Abstract
Cardiovascular researchers are constantly developing new and innovative medical imaging technologies, striving to improve the understanding, diagnosis, and treatment of cardiovascular dysfunction. Combining these sophisticated imaging methods with advancements in image understanding via computational intelligence will continue to advance the frontier of cardiovascular medicine. Recently, researchers have turned to a new class of tissue motion imaging techniques, including displacement encoding with stimulated echoes (DENSE) in cardiac magnetic resonance (cMR) imaging, to directly quantify cardiac displacement and produce accurate spatiotemporal measurements of myocardial strain, twist, and torsion. The associated analysis of DENSE cMR and other tissue motion imagery, however, represents a major bottleneck in the study of intra-myocardial mechanics. In the computational intelligence area of deformable models, this paper develops an automated motion recovery technique termed active trajectory field models (ATFMs) geared towards these new motion imaging protocols, offering quantitative physiological measurements without the pains of manual analyses. This novel generative deformable model exploits both image information and prior knowledge of cardiac motion, utilizing a point distribution model derived from a training set of myocardial trajectory fields to automatically recover cardiac motion from a noisy image sequence. The effectiveness of the ATFM method is demonstrated by quantifying myocardial motion in 2D short-axis murine DENSE cMR image sequences both before and after myocardial infarction, producing results comparable to existing semi-automatic analysis methods.
Keywords: cardiac magnetic resonance imaging (MRI), DENSE, LV function, myocardial tagging, image analysis, deformable models, active models, computational intelligence
I. Introduction
Modern cardiovascular science boasts a rich array of imaging technology within the suite of medical imaging modalities, providing vital physiological insights which aid in the understanding, diagnosis, and treatment of cardiovascular dysfunction. Each imaging technology affords an opportunity to develop associated image analysis software, which strives to automatically quantify cardiac parameters and produce results comparable to existing manual methods while alleviating researchers and clinicians from tedious, time-consuming, and highly variable manual data analyses. As new and more sophisticated imaging techniques are under constant development, automated cardiovascular image analysis will remain an important and vigorous area of research in computational intelligence for many years to come.
One of the most important cardiovascular imaging modalities today is of course cardiac magnetic resonance (cMR) imaging. cMR techniques already offer a wealth of cardiac measurements, including ejection fraction, myocardial mass, myocardial wall thickening, and cardiac perfusion. A promising addition to the cMR toolbox is found in the study of intra-myocardial function, which attempts to quantify tissue motion throughout the cardiac muscle and produce accurate spatiotemporal measurements of myocardial strain, twist, and torsion. Consequently, many cMR researchers have turned to quantitative tissue tracking techniques, such as myocardial tagging [1-3], velocity-encoded phase contrast imaging [4, 5], harmonic phase (HARP) analysis [6], and the more recent displacement encoding with stimulated echoes (DENSE) [7-10], to potentially advance our current understanding of basic cardiovascular science.
Of particular interest is cine DENSE cMR, which offers direct measurement of tissue displacement at a high spatial resolution throughout the course of the cardiac cycle [10]. A recent cine DENSE cMR protocol for murine imaging by Zhong et al [11] achieves spatial and temporal resolutions of 0.2 × 0.2 × 1 mm3 and 6.9 ms, respectively, in a scan time of 6-8 minutes per slice. Even higher resolutions are possible with this imaging technique, albeit at the cost of increased scan time.
DENSE cMR presents a clear opportunity in computational intelligence for the development of novel automated analysis software, as extracting meaningful tissue motion from acquired imagery remains a complex task requiring significant manual interaction [10]. Though existing medical image analysis methods were considered [12-14] in the context of DENSE cMR, it was deemed necessary to develop an application specific algorithm capable of the automatic recovery of meaningful left-ventricular motion from acquired DENSE cMR image sequences.
In this paper, we consider the automated analysis of acquired tissue motion imagery via a novel generative deformable modeling technique we term active trajectory field models (ATFMs). ATFMs are within a class of methods known in the computational intelligence community as deformable models, which include active contours [12], active shape/appearance models [13, 14], active surfaces [15], and splines [16]. The ATFM method introduced here exploits both image information and prior knowledge of the cardiac motion we wish to recover, using a training set of myocardial trajectory fields to automatically recover tissue motion from a noisy image sequence. This technique is able to eliminate our dependence on finite element approximations of complex biomechanical models by instead deriving a myocardial motion model directly from real world training data.
As with active shape/appearance models, the ATFM technique relies on our ability to represent a motion trajectory field as a discrete set of spatiotemporal points. After choosing points in the same manner from every trajectory field within a training set of fields, we can examine the statistics of these labeled point positions and develop a point distribution model describing how the spatiotemporal locations vary. We are then able to move about in this model space by varying a small number of parameters associated with the modes of variation within the training data. To automatically recover motion from a noisy image sequence, we must choose parameter values that best fit model to noisy imagery.
The development of the ATFM motion recovery technique is wrought with challenges, including the preliminary semi-automatic analysis of a training set of complex spatiotemporal trajectory fields, the characterization of variability within the training set, and a solution to a combinatorial optimization problem of searching for the trajectory field of best fit to a noisy image sequence. As DENSE cMR resolutions are well suited for studies that use imaging of transgenic mice to elucidate the roles of individual genes in contractile function, we focus our efforts on left ventricular motion recovery within 2D short-axis murine cine DENSE cMR imagery, discussed in detail in the next section. Though we concentrate on the analysis of this single imaging technique, the ATFM method can be adapted to 2D human DENSE cMR imagery (differing primarily in scale and resolution), other displacement data such as myocardial tagging (differing primarily in resolution), as well as 3D DENSE cMR motion recovery (requiring an extension to 3D space and 3D displacements)
In the next section, we present necessary background information including an in-depth discussion of a state-of-the-art DENSE cMR analysis, as well as an introduction to deformable models and their application to cardiac segmentation problems. We then develop the proposed ATFM method in detail, including a novel preliminary semi-automatic training set analysis, model definition, and the final automated motion field recovery. We demonstrate the effectiveness of the ATFM method by quantifying myocardial motion in 2D short-axis murine DENSE cMR image sequences both before and after myocardial infarction.
II. Background
This section presents two topics essential to the proposed research: a discussion on the typical cine DENSE cMR image analysis method, and a short review of deformable models for cardiac segmentation.
A. Typical Cine DENSE cMR Analysis
A typical magnetic resonance (MR) acquisition consists of a complex valued dataset, and conventionally researchers and clinicians discard all phase information and make use of the magnitude information only. DENSE cMR, however, exploits this phase space by directly encoding tissue displacement into the phase-reconstructed images [7]. DENSE cMR first uses a spatial magnetic field gradient to impart a location-dependent phase shift to the MR signal at the initial cardiac configuration. A similar gradient is applied at subsequent cardiac configurations such that, if no displacement occurred, the initial imparted phase shift would be removed. Any residual phase shift remaining after application of the second gradient pulse directly reflects tissue displacement that occurred during the time between the two gradient pulses. A more detailed discussion of the cine DENSE cMR acquisition method, beyond the scope of this article, can be found in [8].
Fig. 1 illustrates a typical single-frame DENSE cMR acquisition and analysis. We acquire complex DENSE cMR imagery consisting of magnitude (Fig. 1b) and phase (Fig. 1c) information, where phase is directly proportional to displacement in a single direction. In this example, phase is encoded in the horizontal direction. A similar procedure produces phase imagery encoded in the vertical direction (not shown).
Fig. 1.
Single frame murine DENSE cMR analysis. (a) Short-axis anatomical diagram, (b) magnitude image, (c) horizontal encoded phase image, (d) phase-unwrapped image, and (e) displacement field.
Let us now briefly consider the typical transformation of a 2D+time DENSE cMR image acquisition into meaningful tissue motion as described by Spottiswoode et al. [10]. As MR phase is inherently bounded between –π and π, large displacements produce wrapping artifacts, visible at the 3 o'clock position in Fig. 1c. This wrapping effect is corrected by a two-dimensional phase unwrapping technique described in [17], known as quality-guided path following. This technique unwraps the phase image one pixel at a time along a 2D path guided by the phase quality image, proportional to the variance of the partial derivatives of the locally unwrapped phase within a small neighborhood around each pixel. Fig. 1d illustrates unwrapped phase values within the myocardial region of interest, where dark pixels represent movement to the left and bright pixels represent movement to the right.
After unwrapping the vertically encoded phase image (not shown) via a similar procedure, the associated horizontal and vertical displacements are combined to obtain the displacement field illustrated in Fig. 1e. This field consists of noisy vectors with heads located at pixel centers and tails located at the pixel points of origin. A series of these single frame displacement fields measured over the course of the cardiac cycle is concatenated to recover a meaningful myocardial trajectory field, as illustrated in Fig. 2. This generally involves spatial filtering, linear interpolation to determine frame-to-frame motion, measurement of coarse trajectories using these frame-to-frame vectors, and temporal smoothing of the coarse trajectories via Fourier basis functions.
Fig. 2.
Typical murine DENSE cMR trajectory field.
To gain more insight into myocardial function, strain can be directly calculated from the recovered trajectory fields according to the method described in [18]. Consider a single trajectory [x0 (t), y0 (t)] and its nearest neighbor [x1(t), y1(t)] defined by the Euclidean distance between spatial origins. We define the distance between these points at an arbitrary time t as
| (1) |
A two-dimensional deformation gradient tensor F(t) , of size [2×2], maps vectors from the original configuration dx(0) to the current configuration dx(t) and is defined as
| (2) |
Given a trajectory with at least two neighbors, F(t) can be determined via a least squares technique. The associated LaGrangian strain tensor S(t) , of size [2×2], is given as
| (3) |
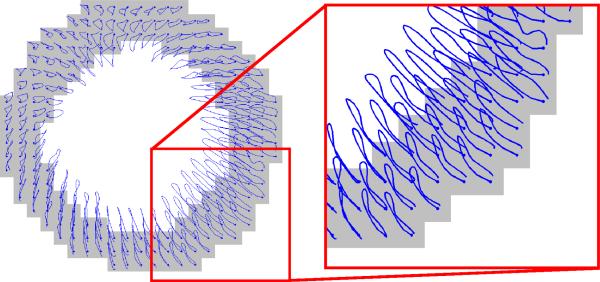
where I is the identity matrix. This 2D Cartesian strain tensor is typically decomposed into its radial and circumferential components, pointing towards the myocardial center and along the circumference of the myocardium, respectively. Fig. 3 illustrates typical end-systolic (full cardiac contraction) strain fields. Note that the average end-systolic radial strain of approximately 0.35 and average end-systolic circumferential strain of approximately -0.15 are in good agreement with previous measurements of strain in the mouse heart [9, 19].
Fig. 3.
End-systolic (left) radial and (right) circumferential strain.
Though this state-of-the-art analysis method provides revealing myocardial trajectory fields, it does suffer from several distinct disadvantages. It first requires a significant amount of user input, as the left ventricle must be manually delineated on at least one frame, and the phase unwrapping algorithm performance is improved when the left ventricle is delineated on all frames. In our lab, manual myocardial segmentation within a typical murine DENSE cMR acquisition of 30 frames may take 5-10 minutes. Performing this segmentation on multiple data sets quickly becomes monotonous and time consuming. Manual segmentation is especially problematic early in the cardiac cycle, as blood encoded with the DENSE cMR pulse sequence remains in the blood pool, virtually eliminating myocardial contrast. Additionally, little effort is directed towards noise compensation among the displacement vectors beyond some spatial filtering and an individual temporal fit to each trajectory. There is a clear opportunity for a more automated solution via advances in computational intelligence.
B. Deformable Models for Cardiac Segmentation
One of the most successful classes of medical image segmentation methods, termed deformable models, attempts to exploit both image information and prior knowledge of the anatomical structure to be delineated. Here, we will consider some of the most relevant prior research on deformable models for myocardial segmentation.
The popularization of deformable models for image segmentation is commonly attributed to the introduction of active contours by Kass et al. [12]. An active contour captures a desired image feature by minimizing a corresponding energy functional, typically the weighted sum of an image-based external energy, attracting the contour to features of interest, and a contour-based internal energy, ensuring a smooth segmentation solution. While active contours have been utilized for cardiac segmentation by a number of researchers [20-22], this method does not allow for the inclusion of any known size, shape, or appearance information.
Cootes et al. [13] introduced a technique termed active shape models (ASMs) that integrates an expected feature shape into the deformable model framework. An ASM measures the variability of a set of training shapes via principle component analysis (PCA), and constrains the segmentation solution to this shape space. Given the proper training set, the corresponding model is able to encapsulate all shape variability within a small subset of principle component weighting parameters. The ASM technique has been used successfully to delineate a number of different anatomical shapes including the myocardium [13, 23], but the location of anatomical features with ASM is driven by often spurious low-level image features.
To improve their segmentation results, Cootes et al. [14] introduced an extension to the ASM technique termed active appearance models (AAMs). This technique considers not only the expected object shape, but also the expected pattern of intensity or color in and around the object termed the object appearance. AAMs belong to a subclass of deformable models termed generative deformable models, as we can generate a synthetic image corresponding to any shape and appearance within the model space. Segmentation via this method proceeds via analysis-by-synthesis, finding the set of model parameters that minimizes the difference between noisy imagery and model-derived synthetic imagery. AAM techniques have shown significant promise in the segmentations of a wide range of cardiac imagery [24-27].
Unfortunately, the aforementioned algorithms are ill-suited to the analysis of cardiac displacement imagery such as DENSE cMR. Myocardial edge contrast is nonexistent early in the cardiac cycle, causing purely edge-based segmentation methods such as active contours and ASMs to fail or require significant correction. AAMs show more promise as they could consider the entire myocardial muscle appearance rather than just the myocardial borders, however we would still require the complex displacement analysis methods discussed in the previous section to recover meaningful myocardial motion from the segmentation solution.
We therefore endeavor to create a novel motion recovery method geared specifically to tissue motion imagery. Of particular interest is the concept of a generative deformable model, as it is a relatively simple matter to generate ideal DENSE cMR phase imagery from a given trajectory field. We hypothesize that it is possible to characterize the variability of a set of training trajectory fields, and then recover cardiac motion via an analysis-by-synthesis technique that minimizes the difference between ideal phase imagery and a noisy DENSE cMR sequence. The ATFM approach described in this paper accomplishes this goal, developing a novel generative deformable model to recover cardiac motion in tissue motion imagery that exploits known intra-myocardial spatiotemporal variability.
III. Methods
This section presents an in-depth description of the proposed ATFM technique, towards the goal of automated motion recovery from tissue displacement imagery. The ATFM method defines a point distribution model characterizing the variability of spatiotemporal myocardial landmarks within a training set of discrete trajectory fields, and then automatically recovers motion from a noisy image sequence by finding the best fit of model to imagery.
The development of this method is not a trivial task, as we must consider the preliminary semi-automatic analysis of a training set of DENSE cMR image sequences, a characterization of variation within recovered trajectory field training set, and a solution to the combinatorial optimization problem of searching a noisy image sequence for the trajectory field of best fit. As previously mentioned, though we focus our efforts on murine DENSE cMR imagery, the ATFM method can be adapted to human data, other tissue motion imaging techniques, as well as 3D motion recovery.
A. Training set analysis
The current state-of-the-art DENSE cMR motion recovery method discussed in Section II.A does not lend itself to the construction of our cardiac point distribution model, as each heart in the training set is sampled at a unique set of spatiotemporal locations. To address this concern, as well improve noise compensation within the acquired datasets, we introduce a novel semi-automatic DENSE cMR analysis method. This method defines a smooth and continuous myocardial trajectory field via two spatiotemporal splines, given phase-unwrapped image sequences and expertly delineated myocardial contours. Note that this novel semi-automatic training set analysis is only a stepping stone towards the fully automated ATFM solution discussed in subsequent sections.
Consider deformation in a single direction. Let {zn , n ∈ [1...N]} represent the finite set of N irregularly spaced spatiotemporal deformations at the Cartesian coordinates (xn , yn) ∈ Ω and at temporal locations tn ∈ [0...1] (varying from the start to the end of the cardiac cycle, respectively). We define the function f (x, y, t) as the unique solution that minimizes
| (4) |
where (λ1,λ2) ∈ [0...1] define the relative weight between energy terms and fdd is the second derivative of f(x, y, t) in the direction d. We apply an additional boundary constraint to our solution that forces trajectories to begin and end at their respective spatial origins by requiring zero valued deformations at these temporal locations.
The first energy term in (4) is derived from a 2D thin plate spline [16, 28], the 2D analog of the 1D cubic spline. This term quantifies the spatial bending energy at each temporal location in the function f (x, y, t) , ensuring a spatially smooth deformation field. The second energy term is derived from a one-dimensional cubic spline [16]. This term quantifies the temporal curvature of our solution, i.e. the sum of squared acceleration across time within the spatial region of interest, ensuring a temporally smooth deformation field when minimized. Our energy functional also includes an L2-norm data constraint, forcing the solution towards the least-squares approximation of the input deformations. Minimizing the weighted sum of these three energy terms produces a spatially and temporally smooth solution that closely matches the input deformation data. We note that although (4) could be formulated in the L1-norm sense, the range and variability of noise present in the displacement data has lent itself to an L2-norm minimization scheme.
To ensure we do not over smooth and lose local tissue motion features, our choice of λ1 and λ2 is governed by the cross-validation technique described in [29]. We divide our entire training set of displacement fields into estimation data and validation data, choosing D random displacements from each field as the validation data. We then search the parameter space for the spatiotemporal splines derived from the estimation data that best fit the validation data.
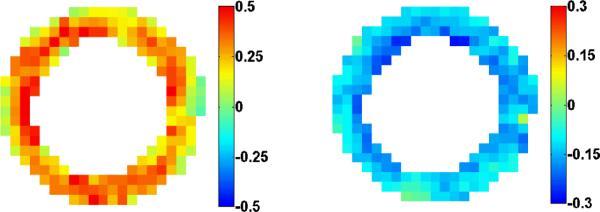
Fig. 4 illustrates a typical spatiotemporal spline fit. The top row shows the original displacement fields, and the bottom row shows the corresponding spline deformation fields. We can additionally calculate strain fields by discretizing this continuous trajectory field and using the method described in Section II.A, as illustrated in Fig. 5.
Fig. 4.
Typical spatiotemporal spline fitting. (top row) Original displacement fields and (bottom row) corresponding spline fields.
Fig. 5.
End-systolic (left) radial and (right) circumferential strain using the spatiotemporal spline method.
B. Model definition
With a training set of continuous spatiotemporal trajectory fields in hand, we may now consider the construction of a trajectory field model for subsequent motion recovery [30]. This is a three step process, wherein we align the training data to a single reference coordinate system, discretize each continuous trajectory field at the same set of myocardial landmarks and cardiac phases, and define a point distribution model that measures the variability among these discrete trajectory fields via PCA.
Let us first consider training data alignment. The center of each resting heart within the training set lies at a different spatial location within its respective image acquisition space. Additionally, each resting heart has a different orientation with respect to this center. Finally, two hearts that differ in size may still exhibit the same motion characteristics. To more accurately characterize motion variability within the training set, we eliminate these differences in cardiac position, orientation, and scale by aligning each resting heart to a single reference coordinate system. We refer to the resulting aligned trajectory field as normalized.
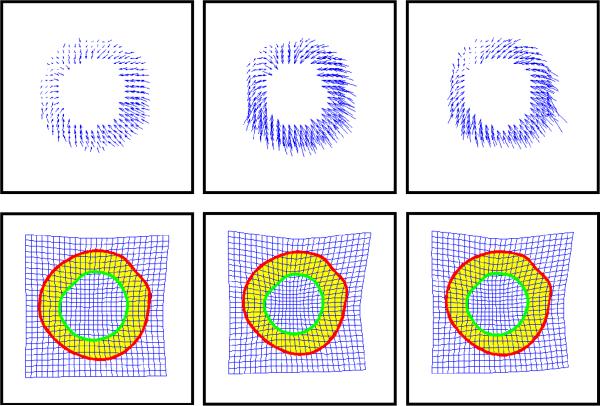
As it is difficult to manually delineate the myocardium in DENSE cMR imagery early in the cardiac cycle, we define the resting heart by projecting expert drawn contours and features (right ventricle insertion points, see Fig. 1a) from the frame of greatest myocardial contrast back to their resting positions via the spatiotemporal spline definition. Alignment of this resting configuration is achieved via a Procrustes analysis [13, 31], with respect to a reference myocardium centered at the spatial origin with right-ventricle insertion points aligned to the vertical axis and unity epicardial radius. Fig. 6 illustrates a typical alignment, transforming a resting configuration and motion field (Fig. 6a-b) to the reference coordinate system (Fig. 6c-d). To aid visualization, the right ventricle insertion points are marked as squares.
Fig. 6.
Alignment of training data to reference coordinate system. (a) Expert segmentation at rest, (b) corresponding trajectory field, (c) aligned resting segmentation, (d) aligned trajectory field.
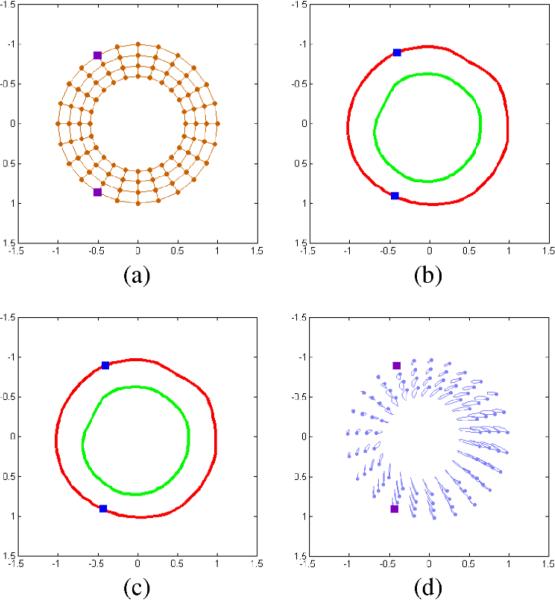
After alignment, the next challenge in model construction is the spatial and temporal discretization of each trajectory field within the training set using an identical set of myocardial- landmarks. To elaborate, if trajectory #10 in the first training field corresponds to the anterior right ventricle insertion point, trajectory #10 in all training fields must correspond to the same anterior right ventricle insertion point. We achieve this trajectory matching by mapping a sampling template to the aligned resting myocardial definition, and recovering the discrete trajectories associated with each of these spatial origins. Fig. 7 illustrates this procedure, using a sampling template (Fig. 7a) to sample the normalized resting myocardial definition (Fig. 7b) and produce a discrete set of resting spatial locations (Fig. 7c) and a corresponding trajectory field (Fig. 7d). Temporal discretization is achieved by sampling each training trajectory field at a known set of cardiac phases.
Fig. 7.
Trajectory field discretization. (a) Sampling template, (b) normalized myocardial borders, (c) discretized myocardial definition, (d) corresponding trajectory field
Given the set of normalized discrete training trajectory fields, we are able to characterize the data variability via principal component analysis (PCA). Let Xijk = [xijk , yijk] represent the x-y position of the ith trajectory at the jth cardiac phase of the kth normalized trajectory field, where i ∈ [1...Ni] , j ∈ [1...Nj] , and k ∈ [1...Nk] . We vectorize the kth trajectory field as
| (5) |
of size [(2 · Ni · Nj) × 1] . The average normalized trajectory field is defined as
| (6) |
and the sample distribution covariance matrix is defined by
| (7) |
The eigenvectors P of the covariance matrix define the modes of variation within the training set, and the corresponding eigenvalues describe the relative significance of each mode. The percentage of variation accounted for by each eigenvector is defined as the corresponding eigenvalue divided by the sum of all eigenvalues.
We can recover any normalized trajectory field within the training set by a linear combination of the average normalized trajectory field with the modes of variation. Moreover, we can approximate any normalized trajectory field within the training data by a linear combination of the average normalized trajectory field with the most significant eigenvectors, termed principle components, as in
| (8) |
where P̂ is a subset of the modes of variation P , and b̂k is set of weighting coefficients corresponding to the kth trajectory field. In practice, we define an amount of variation we wish to approximate (e.g. 95% variation), and use the principle components that account for this level of variation.
These principle components of variation, combined with a rotation, translation, and scale to transform the normalized trajectory fields back to the image space of a new DENSE cMR sequence, define our trajectory field model. We are able to achieve any trajectory field within the search space by varying a small number of parameters, i.e. the principle component weights, orientation, translation, and scale.
C. ATFM motion recovery
Given the aforementioned model space definition, we are able to attempt the automatic recovery of cardiac motion from a newly acquired DENSE cMR sequence. In this section, we will first consider the search criterion which defines the best match between trajectory field and noisy imagery. We then discuss our method of attack to solve the combinatorial optimization problem of locating the best match within the model search space.
We measure the correspondence between a given trajectory field and a noisy image sequence via an analysis-by-synthesis technique. Given any myocardial trajectory field, one can synthesize a set of “ideal” DENSE cMR values which represent the noise-free DENSE cMR data that would have produced the trajectory field in question. This is accomplished by measuring displacement from the spatial origin for each trajectory at each phase of the cardiac cycle, and scaling according to the DENSE cMR encoding parameter. Let Ψ = {[xn(t), yn(t)], n ∈ [1...N]} represent a set of N temporally continuous trajectories. The ideal DENSE cMR value corresponding to each trajectory at some time t are defined as
| (9) |
where and are the x-direction and y-direction ideal values, respectively, and ke is the DENSE cMR encoding parameter.
Let Ix(x, y, t) and Iy(x, y, t) represent the discrete noisy DENSE cMR image sequence encoded in the x-direction and y-direction, respectively, where (x, y) ∈ Ψ and t takes on k ∈ [1...Nk] unique values within the range [0...1] . We define the distance D(·) ∈ [0, ∞] of a given trajectory field Ψ to a noisy DENSE cMR sequence in the L2-norm sense as
| (10) |
To determine the trajectory field of highest similarity to a given noisy DENSE cMR image sequence, we must traverse the combinatorial optimization space of the trajectory field model to locate the field of minimum distance defined by (10). A direct search of the model space for the globally minimum distance is computationally prohibitive. We therefore search the model space via simulated annealing [32], analogous to the metallurgical annealing process wherein hot metal is slowly cooled to form a perfect crystalline structure with minimum free energy. In simulated annealing, we slowly “cool” our model search to locate the point within the model space of minimum energy, where “coolness” reflects the decreasing probability of moving to an inferior solution in terms of an energy measure that quantifies solution quality.
Let Ψ1 represent the current trajectory field state of our model search, with some distance to the noisy DENSE cMR imagery D(Ψ1). We perturb the model parameters associated with this trajectory field, obtaining a new trajectory field Ψ2 with corresponding distance D(Ψ2), and consider changing the state of our system to this new trajectory field. If we accepted only good moves, i.e. when D(Ψ2) < D(Ψ1), we would quickly find a local minima within our search space, but would become trapped in this local minima and unable to discover the global minimum. In simulated annealing we utilize a more sophisticated acceptance condition, i.e. changes in the model state are accepted if
| (11) |
where T is the current system temperature, and U(0,1) is a uniformly distributed random variable between 0 and 1. At high temperatures, all changes have an equal probability of being accepted. As the temperature is lowered, moves that increase the system energy have a lower probability of acceptance, until the algorithm reduces to the greedy method at T = 0 accepting only moves that reduce the system energy.
We begin the annealing process at some initial temperature T0 (in which every possible change to the system state is equally probable), and follow a geometric annealing schedule which reducing the temperature by some constant τ (Tk+1 = τ · Tk) at each temperature until a final temperature Tfinal is reached. At each temperature, we test NC candidate points within the model space. When Tfinal is achieved, the algorithm is essentially a greedy one, for which achieving a local minimum is guaranteed. To provide a small reduction in the search space dimensionality, we define the orientation, position, and scaling of the normalized trajectory field model to the DENSE cMR image space via a small number of user defined myocardial landmarks. The computational complexity of this annealing algorithm scales linearly according to the number of principle components used, the number of candidate point tested at each temperature, and the number of temperatures evaluated.
IV. Results
To demonstrate the effectiveness of the ATFM technique for tissue motion recovery within displacement imagery, we attempted to quantify myocardial motion in 2D short-axis murine DENSE cMR imagery. We considered a dataset containing two distinct murine conditions, healthy mice and mice seven days after induction of an experimental heart attack. The former condition consisted of 13 healthy mice imaged with a standard 2D cine DENSE cMR protocol, obtaining between 20 and 27 short-axis mid-plane images of the cardiac cycle per mouse. The latter condition consisted of six unhealthy mice with an induced myocardial infarction in the anterolateral wall, again imaged with a standard 2D cine DENSE cMR protocol, obtaining between 17 and 21 short-axis mid-plane images of the cardiac cycle per mouse. For each image sequence, a trained technician delineated the endocardial and epicardial borders on every frame and labeled the right ventricle insertion points on the last sequence frame.
In this section, we present our findings. We first illustrate the failings of traditional segmentation methods towards the goal of automated cardiac motion recovery. We then present a physiological comparison of DENSE cMR analysis techniques, evaluating agreement between the traditional semi-automatic DENSE cMR analysis described in Section II.A and the novel ATFM technique presented within this paper.
A. Motion Analysis via Traditional Automated Segmentation
Semi-automatic DENSE cMR analysis can be broken into two distinct steps: 1) the manual delineation of myocardial borders within the magnitude imagery throughout the cardiac cycle by a trained technician, and 2) tissue motion recovery via phase analysis as described in Sections II.A or III.A. This natural division implies a plausible alternative for automated tissue motion recovery, i.e. the delineation of endocardial and epicardial borders via a traditional automated segmentation technique followed by DENSE cMR phase analysis. Many researchers have had significant success with deformable models for cMR segmentation [20, 22-25], and thus we chose to examine such an alternative.
To evaluate the effectiveness of such a system, we considered myocardial segmentation via the classical active contour segmentation technique described in [12]. The active contour was driven by a negative gradient magnitude external force, derived from DENSE cMR magnitude imagery. Endocardial and epicardial borders were separately initialized on each frame of a given image sequence as circles particularly close to the correct myocardial locations. After segmentation, we applied the traditional DENSE cMR phase analysis method described in Section II.A to recover tissue motion.
This traditional segmentation technique was unable to accurately define myocardial borders on every image frame of any healthy DENSE cMR image sequence, which rendered the subsequent trajectory fields meaningless. Across the dataset, endocardial and epicardial active contour segmentation had root mean square errors of 0.87mm and 0.38mm respectively, exceedingly large as compared to the average myocardial wall thickness of ~2mm.
Fig. 8 illustrates two typical segmentation problems. Fig. 8a shows an attempted segmentation early in the cardiac cycle, which fails due to a lack of myocardial contrast. Fig. 8b shows an attempted segmentation at the end of cardiac cycle, which fails as the epicardial border is drawn to the higher edge strength of the endocardial border. These failures lead us to conclude that a more comprehensive tissue motion analysis solution is required.
Fig. 8.
Example active contour segmentation (a) early in the cardiac cycle and (b) late in the cardiac cycle
B. ATFM Motion Recovery Analysis
Let us now consider the evaluation of the novel automated ATFM motion recovery technique. As we desire to automatically reproduce physiologically meaningful measurements of cardiac motion similar to traditional semi-automatic techniques, we compare strain values measured by both the automated ATFM technique and a traditional semi-automatic DENSE cMR analysis. We will quantify agreement between the two methods via correlation and a Bland-Altman analysis [33].
The preliminary analysis of the ATFM training data utilized the same manually delineated myocardial borders and myocardial features of interest as the traditional DENSE cMR analysis. ATFM analysis was performed via a leave-one-out cross-validation study, attempting motion recovery on each noisy DENSE cMR image sequence within the data set using the remaining trajectory fields as training data. Parameters used for a given motion recovery technique remained consistent across the entire dataset.
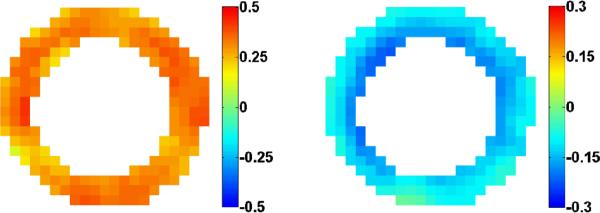
Spatiotemporal strain was calculated at many points within the myocardium as described in Section II.A. For comparison across motion recovery methods, we divide the left-ventricle into six segments, as standardized in [34] and illustrated in Fig. 9a. We quantify and plot the average radial and circumferential strain in each segment throughout the cardiac cycle, as illustrated in Fig. 9b and Fig. 9c respectively.
Fig. 9.
Typical six-segment strain analysis. (a) Anatomical diagram, (b) radial and (c) circumferential segmental strain.
As a quantitative comparison, we sample 10 evenly spaced cardiac phases throughout the cardiac cycle for each left-ventricular segment, resulting in 60 data points per heart. The two methods show good correlation, as illustrated in Fig. 10a-b. The average correlation value of radial strain is 0.83 (p<0.001), and the average correlation value of circumferential strain is 0.86 (p < 0.001). A Bland-Altman analysis of the data, shown in Fig. 10c-d, reveals further similarity between the two methods. We measure an average difference of -0.02 in radial strain (95% confidence interval of –0.16 to 0.11) and an average difference of <0.01 in circumferential strain (95% confidence interval of – 0.05 to 0.07) between the two methods.
Fig. 10.
Strain correlation of traditional and ATFM methods. Radial/circumferential (a-b) correlation and (c-d) Bland-Altman analyses
V. Conclusions
The medical image analysis field continues to benefit from the advent of new and exciting medical imaging techniques. One class of techniques, tissue motion imaging such as DENSE cMR, boasts the ability to quantify tissue motion throughout the cardiac muscle and produce accurate spatiotemporal measurements of myocardial strain, twist, and torsion. Though current manual and semi-automatic analysis methods can generate remarkable trajectory fields, researchers and clinicians would benefit from a fully automated analysis software package.
As an advance in computational intelligence in biomedicine, this paper has presented a novel generative deformable modeling technique, termed active trajectory field models (ATFM), for the automated analysis of acquired tissue motion imagery. The development of this technique was a complex task, requiring a novel preliminary semi-automatic DENSE cMR analysis technique, the alignment and subsequent variability characterization of a complex set of spatiotemporal trajectory fields, and the solution of a combinatorial optimization problem to find the trajectory field of best fit to a given noisy image sequence.
We validated the ATFM method by quantifying myocardial motion in 2D short-axis murine DENSE cMR image sequences both before and after myocardial infarction, producing results comparable to existing semi-automatic analysis methods. Though we focused our efforts on left ventricular motion recovery within 2D short-axis murine cine DENSE cMR imagery, the ATFM method can be adapted to human data, other tissue motion imaging techniques, as well as 3D motion recovery.
Acknowledgement
The authors would like to thank several researchers at the University of Virginia for their continued support: Dr. B. A. French of the Depts. of Biomedical Engineering, Radiology and Medicine; Dr. J.A. Hossack of the Dept. of Biomedical Engineering; and Dr. C. M. Kramer of the Depts. of Radiology and Medicine.
This work was supported in part by the National Institutes of Health (NIH) under Grant EB 001826 and Grant R01 EB 001763.
Biographies
 Andrew D. Gilliam (M '05) received the B.S. degree in electrical engineering from the University of Virginia in 2002, the M.S. degree in electrical engineering from the University of Illinois at Urbana-Champaign in 2004, and the Ph.D. degree from the University of Virginia in 2008.
Andrew D. Gilliam (M '05) received the B.S. degree in electrical engineering from the University of Virginia in 2002, the M.S. degree in electrical engineering from the University of Illinois at Urbana-Champaign in 2004, and the Ph.D. degree from the University of Virginia in 2008.
He is currently an independent image analysis consultant. His research interests include cardiac MRI and echocardiographic image analysis and image analysis software development. He currently resides in Providence, Rhode Island with his wife.
 Frederick H. Epstein received the B.A. and B.S. degrees in mathematics and physics, respectively, from the University of Rochester in 1988. Subsequently, he received the M.S. degree in engineering physics and the Ph.D. degree in biomedical engineering from the University of Virginia in 1990 and 1993, respectively.
Frederick H. Epstein received the B.A. and B.S. degrees in mathematics and physics, respectively, from the University of Rochester in 1988. Subsequently, he received the M.S. degree in engineering physics and the Ph.D. degree in biomedical engineering from the University of Virginia in 1990 and 1993, respectively.
He was a Senior Engineer and Research Scientist at GE Medical Systems from 1994-1999, and a staff scientist at the National Heart, Lung, and Blood Institute from 1999-2000. He returned to the University of Virginia as an Associate Professor of Radiology and Biomedical Engineering in 2000.
In 2005, Dr. Epstein was named an Established Investigator of the American Heart Association, and in 2007 he was inducted into the University of Virginia's Academy of Distinguished Educators. Dr. Epstein is currently the Chair of the Science Committee of the Society for Cardiovascular Magnetic Resonance, and the Chair of the Cardiac MR Study Group of the International Society for Magnetic Resonance in Medicine. His research interests include MRI of cardiac function and perfusion, as well as molecular and cellular MRI in heart disease. Dr. Epstein resides in Charlottesville, Virginia with his wife and two children.
 Scott T. Acton (SM '99) received the Graduate degree from Oakton High School, Vienna, VA, in 1984, the B.S. degree in electrical engineering from Virginia Tech, Blacksburg, in 1988, and the M.S. and Ph.D. degrees in electrical and computer engineering from the University of Texas at Austin, Austin, in 1990 and 1993, respectively.
Scott T. Acton (SM '99) received the Graduate degree from Oakton High School, Vienna, VA, in 1984, the B.S. degree in electrical engineering from Virginia Tech, Blacksburg, in 1988, and the M.S. and Ph.D. degrees in electrical and computer engineering from the University of Texas at Austin, Austin, in 1990 and 1993, respectively.
He was in industry with AT&T, Oakton, VA; the MITRE Corporation, McLean, VA; and Motorola, Inc., Phoenix, AZ. He was also with Oklahoma State University, Stillwater. He is currently a Professor of electrical and computer engineering, and biomedical engineering at the University of Virginia (UVa), Charlottesville. During 2007–2008, he was on sabbatical in Santa Fe, NM. His current research interests include anisotropic diffusion, basketball, active models, biomedical segmentation problems, and biomedical tracking problems.
Prof. Acton is an Associate Editor for the IEEE Transactions on Image Processing. He was also an Associate Editor of the IEEE Signal Processing Letters. He was the 2004 Technical Program Chair and the 2006 General Chair for the Asilomar Conference on Signals, Systems and Computers. At UVa, he was named a Virginia Scholar, the Outstanding New Teacher in 2002, a Faculty Fellow in 2003, and the Walter N. Munster Chair for Intelligence Enhancement in 2003.
Footnotes
Color versions of one or more of the figures in this paper are available online at http://ieeexplore.ieee.org.
Contributor Information
Andrew D. Gilliam, C. L. Brown Department of Electrical and Computer Engineering, University of Virginia, Charlottesville, VA 22904 USA. He resides in Providence, RI 02906 USA (drew.gilliam@virginia.edu)..
Frederick H. Epstein, Department of Radiology and the Department of Biomedical Engineering, University of Virginia, Charlottesville,VA22904USA (fredepstein@virginia.edu).
Scott T. Acton, C. L. Brown Department of Electrical and Computer Engineering and the Department of Biomedical Engineering, University of Virginia, Charlottesville, VA 22904 USA (acton@virginia.edu)..
REFERENCES
- 1.Axel L, Dougherty L. MR imaging of motion with spatial modulation of magnetization. Radiology. 1989;171:841–845. doi: 10.1148/radiology.171.3.2717762. [DOI] [PubMed] [Google Scholar]
- 2.Axel L, Dougherty L. Heart wall motion: improved method of spatial modulation of magnetization for MR imaging. Radiology. 1989;172:349–350. doi: 10.1148/radiology.172.2.2748813. [DOI] [PubMed] [Google Scholar]
- 3.Zerhouni. EA, Parish DM, Rogers WJ, Yang A, Shapiro EP. Human heart: tagging with MR imaging--a method for noninvasive assessment of myocardial motion. Radiology. 1988;169:59–63. doi: 10.1148/radiology.169.1.3420283. [DOI] [PubMed] [Google Scholar]
- 4.Dijk PV. Direct cardiac NMR imaging of heart wall and blood flow velocity. Journal of Computer Assisted Tomography. 1984;8:429–436. doi: 10.1097/00004728-198406000-00012. [DOI] [PubMed] [Google Scholar]
- 5.Bryant D, Payne J, Firmin D, Longmore D. Measurement of flow with NMR imaging using gradient pulse and phase difference technique. Journal of Computer Assisted Tomography. 1984;8:588–593. doi: 10.1097/00004728-198408000-00002. [DOI] [PubMed] [Google Scholar]
- 6.Osman NF, McVeigh ER, Prince JL. Imaging heart motion using harmonic phase MRI. Medical Imaging, IEEE Transactions on. 2000;19:186. doi: 10.1109/42.845177. [DOI] [PubMed] [Google Scholar]
- 7.Aletras AH, Ding S, Balaban RS, Wen H. DENSE: Displacement Encoding with Stimulated Echoes in Cardiac Functional MRI. Journal of Magnetic Resonance. 1999;137:247. doi: 10.1006/jmre.1998.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim D, Gilson WD, Kramer CM, Epstein FH. Myocardial Tissue Tracking with Two-dimensional Cine Displacement-encoded MR Imaging: Development and Initial Evaluation. Radiology. 2004;230:862–871. doi: 10.1148/radiol.2303021213. [DOI] [PubMed] [Google Scholar]
- 9.Gilson WD, Yang Z, French BA, Epstein FH. Measurement of myocardial mechanics in mice before and after infarction using multislice displacement-encoded MRI with 3D motion encoding. Am J Physiol Heart Circ Physiol. 2005;288:H1491–1497. doi: 10.1152/ajpheart.00632.2004. [DOI] [PubMed] [Google Scholar]
- 10.Spottiswoode BS, Zhong X, Hess AT, Kramer CM, Meintjes EM, Mayosi BM, Epstein FH. Tracking Myocardial Motion From Cine DENSE Images Using Spatiotemporal Phase Unwrapping and Temporal Fitting. Medical Imaging, IEEE Transactions on. 2007;26:15. doi: 10.1109/TMI.2006.884215. [DOI] [PubMed] [Google Scholar]
- 11.Zhong X, Janiczek R, French B, Roy R, Kramer C, Meyer C, Epstein F. Spiral Cine DENSE MRI at 7T for Quantification of Regional Function in the Mouse Heart. Proceedings 16th Scientific Meeting, International Society for Magnetic Resonance in Medicine. 2008:580. [Google Scholar]
- 12.Kass M, Witkin A, Terzopoulos D. Snakes: Active Contour Models. International Journal of Computer Vision. 1988;1:321–331. [Google Scholar]
- 13.Cootes TF, Taylor CJ, Cooper DH, Graham J. Active Shape Models-Their Training and Application. Computer Vision and Image Understanding. 1995;61:38. [Google Scholar]
- 14.Cootes TF, Edwards GJ, Taylor CJ. Active appearance models. Pattern Analysis and Machine Intelligence, IEEE Transactions on. 2001;23:681. [Google Scholar]
- 15.Terzopoulos D, Witkin A, Kass M. Constraints on deformable models-recovering 3D shape and nonrigid motion. Artificial Intelligence. 1988;36:91–123. [Google Scholar]
- 16.Wahba G. Spline models for observational data. Society for Industrial and Applied Mathematics (SIAM); Philadelphia: 1990. [Google Scholar]
- 17.Ghiglia DC, Pritt MD. Two-Dimensional Phase Unwrapping: Theory, Algorithms and Software. Wiley-Interscience; New York: 1998. [Google Scholar]
- 18.Truesdell C, Noll W. The non-linear field theories of mechanics. Springer; New York: 2004. [Google Scholar]
- 19.Zhou R, Pickup S, Glickson JD, Scott CH, Ferrari VA. Assessment of global and regional myocardial function in the mouse using cine and tagged MRI. Magnetic Resonance in Medicine. 2003;49:760–764. doi: 10.1002/mrm.10423. [DOI] [PubMed] [Google Scholar]
- 20.Pluempitiwiriyawej C, Moura JMF, Yi-Jen Lin W, Chien H. STACS: new active contour scheme for cardiac MR image segmentation. Medical Imaging, IEEE Transactions on. 2005;24:593. doi: 10.1109/TMI.2005.843740. [DOI] [PubMed] [Google Scholar]
- 21.McInerney T, Terzopoulos D. A dynamic finite element surface model for segmentation and tracking in multidimensional medical images with application to cardiac 4D image analysis. Computerized Medical Imaging and Graphics. 1995;19:69. doi: 10.1016/0895-6111(94)00040-9. [DOI] [PubMed] [Google Scholar]
- 22.Chalana V, Linker DT, Haynor DR, Yongmin K. A multiple active contour model for cardiac boundary detection on echocardiographic sequences. Medical Imaging, IEEE Transactions on. 1996;15:290. doi: 10.1109/42.500138. [DOI] [PubMed] [Google Scholar]
- 23.Cootes TF, Hill A, Taylor CJ, Haslam J. Use of active shape models for locating structures in medical images. Image and Vision Computing. 1994;12:355. [Google Scholar]
- 24.Mitchell SC, Lelieveldt BPF, van der Geest RJ, Bosch HG, Reiver JHC, Sonka M. Multistage hybrid active appearance model matching: segmentation of left and right ventricles in cardiac MR images. Medical Imaging, IEEE Transactions on. 2001;20:415. doi: 10.1109/42.925294. [DOI] [PubMed] [Google Scholar]
- 25.Mitchell SC, Bosch JG, Lelieveldt BPF, van der Geest RJ, Reiber JHC, Sonka M. 3-D active appearance models: segmentation of cardiac MR and ultrasound images. Medical Imaging, IEEE Transactions on. 2002;21:1167. doi: 10.1109/TMI.2002.804425. [DOI] [PubMed] [Google Scholar]
- 26.Bosch JG, Mitchell SC, Lelieveldt BPF, Nijland F, Kamp O, Sonka M, Reiber JHC. Automatic segmentation of echocardiographic sequences by active appearance motion models. Medical Imaging, IEEE Transactions on. 2002;21:1374. doi: 10.1109/TMI.2002.806427. [DOI] [PubMed] [Google Scholar]
- 27.Beichel R, Bischof H, Leberl F, Sonka M. Robust active appearance models and their application to medical image analysis. Medical Imaging, IEEE Transactions on. 2005;24:1151. doi: 10.1109/TMI.2005.853237. [DOI] [PubMed] [Google Scholar]
- 28.Bookstein FL. Principal warps: thin-plate splines and the decomposition of deformations. Pattern Analysis and Machine Intelligence, IEEE Transactions on. 1989;11:567. [Google Scholar]
- 29.Acton ST, Bovik AC. Piecewise and local image models for regularized image restoration using cross-validation. Image Processing, IEEE Transactions on. 1999;8:652. doi: 10.1109/83.760313. [DOI] [PubMed] [Google Scholar]
- 30.Gilliam AD, Acton ST. Murine Spatiotemporal Cardiac Segmentation. Asilomar Conference on Signals, Systems and Computers (ACSSC) 2007 [Google Scholar]
- 31.Gower JC, Dijksterhuis GB. Procrustes problems. Oxford University Press; New York: 2004. [Google Scholar]
- 32.Aarts E, Korst J. Simulated annealing and Boltzmann machines: a stochastic approach to combinatorial optimization and neural computing. John Wiley & Sons, Inc.; 1989. [Google Scholar]
- 33.Bland J, Altman D. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;i:307–310. [PubMed] [Google Scholar]
- 34.Cerqueira MD, Weissman NJ, Dilsizian V, Jacobs AK, Kaul S, Laskey WK, Pennell DJ, Rumberger JA, Ryan T, Verani MS. Standardized Myocardial Segmentation and Nomenclature for Tomographic Imaging of the Heart: A Statement for Healthcare Professionals From the Cardiac Imaging Committee of the Council on Clinical Cardiology of the American Heart Association. Circulation. 2002;105:539–542. doi: 10.1161/hc0402.102975. [DOI] [PubMed] [Google Scholar]