Abstract
Mitochondria must uptake some phospholipids from the endoplasmic reticulum (ER) for the biogenesis of their membranes. They convert one of these lipids, phosphatidylserine, to phosphatidylethanolamine, which can be re-exported via the ER to all other cellular membranes. The mechanisms underlying these exchanges between ER and mitochondria are poorly understood. Recently, a complex termed ER–mitochondria encounter structure (ERMES) was shown to be necessary for phospholipid exchange in budding yeast. However, it is unclear whether this complex is merely an inter-organelle tether or also the transporter. ERMES consists of four proteins: Mdm10, Mdm34 (Mmm2), Mdm12 and Mmm1, three of which contain the uncharacterized SMP domain common to a number of eukaryotic membrane-associated proteins. Here, we show that the SMP domain belongs to the TULIP superfamily of lipid/hydrophobic ligand-binding domains comprising members of known structure. This relationship suggests that the SMP domains of the ERMES complex mediate lipid exchange between ER and mitochondria.
Contact: andrei.lupas@tuebingen.mpg.de
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Mitochondria are organelles of endosymbiotic origin, found in virtually all eukaryotic organisms. They are the main generators of adenosine triphosphate (ATP)—the energy currency of the cell. Additionally, they participate in a series of other important processes, including apoptosis, amino acid and lipid metabolism, iron–sulphur cluster assembly and the regulation of calcium levels within the cell (Lill and Kispal, 2000; McBride et al., 2006). However, only a small fraction of the biopolymers required to carry out these functions is synthesized in the mitochondria, the rest must be imported from the outside. For example, they only produce some of the phospholipids that make up their membranes, whereas the remainder originates from the endoplasmic reticulum (ER). Interestingly, mitochondria not only import phospholipids but also export a particular one, phosphatidylethanolamine (PtdEtn), to the ER, where it is methylated to form the phospholipid phosphatidylcholine (Voelker, 2003). Mitochondria synthesize PtdEtn by decarboxylating phosphatidylserine, a phospholipid imported from the ER. The mechanisms responsible for the influx and efflux of phospholipids are unclear.
Unlike most organelles, mitochondria do not exchange phospholipids via vesicular transport. Previous studies have suggested that this exchange takes place via ER–mitochondria associations (Achleitner et al., 1999; Voelker, 2003). More recently, Kornmann et al. (2009) identified a complex, the ER–mitochondria encounter structure (ERMES), that acts as a molecular tether between ER and mitochondria in Saccharomyces cerevisiae and is required for efficient inter-organelle phospholipid exchange. However, it remained unclear whether ERMES merely tethers these organelles together, thereby aligning proteins that carry out the actual transport, or also recruits and transfers phospholipids itself.
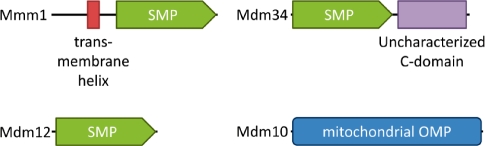
ERMES comprises four proteins (Fig. 1): the mitochondrial outer membrane protein Mdm10, the putative outer membrane protein Mdm34 (Mmm2), the ER-resident Mmm1 and the cytosolic Mdm12 (Kornmann et al., 2009). These proteins have also been implicated in other mitochondrial functions, including morphology maintenance (Okamoto and Shaw, 2005) and protein import (Meisinger et al., 2004). Two of the ERMES components, Mmm1 and Mdm12, were reported to contain the uncharacterized SMP domain (synaptotagmin-like, mitochondrial and lipid-binding proteins), which is also present in a number of other eukaryotic membrane-associated proteins (Lee and Hong, 2006). SMP domain-containing proteins have been classified into four broad groups: C2 domain synaptotagmin-like, PH domain-containing HT-008, PDZK8 and mitochondrial protein families (Lee and Hong, 2006). The functions of these proteins are poorly understood.
Fig. 1.
Domain organization of the four ERMES proteins. All ERMES proteins, except the mitochondrial outer membrane protein (OMP) Mdm10, contain an SMP domain. The SMP domain in Mdm34 was discovered in this study.
In this study, we show that the SMP domain belongs to a superfamily of lipid/hydrophobic ligand-binding domains of known structure, which we call TULIP for tubular lipid-binding proteins, and propose a role for it in cellular phospholipid traffic.
2 METHODS
All sequence similarity searches were carried out in the MPI bioinformatics toolkit (http://toolkit.tuebingen.mpg.de; Biegert et al., 2006) using HHpred (Söding et al., 2005) and HHsenser (Söding et al., 2006) with default settings. HHpred searches were performed against a database comprising PDB70 (protein databank structures, as available on the 15th of April 2010, clustered at 70% sequence identity) and genomes of phylogenetically diverse organisms (Arabidopsis thaliana, Caenorhabditis elegans, Drosophila melanogaster, Homo sapiens and S.cerevisiae). Representatives of the four SMP domain-containing groups from the aforementioned organisms were chosen as seeds for the searches in Figures 2 and 3, based on their presence in the core of their respective clusters in the sequence cluster map (Fig. 5).
Fig. 2.
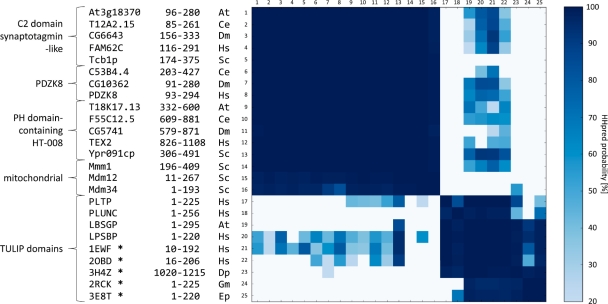
Pairwise HMM comparison of SMP and TULIP domains. Representatives of the four SMP domain-containing groups and of TULIP domains were chosen from Arabidopsis thaliana (At), Caenorhabditis elegans (Ce), Drosophila melanogaster (Dm), Dermatophagoides pteronyssinus (Dp), Epiphyas postvittana (Ep), Galleria mellonella (Gm), Homo sapiens (Hs) and Saccharomyces cerevisiae (Sc). Group and protein names of these representatives, along with domain boundaries and names of source species are indicated (from left to right). HHpred was used to perform pairwise HMM comparisons between them. Cell color indicates the HHpred probability of the match as depicted in the scale on the right; probabilities <20% are shown as white cells. Proteins with known structures are marked with an asterisk. The abbreviations of the hydrophobic ligand binders are explained in the text.
Fig. 3.
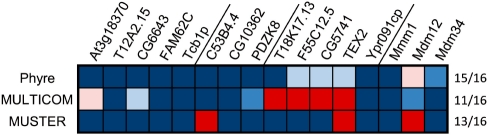
Fold predictions for SMP domains. The highest scoring PDB matches for the 16 representative SMP sequences in Figure 2 were collected with three of the top-scoring prediction servers in CASP8. Top matches to TULIP domains are shown in blue and to any other structure in red. The color saturation is scaled linearly between the maximum and minimum scores returned by the respective method. The value ranges corresponding to pale (low confidence), medium and dark (high confidence) saturation are: Phyre-estimated precision 0–33, 34–66 and 67–100, MULTICOM e-value 7.4–5, 5.1–2.6, 2.5–0, MUSTER Z-score 0–1.8, 1.9–3.5, 3.6–5.3. The number of matches to TULIP domains against the total is shown in the right-hand column.
Fig. 5.
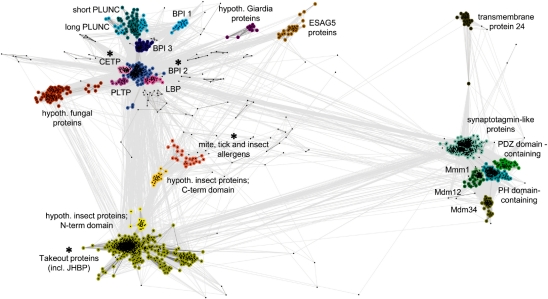
Cluster map of the TULIP domain superfamily. We searched for relatives of SMP domains, Takeout proteins, dust mite allergens and N-terminal domains of BPI-like proteins in the non-redundant database using HHsenser and clustered them in CLANS based on their all-against-all pairwise similarities as measured by BLAST P-values. Dots represent sequences. Sequences within one group are indicated by the same color; sequences that could not be assigned to a group are not colored. Line coloring reflects BLAST P-values; the darker a line, the lower the P-value. Protein families containing members with known structure are indicated with an asterisk. The lipid-binding proteins cluster (LBP) also comprises LPSBP and LPSBP. Abbreviations are as in the text. Accession details for representatives of all clusters are provided in Supplementary Table 1.
To identify sequences for cluster analysis, we searched the non-redundant protein sequence database (nr) at NCBI for homologs of the SMP domain from the yeast protein Mmm1 (residues 196–409), the N-terminal domain of human cholesteryl ester transfer protein (2OBD, residues 16–206), the Takeout 1 protein from Epiphyas postvittana (3E8T), and the dust mite allergen Der p 7 (3H4Z) using HHsenser. We pooled the permissive sets returned by HHsenser to obtain 2033 sequences, which we clustered by their pairwise BLAST P-values (Altschul et al., 1990) in CLANS (Frickey and Lupas, 2004). Clustering was done to equilibrium in 2D at a P-value cutoff of e-4 using default settings.
3 RESULTS
The number of structural solutions available to a polypeptide chain is limited, making protein structures multiply convergent (Cheng et al., 2008; Krishna and Grishin, 2004; Salem et al., 1999), while the combinatorial possibilities in sequence space are nearly endless. For this reason, sequence similarity is considered the primary marker of homology. We thus used sensitive sequence comparisons, as implemented in HHpred, to find homologs of the SMP domain in a database concatenating several complete genomes with PDB70 (see ‘Methods’ section). The search was seeded with the SMP domain from Mmm1. The best hits were to other proteins that have previously been described to contain this domain (Fig. 2). In addition, we detected a hitherto unknown SMP domain in the ERMES protein Mdm34. This protein has been reported as an integral outer membrane protein (Youngman et al., 2004), but we were unable to identify any sequence motifs in it that would indicate membrane insertion. HHpred searches with other representatives and reciprocal searches with Mdm34 confirmed the presence of an SMP domain, raising the number of SMP domains in ERMES to three (Fig. 1).
We also found statistically significant matches to many eukaryotic proteins from the bactericidal/permeability-increasing protein-like (BPI-like) family (Fig. 2), including two with known structures: BPI (1EWF) and cholesteryl ester transfer protein (CETP; 2OBD). Other members of this family are lipopolysaccharide-binding protein (LPSBP), lipid-binding serum glycoprotein (LBSGP), phospholipid transfer protein (PLTP) and long and short paralogs of palate, lung and nasal epithelium carcinoma-associated protein (PLUNC). Some of these proteins have been shown to bind lipids, e.g. CETP facilitates lipid transport between different lipoproteins (Qiu et al., 2007).
BPI and CETP have similar structures, each containing two tandem domains that adopt the same fold, comprising a long α-helix wrapped in a highly curved anti-parallel β-sheet. All BPI-like proteins contain these two domains, the only exception being short PLUNC, which has only one. The domains show little sequence identity (<15%) and sequence comparisons do not yield significant matches between them. Instead, the C-terminal domain only shows matches to the Aha1 protein, a co-chaperone of Hsp90 in eukaryotes which shares the same fold (1USU; d.83.2). Nevertheless, the N- and C-terminal domains of BPI-like proteins are thought to have a common ancestry based on their structural similarity (Kleiger et al., 2000). The Structural Classification of Proteins database (SCOP; Murzin et al., 1995) also considers them to be homologous and classifies them into the same family (d.83.1.1).
HHpred searches with SMP domains yielded many statistically significant matches to the N-terminal domain of BPI-like proteins (Fig. 2), but not to the C-terminal domain. We confirmed these findings with reciprocal searches using both domains of BPI-like proteins. From the statistical significance of these matches, we conclude that SMP domains are homologous to BPI-like proteins and therefore predict that they share the same tubular fold and lipid-binding properties.
Further searches with the N-terminal domain of BPI-like proteins retrieved three more proteins of known structure: dust mite allergen Der p 7 (3H4Z), a juvenile hormone-binding protein from Galleria mellonella (JHBP, 2RCK), and a Takeout 1 protein from Epiphyas postvittana (3E8T). These proteins are exclusively found in arthropods and are involved in binding hydrophobic ligands. They are composed of a single domain homologous to the N-terminal domain of BPI-like proteins (Supplementary Fig. S1), a relationship that has been described previously (Hamiaux et al., 2009; Kolodziejczyk et al., 2008; Mueller et al., 2010). In view of their similarities in sequence and structure, we propose to group the arthropod proteins together with the BPI-like family into the TULIP superfamily.
To confirm the membership of SMP domains in the TULIP superfamily, we generated fold predictions for 16 representative sequences using the servers Phyre (Kelley and Sternberg, 2009), MULTICOM (Wang et al., 2010) and MUSTER (Wu and Zhang, 2008), all of which performed very well in the most recent Critical Assessment of Structure Prediction Experiment, CASP8 (Kryshtafovych et al., 2009). All three methods yielded many highest-scoring matches to TULIP domains (Fig. 3 and Supplementary Fig. S2). Additionally, we queried the fold prediction metaserver I-TASSER (Roy et al., 2010), which was the top performing server in CASP 7 & 8 (Zhang, 2007, 2009). This server returned a TULIP domain as the top match for 10 of 16 queries and as one of the top three matches for all but one query (Supplementary Fig. S3). These matches included both BPI-like and Takeout-like proteins. A structure-assisted multiple sequence alignment of SMP domains to TULIP domains of known structure highlights the basis for these matches (Fig. 4). All sequences have similar length, distribution of (predicted) secondary structure, and pattern of hydrophobic residues. However, there are no conserved sequence motifs, unsurprisingly as such motifs are not even detectable within individual families (Beamer et al., 1997; Kolodziejczyk et al., 2008).
Fig. 4.
Multiple sequence alignment of TULIP domains. An alignment comprising representatives of the SMP domain family and TULIP domains of known structure is shown. Sequences are labeled as in Figure 2. The alignment was generated by a three-step approach. First, a multiple alignment of SMP sequences was obtained using HHpred in local maximum accuracy (MAC) alignment mode. Second, a structure-based sequence alignment of TULIP domain structures was derived from a multiple structure superimposition calculated using MAMMOTH-mult (Lupyan et al., 2005). In the final step, these two alignments were merged manually using as guide an alignment between 1EWF and Mmm1 obtained with HHpred. The α-helices are shown in red and β-strands in blue. Secondary structure predictions for the SMP domains were performed with Ali2D (Biegert et al., 2006). Numbers in parentheses represent length of omitted segments. Positions in the alignments that are highly conserved or strongly hydrophobic are shown in boldface. Residues that could not be aligned in structure or sequence are shown in lower case.
To explore the relative positions in sequence space of proteins of the TULIP superfamily, we searched for homologs of SMP domains, N-terminal domains of BPI-like proteins, as well as allergens and Takeout proteins in the nr database using HHsenser, and clustered the obtained sequences in CLANS (see ‘Methods’ section). The resulting cluster map (Fig. 5) shows three distinct but connected regions corresponding to SMP, BPI and Takeout-like domain families, confirming the proposed homology between them. In addition to the SMP groups described by Lee and Hong (2006) and the group of Mdm34 proteins described in this article, the clustering revealed a further SMP group, the uncharacterized transmembrane 24 proteins. It also yielded a number of additional groups of BPI-like proteins, including the expression site-associated gene 5 proteins (ESAG5) from trypanosomes, whose homology to BPI-like proteins has been reported previously (Barker et al., 2008). BPI and Takeout-like domains are connected by the arthropod allergens, one form of which is unique in containing tandem domains with clear sequence similarity, indicating a domain duplication that occurred in insects (yellow and orange clusters in Fig. 5).
4 CONCLUSIONS
In this study, we have shown that the SMP domain belongs to the TULIP domain superfamily, a large group of proteins that bind lipids and other hydrophobic ligands within a central, tubular cavity (Fig. 6). In several cases (CETP, PLTP), members of this superfamily are known to exploit this binding activity in order to mediate lipid trafficking. Given the extensive lipid exchange between the ER and the mitochondrial outer membrane and the location of the ERMES complex as a connector between them, it is attractive to consider that this exchange is mediated by the SMP domains of the ERMES subunits. As the ERMES complex does not include a nucleotidase that could energize this process, we propose that it proceeds along an affinity gradient, amounting to facilitated diffusion. Although this could be envisaged as resulting from many short, structurally unspecific contacts between the SMP domains (‘kiss-and-run’ mechanism), we prefer to consider that the domains assemble into structurally well-defined complexes, which establish a lipophilic, tubular path between the two membranes. Since the stoichiometry of subunits within the ERMES complex is currently unknown, it is however not possible at this time to judge on whether 1 : 1 : 1 or some other ratio would most appropriately describe the composition of such complexes.
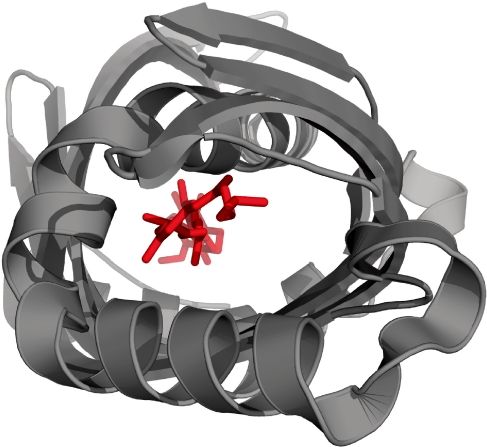
Fig. 6.
View along the ligand-binding tunnel of a Takeout protein (3E8T, residues 5–211). The ligand is shown as red sticks.
Funding: This work was supported by institutional funds from the Max-Planck-Society.
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Achleitner G, et al. Association between the endoplasmic reticulum and mitochondria of yeast facilitates interorganelle transport of phospholipids through membrane contact. Eur. J. Biochem. 1999;264:545–553. doi: 10.1046/j.1432-1327.1999.00658.x. [DOI] [PubMed] [Google Scholar]
- Altschul SF, et al. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Barker AR, et al. Bioinformatic insights to the ESAG5 and GRESAG5 gene families in kinetoplastid parasites. Mol. Biochem. Parasitol. 2008;162:112–122. doi: 10.1016/j.molbiopara.2008.08.003. [DOI] [PubMed] [Google Scholar]
- Beamer LJ, et al. Crystal structure of human BPI and two bound phospholipids at 2.4 angstrom resolution. Science. 1997;276:1861–1864. doi: 10.1126/science.276.5320.1861. [DOI] [PubMed] [Google Scholar]
- Biegert A, et al. The MPI Bioinformatics Toolkit for protein sequence analysis. Nucleic Acids Res. 2006;34:W335–W339. doi: 10.1093/nar/gkl217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, et al. MALISAM: a database of structurally analogous motifs in proteins. Nucleic Acids Res. 2008;36:D211–D217. doi: 10.1093/nar/gkm698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frickey T, Lupas A. CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics. 2004;20:3702–3704. doi: 10.1093/bioinformatics/bth444. [DOI] [PubMed] [Google Scholar]
- Hamiaux C, et al. Crystal structure of Epiphyas postvittana takeout 1 with bound ubiquinone supports a role as ligand carriers for takeout proteins in insects. J. Biol. Chem. 2009;284:3496–3503. doi: 10.1074/jbc.M807467200. [DOI] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protocols. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Kleiger G, et al. The 1.7 A crystal structure of BPI: a study of how two dissimilar amino acid sequences can adopt the same fold. J. Mol. Biol. 2000;299:1019–1034. doi: 10.1006/jmbi.2000.3805. [DOI] [PubMed] [Google Scholar]
- Kolodziejczyk R, et al. Insect juvenile hormone binding protein shows ancestral fold present in human lipid-binding proteins. J. Mol. Biol. 2008;377:870–881. doi: 10.1016/j.jmb.2008.01.026. [DOI] [PubMed] [Google Scholar]
- Kornmann B, et al. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009;325:477–481. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna SS, Grishin NV. Structurally analogous proteins do exist! Structure. 2004;12:1125–1127. doi: 10.1016/j.str.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Kryshtafovych A, et al. Protein structure prediction center in CASP8. Proteins. 2009;77(Suppl. 9):5–9. doi: 10.1002/prot.22517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Hong W. Diverse membrane-associated proteins contain a novel SMP domain. FASEB J. 2006;20:202–206. doi: 10.1096/fj.05-4581hyp. [DOI] [PubMed] [Google Scholar]
- Lill R, Kispal G. Maturation of cellular Fe-S proteins: an essential function of mitochondria. Trends Biochem. Sci. 2000;25:352–356. doi: 10.1016/s0968-0004(00)01589-9. [DOI] [PubMed] [Google Scholar]
- Lupyan D, et al. A new progressive-iterative algorithm for multiple structure alignment. Bioinformatics. 2005;21:3255–3263. doi: 10.1093/bioinformatics/bti527. [DOI] [PubMed] [Google Scholar]
- McBride HM, et al. Mitochondria: more than just a powerhouse. Curr. Biol. 2006;16:R551–R560. doi: 10.1016/j.cub.2006.06.054. [DOI] [PubMed] [Google Scholar]
- Meisinger C, et al. The mitochondrial morphology protein Mdm10 functions in assembly of the preprotein translocase of the outer membrane. Dev. Cell. 2004;7:61–71. doi: 10.1016/j.devcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Mueller GA, et al. The structure of the dust mite allergen Der p 7 reveals similarities to innate immune proteins. J. Allergy Clin. Immunol. 2010;125:909–917. doi: 10.1016/j.jaci.2009.12.016. e904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzin AG, et al. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Shaw JM. Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu. Rev. Genet. 2005;39:503–536. doi: 10.1146/annurev.genet.38.072902.093019. [DOI] [PubMed] [Google Scholar]
- Qiu X, et al. Crystal structure of cholesteryl ester transfer protein reveals a long tunnel and four bound lipid molecules. Nat. Struct. Mol. Biol. 2007;14:106–113. doi: 10.1038/nsmb1197. [DOI] [PubMed] [Google Scholar]
- Roy A, et al. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protocols. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem GM, et al. Correlation of observed fold frequency with the occurrence of local structural motifs. J. Mol. Biol. 1999;287:969–981. doi: 10.1006/jmbi.1999.2642. [DOI] [PubMed] [Google Scholar]
- Söding J, et al. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söding J, et al. HHsenser: exhaustive transitive profile search using HMM-HMM comparison. Nucleic Acids Res. 2006;34:W374–W378. doi: 10.1093/nar/gkl195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker DR. New perspectives on the regulation of intermembrane glycerophospholipid traffic. J. Lipid Res. 2003;44:441–449. doi: 10.1194/jlr.R200020-JLR200. [DOI] [PubMed] [Google Scholar]
- Wang Z, et al. MULTICOM: a multi-level combination approach to protein structure prediction and its assessments in CASP8. Bioinformatics. 2010;26:882–888. doi: 10.1093/bioinformatics/btq058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Zhang Y. MUSTER: Improving protein sequence profile-profile alignments by using multiple sources of structure information. Proteins. 2008;72:547–556. doi: 10.1002/prot.21945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman MJ, et al. Mmm2p, a mitochondrial outer membrane protein required for yeast mitochondrial shape and maintenance of mtDNA nucleoids. J. Cell Biol. 2004;164:677–688. doi: 10.1083/jcb.200308012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. Template-based modeling and free modeling by I-TASSER in CASP7. Proteins. 2007;69(Suppl. 8):108–117. doi: 10.1002/prot.21702. [DOI] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER: fully automated protein structure prediction in CASP8. Proteins. 2009;77(Suppl. 9):100–113. doi: 10.1002/prot.22588. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.