Abstract
Motivation: Splice variation plays important roles in evolution and cancer. Different splice variants of a gene may be characteristic of particular cellular processes, subcellular locations or organs. Although several genomic projects have identified splice variants, there have been no large-scale computational studies of the relationship between number of splice variants and biological function. The Gene Ontology (GO) and tools for leveraging GO, such as GoMiner, now make such a study feasible.
Results: We partitioned genes into two groups: those with numbers of splice variants ≤b and >b (b=1,…, 10). Then we used GoMiner to determine whether any GO categories are enriched in genes with particular numbers of splice variants. Since there was no a priori ‘appropriate’ partition boundary, we studied those ‘robust’ categories whose enrichment did not depend on the selection of a particular partition boundary. Furthermore, because the distribution of splice variant number was a snapshot taken at a particular point in time, we confirmed that those observations were stable across successive builds of GenBank. A small number of categories were found for genes in the lower partitions. A larger number of categories were found for genes in the higher partitions. Those categories were largely associated with cell death and signal transduction. Apoptotic genes tended to have a large repertoire of splice variants, and genes with splice variants exhibited a distinctive ‘apoptotic island’ in clustered image maps (CIMs).
Availability: Supplementary tables and figures are available at URL http://discover.nci.nih.gov/OG/supplementaryMaterials.html. The Safari browser appears to perform better than Firefox for these particular items.
Contact: barry@discover.nci.nih.gov
1 INTRODUCTION
Alternative splicing generates enhanced diversity in the transcriptome relative to the genome, and various reports have suggested that the percentage of genes exhibiting alternative splicing may be as high as 94% (Boue et al., 2003; Lee and Roy, 2004; Wang et al., 2008). In the present study, we found alternative splicing for ∼84% of those human genes that had HGNC symbols (Ashburner et al., 2000a; Gene Ontology Consortium, 2006; Little, 1998; McKusick, 1989; Wain et al., 2002). Numerous reviews describe alternative splicing in general (Black, 2000; Breitbart et al., 1987; Graveley, 2001; Modrek and Lee, 2002), mechanisms of alternative splicing (Black, 2003; Smith et al., 1989), and the roles played by alternative splicing in particular biological processes and diseases (Black, 1998; Black and Grabowski, 2003; Blencowe, 2000; Burgess et al., 1999; Caceres and Kornblihtt, 2002; Cooper and Mattox, 1997; Garcia-Blanco et al., 2004; Grabowski and Black, 2001; Jiang and Wu, 1999; Schutt and Nothiger, 2000; Xu et al., 2002). Splice isoforms can have different degrees of activity (Zhang et al., 2006) or can perform radically different functions (Fernandez-Real et al., 2006). Splice variation plays important roles in cancer and in evolution (Kriventseva et al., 2003).
Although several genomic studies have attempted to identify splice variants (Carninci et al., 2005; Tress et al., 2007), we are not aware of any computational studies in which the global relationship between the number of splice variants and biological function of a gene has been analyzed. The Gene Ontology (GO; Ashburner et al., 2000b; Gene Ontology Consortium, 2006) and tools like GoMiner (Zeeberg et al., 2003) and High-Throughput GoMiner (HTGM; Zeeberg et al., 2005) now make such studies feasible.
Using those resources, we attempted to determine whether any GO categories were enriched in classes of genes with particular ranges of splice variant number. That is, we tested the null hypothesis that there is no correlation between the number of characterized splice variants and the GO classification, starting with no a priori expectation that there would be even one such category. Neither did we have any expectation as to whether any enriched categories would be related to one another nor whether they would reflect any particular biological process(es).
In fact, enriched categories did appear. Many of those categories were closely related to one another, and categories relevant to cancer were particularly prominent among them. Specifically, we found a particularly strong global relationship for genes involved in apoptosis, in that genes related to apoptosis and signaling tended to have large repertoires of splice variants. A specialized form of clustered image map (CIM) highlighted GO categories with relatively high numbers of splice variants in the form of distinctive ‘apoptotic islands.’
2 METHODS
2.1 Data acquisition
Data for computing the number of splice variants per gene were acquired from the Evidence Viewer Database (EVDB; Kahn et al., 2006) and http://www.tigerteamconsulting.com/SpliceCenter/FAQ_Database.jsp. EVDB is an exhaustive, non-redundant relational database of all known genes and their splice variants that we have developed, based on the NCBI Gene Evidence Viewer. EVDB allows high-throughput querying of splice variant data. The complete description of EVDB and the methods used to create it are given in Kahn et al. (2006). The technical details of the construction of EVDB for builds 35 and 36 are given in Kahn et al. (2006) and http://www.tigerteamconsulting.com/SpliceCenter/FAQ_Database.jsp, respectively. Supplementary Table S1 shows the versions of the relevant data sources that were used.
2.2 High-Throughput GoMiner (HTGM)
GoMiner (Zeeberg et al., 2003) is a tool for biological interpretation of ‘omic’ data, including data from gene expression microarrays. It leverages the GO database (http://www.geneontology.org/GO.downloads.database.shtml) to identify ‘biological processes,’ ‘molecular functions,’ and ‘cellular components’ represented in a list of genes. HTGM (Zeeberg et al., 2005), which was used for many of the analyses reported here, is an enhancement of GoMiner that efficiently performs the computationally challenging task of automated batch processing of an arbitrary number of such gene lists. A GO category is considered to be enriched if the number of changed genes that HTGM assigned to it is statistically significantly greater than the number expected by chance. A category is considered to be significant if its false discovery rate (FDR) is less than or equal to a given threshold (typically 0.10). Briefly, GoMiner computes a one-tail Fisher Exact P-value that is based on a 2 × 2 contingency table representing ‘in GO category’ and ‘not in GO category’ versus ‘in partition’ and ‘not in partition’. The FDR is estimated by a comparison of the distribution of P-values for the real data and for multiple determinations of randomized data. See Zeeberg et al. (2003, 2005) for detailed discussions of GoMiner and HTGM, including calculations of statistical significance. The parameters used in all HTGM analyses are listed in Supplementary Table S2. Only genes that had HGNC symbols and GO annotations were used in our studies.
2.3 Clustering with genesis
CIMs were first introduced in Weinstein et al. (1997) and were produced here with the Genesis program (Sturn et al., 2002). We selected the Euclidean distance metric and average linkage for hierarchal clustering. To facilitate visualization, we implemented a recently added feature of GoMiner that removes large, generic categories from all CIMs.
2.4 Directed acyclic graph (DAG) representation of the robust categories
A leaf category is defined as a robust category that is not the parent of another robust category. A DAG segment is constructed for each leaf category, with the leaf category as the sole leaf node of that segment. The GO database is directly queried by SQL to determine the DAG structure for each leaf category. Although the vertical position of each node in the segment is pre-determined, the horizontal position is arbitrary. The internal layout for each segment is optimized by interchanging the horizontal position of the nodes to generate the shortest overall length of connecting lines.
3 RESULTS AND DISCUSSION
3.1 Partitioning genes according to splice variant number
GenBank is continually evolving, and there is uncertainty about the exact number of splice variants for any given gene. Therefore, cumulative classes may be more robust than individual classes for investigating the biological meaning of splice variant number. Cumulative gene classes were formed by taking the union of individual gene classes: i.e. genes were partitioned into two groups:
those with a number of splice variants ≤b
those with a number of splice variants >b
where b is the partition boundary (b = 1,…, 10). A partitioning was represented as
where M is the maximum number of variants per gene. For human genome builds 35 and 36, M is 73 and 66, respectively. For example, we represent gene sets formed from build 36 using the partitioning value of 7 splice variants as {1,…, 7}, {8,…, 66}.
3.2 Stability of representation of splice variants/gene by GenBank mRNA sequences corresponding to human genome builds 35 and 36
We searched for partitionings stable across human genome builds 35 and 36 by defining a set of genes G that is the intersection of genes in builds 35 and 36. Each gene in G fell into one of four classes, as defined in Table 1. For a given partitioning, (ll + hh)/T = 1.0 would represent perfect stability. In practice, the stability was somewhat lower than 1.0. For instance, that ratio achieved 0.984 for b = 10 (Supplementary Table S4).
Table 1.
Four classes for genes in common in human genome builds 35 and 36
| Classes | Definition |
|---|---|
| ll | Genes that fall into the lower partition in both builds |
| lh | Genes that fall into the higher partition for build 36 and the lower partition for build 35 |
| hl | Genes that fall into the lower partition for build 36 and the higher partition for build 35 |
| hh | Genes that fall into the higher partition in both builds |
| T | ll + lh + hl + hh |
In selecting a partitioning, we needed to consider both the stability and suitability as input to GoMiner. If the gene set were too small or too large, then it would not be suitable. A set that was too small would not provide adequate statistical power. Generally, the set should contain at least 200 genes that map to the GO database. On the other hand, a set that was too large would result in all categories being heavily populated with genes, and it would be impossible to detect meaningful enrichment. All three stability/GoMiner requirements
(ll + hh)/T ≥ 0.90
(hl + hh)/T ≤ 0.50
hl + hh ≥200
were met by partitions 6 through 10 (Supplementary Table S4).
3.3 Summary statistics for build 36
The numbers of genes that survived successive processing steps are shown in Supplementary Table S5. The total number of genes represented was 19 215 (the corresponding number for build 35 was 16 893). A total of 7201 were represented by HGNC symbols and had mappings in the GO database. A total of 1285 genes mapped to partition {8,…, 66}, for example. 835 of the 1285 had mappings in the GO database.
3.4 Robustness of GO categories
A robust GO category is one that GoMiner deemed to be statistically significantly enriched through a broad range of partitions. A combination CIM (Supplementary Fig. S1) provided a convenient visualization of the comparison of the degree of robustness of categories in lower partitions across builds 35 and 36. A few of the significant categories that involved sensory perception appeared in both, but none of those categories were considered to be robust within or across builds. It was surprising that sensory perception categories would be associated with low partitions. For example, it is well known that olfactory receptors enjoy considerable degree of alternate splicing (Young et al., 2003). Yet, our search of GenBank revealed that independent records for the different variants had not been deposited. We expect that the apparent low number of splice variants is an artifact resulting from poor annotation of sensory perception genes in GenBank.
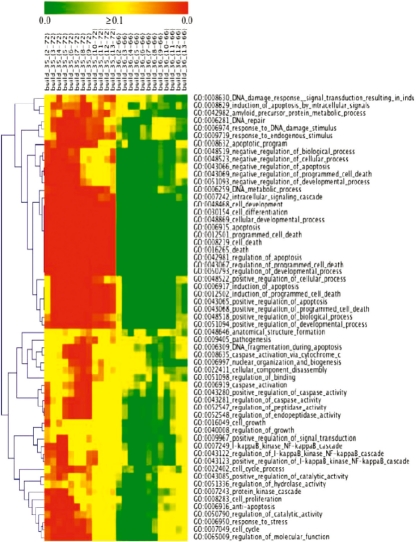
In contrast, there were a substantial number of robust categories in higher partitions (Fig. 1, Supplementary Figs S2 and S3, and Table S5). A total of 835 genes mapped to a significant GO category in partition {8,…, 66} (Supplementary Table S5). Of those 835 genes, 799 mapped to a category that was deemed by GoMiner to be statistically significant. Approximately half (404) of those genes mapped to a significant category that was robust. There were a total of 59 robust categories, of which 55 occurred in partition {8,…, 66}.
Fig. 1.
Integrative CIM of robust (i.e. significant for at least 12 partition boundaries in the higher partition) categories versus partition boundaries for builds 35 and 36. The color scales represent the FDRs for builds 35 (red) and 36 (green). This CIM is also available as Supplementary Fig. S3.
3.5 Categories-versus-genes CIMs
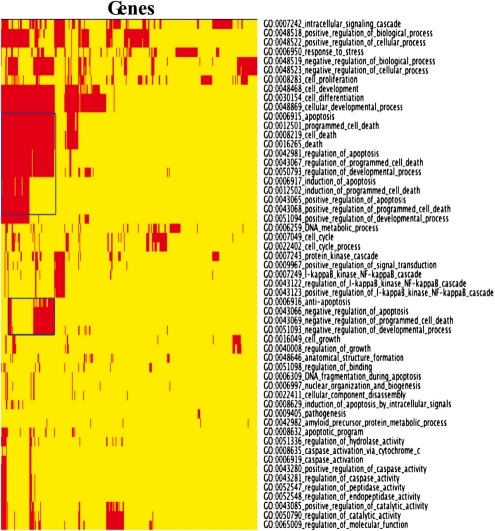
The robust categories-versus-genes CIM for partition {8,…, 66} exhibited a distinct apoptotic island (Fig. 2 and Supplementary Fig. S4). The identity of the genes associated with that feature and with other features can be determined from the full-size CIM (Supplementary Fig. S4). The full-size CIM in Supplementary Figure S4 can also be used to examine cross-talk between categories. For example, many genes appear in common between GO:0006915_apoptosis and GO:0007242_intracellular_signaling_cascade (Supplementary Table S6).
Fig. 2.
Thumbnail CIM of robust categories versus genes for build 36, partition {8,…, 66}. The apoptotic island is outlined with blue rectangles. The full-size CIM can be viewed as Supplementary Figure S4.
3.6 DAG representation of the robust categories
The CIM visualizations does not explicitly represent the hierarchical arrangement of categories that is present in the GO DAG structure. We have observed that a single complete DAG representation is useful for conveying the complexity of the arrangement, but it is too complex to be very informative to the human visual system. Consequently, we have developed a novel piecewise DAG representation of the 59 robust categories that are depicted in Supplementary Figure S5. Each DAG segment was derived from a single robust category, which was the sole leaf node in that segment. If a robust category appeared as the parent of another robust category, that robust parent category was not used as a leaf node elsewhere. The robust and non-robust categories that appeared in a DAG segment are rendered in red and blue, respectively. The DAG segment representation contain a modest degree of redundancy because some segments displays the same nodes. We feel that the slight redundancy is justified because of the enhanced clarity of the representation.
3.7 An alternative complementary perspective of category enrichment
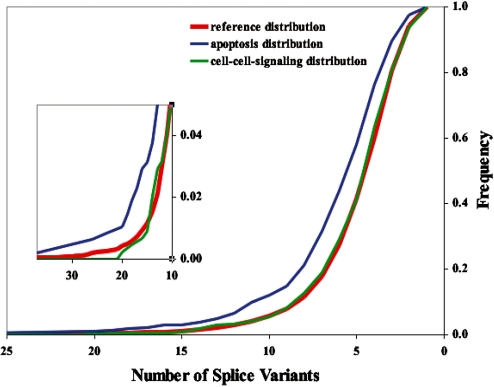
An alternate approach that complements traditional GoMiner category enrichment is analysis of the distribution of splice variant number for genes mapping to a category. The probability distribution (Fig. 3 and Supplementary Fig. S6) indicate a shift to higher splice variant number for genes in the apoptosis category (i.e. significant in GoMiner) compared with the reference distribution (i.e. the biological_process category). The control category cell–cell signaling (i.e. non-significant in GoMiner) does not exhibit any such shift.
Fig. 3.
Cumulative probability distributions (CPDs) for the frequency of splice variant numbers in the reference category (biological process; 7201 genes), apoptosis (477 genes) and cell–cell signaling (440 genes). The CPD was taken in the ‘reverse’ direction, from high to low number of splice variants, to illustrate more effectively the enrichment in high numbers of splice variants for apoptosis. The CPD for apoptosis was significantly different from the reference category (K-S, P < 0.0001, computed for the normal direction for the CPD), whereas that for cell–cell signaling was not (K-S, P = 0.92). The expanded inset emphasizes the dramatic enrichment for apoptosis in the high-splice variant-number tail of the distribution, in contrast to the behavior of the control category, cell–cell signaling.
The shift in the probability distribution visually reflects the underlying dramatic quantitative enrichment of the apoptosis category. The reference distribution contains a total of 7201 genes of which 835 have 8 or more splice variants (Supplementary Table S5). Thus the reference distribution exhibits a ratio of 835/7201 = 0.116. The apoptosis category contains a total of 477 genes (data not shown), so the expected number of genes with 8 or more splice variants was 477*0.116 ≈ 55. But the apoptosis category actually includes 101 genes (data not shown) with 8 or more splice variants, an excess of 101 − 55 = 46, corresponding to a ratio of 101/477 = 0.212 or a 1.826-fold enrichment relative to the reference (GoMiner P-value ≈ 10−9.42 and FDR ≤ 10−6). Thus, apoptosis includes almost twice the expected number of genes with 8 or more splice variants, and that excess is reflected visually in Figure 3.
4 CONCLUSIONS
The current analysis was originally intended purely as a genomics study to correlate functional categories with number of splice isoforms. The observations unexpectedly turned out to have strong relevance for cancer-interesting processes, especially apoptosis. We will summarize the major findings and then examine the implications for apoptosis in more detail.
When displayed in a CIM, genes with a high number of splice variants, class {8,…, 66}, form a distinctive ‘apoptotic island’ (Fig. 2 and Supplementary Fig. S4).
Forty-one genes fall into both apoptosis and intracellular signaling (Fig. 2, Supplementary Fig. S4 and Table S6). Those genes provide a mechanism for ‘cross-talk’. Apoptotic and intracellular signaling categories are well studied and it is not a surprise that a number of genes are shared (see for example, Wu et al., 2006). The potential for the CIM to uncover such relationships will be of particular value in less well-studied disease states.
In fact, several of the shared genes are central to both apoptosis and signaling. Their modes of alternate splicing have consequently been studied in some detail. For example, Benedict et al. (2000) studied alternate splicing of Apaf-1. Alternative splicing can create an NH2-terminal 11-amino acid insert between the caspase recruitment domain and ATPase domains or an additional COOH-terminal WD-40 repeat. Apaf-1XL contains both the NH2-terminal and COOH-terminal inserts and is the major RNA form expressed in all tissues tested. Apaf-1LN contains the NH2-terminal insert, but lacks the additional WD-40 repeat. Only those isoforms with the additional WD-40 repeat activated procaspase-9 in vitro in response to cytochrome c and dATP, whereas the NH-terminal insert was not required for that activity.
Merdzhanova et al. (2008) studied the upregulation of SC35 by E2F1. They found that DNA-damaging agents stabilize E2F1 and induce its transcriptional activity. Overexpression of SC35 alters the splicing of caspase-2 mRNA, favoring expression of the pro-apoptotic isoform accumulation. E2F1 requires SC35 to switch the alternative splicing profile of various apoptotic genes such as c-flip, caspases-8, caspases-9 and Bcl-x towards the expression of pro-apoptotic splice variants.
Jiang and Wu (1999) aptly summarized the state of the field as it stood in 1999:
Expression and function of a large number of genes involved in PCD [programmed cell death] are regulated by alternative splicing, including death receptors and intracellular components of the death machinery. Alternative splicing affects not only intracellular distribution but also functional activity of these death regulators, providing a fine-tuning mechanism in modulating a presumably tightly controlled process of cell death.
Our current results, which are based on the much larger number of GenBank records that are available now as compared with the number available to Jiang and Wu (1999), are consistent with that summary statement.
We speculate that the relationship, uncovered in our studies, between the structure of the human genome and the life and death of a cell, is a fundamental property of the cell. That is, the locations of nucleotide sequences dictating the regulation of the number of alternate splice forms in the human genome (‘structure’) are intimately related to apoptosis (‘function’). With regard to splice variants, there are two competing ‘drives’: robustness, which will be enhanced as a component becomes simpler, and diversity to allow fine-tuning in different tissues, stages of development or states of health and disease.
The concrete manifestation of robustness is one transcript per gene, whereas the concrete manifestation of diversity is multiple or a high number of transcripts per gene. The negative aspects of those two attributes are inflexibility for the former and an increase in errors due to complexity [e.g. susceptibility to lethal mutation in splice site signal sequences (Rogan et al., 1998; Schneider, 2005)] in the latter. We speculate that all genes would have multiple transcripts to achieve greater flexibility if susceptibility to mutation were not of over-riding importance. Therefore, only those genes that must have multiple transcripts do so. In such cases, the need for flexibility apparently over-rides the ‘prudence’ of avoiding susceptibility to mutation.
Supplementary Table S5 shows that the majority of genes do, in fact, exhibit at least two known splice forms: 16 117/19 215 = 0.84 overall, and 6793/7201 = 0.94 for those genes in human genome build 36 that have HGNC symbols and that are represented in the GO database. In the context of our speculation, that observation would imply that the flexibility afforded by fine-tuning is essential.
Funding: Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Conflict of Interest: none declared.
REFERENCES
- Ashburner M, et al. Gene Ontology: tool for the unification of biology. Nat. Genet. 2000a;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000b;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedict MA, et al. Expression and functional analysis of Apaf-1 isoforms. J. Biol. Chem. 2000;275:8461–8468. doi: 10.1074/jbc.275.12.8461. [DOI] [PubMed] [Google Scholar]
- Black DL. Splicing in the inner ear: a familiar tune, but what are the instruments? Neuron. 1998;20:165–168. doi: 10.1016/s0896-6273(00)80444-4. [DOI] [PubMed] [Google Scholar]
- Black DL. Protein diversity from alternative splicing: a challenge for bioinformatics and post-genome biology. Cell. 2000;103:367–370. doi: 10.1016/s0092-8674(00)00128-8. [DOI] [PubMed] [Google Scholar]
- Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Black DL, Grabowski PJ. Alternative pre-mRNA splicing and neuronal function. Prog. Mol. Subcell. Biol. 2003;31:187–216. doi: 10.1007/978-3-662-09728-1_7. [DOI] [PubMed] [Google Scholar]
- Blencowe BJ. Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem. Sci. 2000;25:106–110. doi: 10.1016/s0968-0004(00)01549-8. [DOI] [PubMed] [Google Scholar]
- Boue S, et al. Alternative splicing and evolution. Bioessays. 2003;25:1031–1034. doi: 10.1002/bies.10371. [DOI] [PubMed] [Google Scholar]
- Breitbart RE, et al. Alternative splicing: a ubiquitous mechanism for the generation of multiple protein isoforms from single genes. Annu. Rev. Biochem. 1987;56:467–495. doi: 10.1146/annurev.bi.56.070187.002343. [DOI] [PubMed] [Google Scholar]
- Burgess RW, et al. Alternatively spliced isoforms of nerve- and muscle-derived agrin: their roles at the neuromuscular junction. Neuron. 1999;23:33–44. doi: 10.1016/s0896-6273(00)80751-5. [DOI] [PubMed] [Google Scholar]
- Caceres JF, Kornblihtt AR. Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet. 2002;18:186–193. doi: 10.1016/s0168-9525(01)02626-9. [DOI] [PubMed] [Google Scholar]
- Carninci P, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1555–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Cooper TA, Mattox W. The regulation of splice-site selection, and its role in human disease. Am. J. Hum. Genet. 1997;61:259–266. doi: 10.1086/514856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Real JM, et al. An alternative spliced variant of circulating soluble tumor necrosis factor-{alpha} receptor-2 is paradoxically associated with insulin action. Eur. J. Endocrinol. 2006;154:723–730. doi: 10.1530/eje.1.02145. [DOI] [PubMed] [Google Scholar]
- Garcia-Blanco MA, et al. Alternative splicing in disease and therapy. Nat. Biotechnol. 2004;22:535–546. doi: 10.1038/nbt964. [DOI] [PubMed] [Google Scholar]
- Gene Ontology Consortium. The Gene Ontology (GO) project in 2006. Nucleic Acids Res. 2006;34:D322–326. doi: 10.1093/nar/gkj021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowski PJ, Black DL. Alternative RNA splicing in the nervous system. Prog. Neurobiol. 2001;65:289–308. doi: 10.1016/s0301-0082(01)00007-7. [DOI] [PubMed] [Google Scholar]
- Graveley BR. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- Jiang ZH, Wu JY. Alternative splicing and programmed cell death. Proc. Soc. Exp. Biol. Med. 1999;220:64–72. doi: 10.1046/j.1525-1373.1999.d01-11.x. [DOI] [PubMed] [Google Scholar]
- Kahn AB, et al. SpliceMiner: a high-throughput database implementation of the NCBI Evidence Viewer for microarray splice variant analysis. BMC Bioinform. 2006;1:1. doi: 10.1186/1471-2105-8-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriventseva EV, et al. Increase of functional diversity by alternative splicing. Trends Genet. 2003;19:124–128. doi: 10.1016/S0168-9525(03)00023-4. [DOI] [PubMed] [Google Scholar]
- Lee C, Roy M. Analysis of alternative splicing with microarrays: successes and challenges. Genome Biol. 2004;5:231. doi: 10.1186/gb-2004-5-7-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little P. Human genome annotation—a possible role for HUGO? Human Genome Organisation. Nat. Genet. 1998;19:222. doi: 10.1038/896. [DOI] [PubMed] [Google Scholar]
- McKusick VA. HUGO news. The Human Genome Organisation: history, purposes, and membership. Genomics. 1989;5:385–387. doi: 10.1016/0888-7543(89)90077-3. [DOI] [PubMed] [Google Scholar]
- Merdzhanova G, et al. E2F1 controls alternative splicing pattern of genes involved in apoptosis through upregulation of the splicing factor SC35. Cell Death Diff. 2008;15:1815–1823. doi: 10.1038/cdd.2008.135. [DOI] [PubMed] [Google Scholar]
- Modrek B, Lee C. A genomic view of alternative splicing. Nat. Genet. 2002;30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- Rogan PK, et al. Information analysis of human splice site mutations. Hum. Mutat. 1998;12:153–171. doi: 10.1002/(SICI)1098-1004(1998)12:3<153::AID-HUMU3>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Schneider T. Medical Applications of Sequence Walkers: ABCR Mutation G863A. 2005 Available at: http://www.ccrnp.ncifcrf.gov/∼toms/g863a.html. [Google Scholar]
- Schutt C, Nothiger R. Structure, function and evolution of sex-determining systems in Dipteran insects. Development. 2000;127:667–677. doi: 10.1242/dev.127.4.667. [DOI] [PubMed] [Google Scholar]
- Smith CW, et al. Alternative splicing in the control of gene expression. Annu. Rev. Genet. 1989;23:527–577. doi: 10.1146/annurev.ge.23.120189.002523. [DOI] [PubMed] [Google Scholar]
- Sturn A, et al. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18:207–208. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- Tress ML, et al. The implications of alternative splicing in the ENCODE protein complement. Proc. Natl Acad. Sci. USA. 2007;104:5495–5500. doi: 10.1073/pnas.0700800104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wain HM, et al. Guidelines for human gene nomenclature. Genomics. 2002;79:464–470. doi: 10.1006/geno.2002.6748. [DOI] [PubMed] [Google Scholar]
- Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein JN, et al. An information-intensive approach to the molecular pharmacology of cancer. Science. 1997;275:343–349. doi: 10.1126/science.275.5298.343. [DOI] [PubMed] [Google Scholar]
- Wu Y, et al. The vascular endothelial growth factor receptor (VEGFR-1) supports growth and survival of human breast carcinoma. Int. J. Cancer. 2006;119:1519–1529. doi: 10.1002/ijc.21865. [DOI] [PubMed] [Google Scholar]
- Xu Q, et al. Genome-wide detection of tissue-specific alternative splicing in the human transcriptome. Nucleic Acids Res. 2002;30:3754–3766. doi: 10.1093/nar/gkf492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JM, et al. Odorant receptor expressed sequence tags demonstrate olfactory expression of over 400 genes, extensive alternate splicing and unequal expression levels. Genome Biol. 2003;4:R71. doi: 10.1186/gb-2003-4-11-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeberg BR, et al. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 2003;4:R28. doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeberg BR, et al. High-Throughput GoMiner, an ‘industrial-strength’ integrative gene ontology tool for interpretation of multiple-microarray experiments, with application to studies of Common Variable Immune Deficiency (CVID) BMC Bioinform. 2005;6:168. doi: 10.1186/1471-2105-6-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, et al. Alternatively spliced FGFR-1 isoforms differentially modulate endothelial cell activation of c-YES. Arch. Biochem. Biophys. 2006;450:50–62. doi: 10.1016/j.abb.2006.03.017. [DOI] [PubMed] [Google Scholar]