Abstract
Summary: A tool to predict the effect that newly discovered genomic variants have on known transcripts is indispensible in prioritizing and categorizing such variants. In Ensembl, a web-based tool (the SNP Effect Predictor) and API interface can now functionally annotate variants in all Ensembl and Ensembl Genomes supported species.
Availability: The Ensembl SNP Effect Predictor can be accessed via the Ensembl website at http://www.ensembl.org/. The Ensembl API (http://www.ensembl.org/info/docs/api/api_installation.html for installation instructions) is open source software.
Contact: wm2@ebi.ac.uk; fiona@ebi.ac.uk
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
As costs of resequencing and genotyping fall, increasing amounts of variation data are being produced that cannot be annotated effectively without access to considerable computational resources and genomic annotation databases. Often the most valuable information to know about a variant is the effect the observed alleles have on transcripts, which may aid selection of variations for genotyping studies and in turn have a part to play in the discovery of new drug targets and other biologically significant loci. Deriving this information manually is laborious and error-prone, impractical for large sets of data and impossible without access to suitable genomic annotation resources. Of critical interest is the existence of any novel variant positions within a dataset, and what information is available for known variant loci. Although these answers are available through dbSNP (Sherry et al., 1999), the process of submitting the data to the NCBI to be processed and annotated can often take months and requires the data to be made public.
Even before the developments reported in this article, Ensembl (Flicek et al., 2010) could also be used to derive similar annotation by setting up a full local Ensembl database containing the variant information and running scripts from the Ensembl Variation database production pipeline. Other tools available for the annotation of single nucleotide polymorphisms (SNPs) in humans are comprehensively reviewed in Karchin (2008).
Existing methods of deriving the effects of variants can be limiting: many present too high a hurdle in terms of timeframe, privacy or ease of use; others are species limited. To address this, Ensembl has been extended to include an easy to use web-based tool for deriving variation consequences, as well as programmatic access to the same functionality using the Ensembl Perl API.
2 IMPLEMENTATION
2.1 SNP Effect Predictor
The Ensembl project provides access to genomic annotation for numerous species via its web-based genome browser, as well as programmatic access via the object-oriented Perl API. The SNP Effect Predictor tool on the Ensembl website, accessed via the ‘Manage your data’ link on any species-specific Ensembl page (e.g. http://www.ensembl.org/Homo_sapiens/), uses the API calls described below to provide access to consequence prediction functionality without the need for writing code. The SNP Effect Predictor can be used for all species within Ensembl, including those with no existing variation dataset.
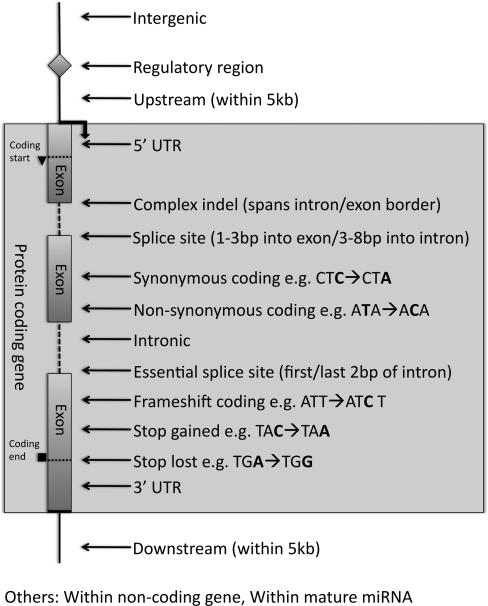
Users upload lists of variant positions and alleles via a HTML form page. Input for each variant consists simply of a chromosome (or contig name in the absence of assembled chromosomes), start and end coordinates, strand designation and a set of alleles. Users can then select text or HTML formatted output, the latter incorporating hyperlinks to loci, transcripts and genes in the Ensembl genome browser. The output includes: Ensembl stable identifiers for the relevant transcript and gene; transcript-relative coordinates; possible amino acids; and the identifier of any existing variants that are co-located with the user-defined variant. Since a variant may co-locate with more than one transcript, one line of output is provided for each instance of co-location. Consequence types predicted by Ensembl are shown in transcript context in Figure 1, with further detail provided at (http://www.ensembl.org/info/docs/variation/index.html).
Fig. 1.
Consequence types predicted by Ensembl in the context of transcript structure. The other types shown apply to non-protein coding genes.
User uploaded variations can subsequently be viewed in the context of their location on the Ensembl browser, with each uploaded file given its own track on the browser's location view.
2.2 Ensembl API
The Ensembl API can be installed on any operating system that supports Perl and MySQL, and can be configured to use any combination of local or remote databases. The Ensembl Variation API (Chen et al., 2010; Rios et al., 2010) exists to retrieve variation data such as SNPs, insertions and deletions from Ensembl databases. Entities such as variants are represented as objects, created by adaptors that act as factories for generating specific objects. Example code demonstrating the use of the API to derive consequences for a list of variant positions is shown in Supplementary Figure 1. Documentation on the API is found at http://www.ensembl.org/info/docs/Pdoc/ensembl-variation/index.html.
Given a variant position, the API retrieves overlapping transcripts from the Ensembl Core database and determines where in the transcript structure the variant falls. If the variant falls within an exon, new codons for each variant allele are derived and compared to the reference codon. The location of the variant relative to regulatory regions is also assessed using the Ensembl Functional Genomics database where available. The results, including amino acid changes and relative positions in the cDNA and peptide sequences, are stored in the resulting transcript variation objects, along with one or more named consequence types.
At present Ensembl provides only a Perl API, but enabled by the open source nature of the project Python (PyCogent, http://pycogent.sourceforge.net/examples/query_ensembl.html; PyGr, http://code.google.com/p/pygr/wiki/PygrOnEnsembl) APIs have been created. As yet, these do not encompass the full scope of Ensembl, and hence do not include consequence prediction functionality.
3 RESULTS
The SNP Effect Predictor tool can be used to quickly and accurately predict the effects of variants on Ensembl-annotated transcripts. Up to 750 variant loci can be uploaded in a file to http://www.ensembl.org/, with the time taken to return results scaling linearly with the number of variants uploaded within a species; calculation time will also vary by species depending on the number of transcripts. A file containing 750 variants in Homo sapiens takes ∼35 s to return results; an equivalent calculation in Danio rerio takes 20 s. Users with more than 750 variants may download a standalone script to run locally that produces identical results. The script can be configured to connect to both the public Ensembl database as well as any combination of local and remote databases. A wider range of input file formats is also supported, including the commonly used pileup variant format.
The provision of a simple web interface to powerful algorithms that transparently process large data volumes is a valuable asset to users without computing expertise, and also to those who need a quick and easy way to retrieve annotation for novel variants. Having this tool integrated with the extensive, rich annotation available on the Ensembl website will facilitate interpretation and analysis of the data.
Direct use of the Ensembl Variation API enables users to incorporate consequence prediction into their variation software and pipelines, providing predicted consequences for an unlimited number of variants. By optimizing code and database access times it is possible to retrieve consequences for 1000 distinct variants in H.sapiens in <30 s; for D.rerio this takes <15 s.
The flexibility of the Ensembl API means that consequences can be predicted for any species with an Ensembl gene set, or using any valid Ensembl database on users' own systems. Using these features in coalition with others in the API enables the creation of advanced pipelines that can produce biologically important information from high-throughput experimental data. Such information is invaluable both as a screening system for variants and as an aid in the study of phenotypically linked variants.
Funding: Wellcome Trust; European Community's Seventh Framework Programme (FP7/2007-2013) under grant agreement number 200754 – the GEN2PHEN project.
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Chen Y, et al. Ensembl variation resources. BMC Genomics. 2010;11:238. doi: 10.1186/1471-2164-11-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, et al. Ensembl's 10th year. Nucleic Acids Res. 2010;38:D557–D562. doi: 10.1093/nar/gkp972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karchin R. Next generation tools for the annotation of human SNPs. Brief Bioinformatics. 2008;10:35–52. doi: 10.1093/bib/bbn047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios D, et al. A database and API for variation, dense genotyping and resequencing data. BMC Bioinformatics. 2010;11:293. doi: 10.1186/1471-2105-11-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, et al. dbSNP—database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res. 1999;9:677–679. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.