Osterhage J., et al. High dose vitamin E therapy in amyotrophic lateral sclerosis as add-on therapy to riluzole: results of a placebo-controlled double-blind study. J. Neural. Transm. 2005;112:649–660. doi: 10.1007/s00702-004-0220-1. doi:10.1007/s00702-004-0220-1. [DOI] [PubMed] [Google Scholar]
- 41.Millea P.J. N-acetylcysteine: multiple clinical applications. Am. Fam. Physician. 2009;80:265–269. [PubMed] [Google Scholar]
- 42.Ito H., Daido S., Kanzawa T., Kondo S., Kondo Y. Radiation-induced autophagy is associated with LC3 and its inhibition sensitizes malignant glioma cells. Int. J. Oncol. 2005;26:1401–1410. [PubMed] [Google Scholar]
- 43.Crossthwaite A.J., Hasan S., Williams R.J. Hydrogen peroxide-mediated phosphorylation of ERK1/2, Akt/PKB and JNK in cortical neurones: dependence on Ca(2+) and PI3-kinase. J. Neurochem. 2002;80:24–35. doi: 10.1046/j.0022-3042.2001.00637.x. doi:10.1046/j.0022-3042.2001.00637.x. [DOI] [PubMed] [Google Scholar]
- 44.Franceschini N., Kirschfeld K. In vivo optical study of photoreceptor elements in the compound eye of Drosophila. Kybernetik. 1971;8:1–13. doi: 10.1007/BF00270828. doi:10.1007/BF00270828. [DOI] [PubMed] [Google Scholar]
- 45.Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., Schilling T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 46.Williams A., Sarkar S., Cuddon P., Ttofi E.K., Saiki S., Siddiqi F.H., Jahreiss L., Fleming A., Pask D., Goldsmith P., et al. Novel targets for Huntington's disease in an mTOR-independent autophagy pathway. Nat. Chem. Biol. 2008;4:295–305. doi: 10.1038/nchembio.79. doi:10.1038/nchembio.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wyttenbach A., Swartz J., Kita H., Thykjaer T., Carmichael J., Bradley J., Brown R., Maxwell M., Schapira A., Orntoft T.F., et al. Polyglutamine expansions cause decreased CRE-mediated transcription and early gene expression changes prior to cell death in an inducible cell model of Huntington's disease. Hum. Mol. Genet. 2001;10:1829–1845. doi: 10.1093/hmg/10.17.1829. doi:10.1093/hmg/10.17.1829. [DOI] [PubMed] [Google Scholar]