Abstract
Specification of the peripheral optic cup by Wnt signaling is critical for formation of the ciliary body/iris. Identification of marker genes for this region during development provides a starting point for functional analyses. During transcriptional profiling of single cells from the developing eye, two cells were identified that expressed genes not found in most other single cell profiles. In situ hybridizations demonstrated that many of these genes were expressed in the peripheral optic cup in both early mouse and chicken development, and in the ciliary body/iris at subsequent developmental stages. These analyses indicate that the two cells probably originated from the developing ciliary body/iris. Changes in expression of these genes were assayed in embryonic chicken retinas when canonical Wnt signaling was ectopically activated by CA-β-catenin. Twelve ciliary body/iris genes were identified as upregulated following induction, suggesting they are excellent candidates for downstream effectors of Wnt signaling in the optic cup.
Keywords: ciliary body, iris, canonical Wnt signaling, peripheral optic cup, single cell profiling
INTRODUCTION
Formation of the eye is a complex and coordinated process that results from the interaction of cells from several distinct sources during the early embryonic period (Chow and Lang, 2001). Among these cellular interactions, neuroectoderm-derived cells in the eye field, located at the anterior/ventral sides of the neural tube, undergo extensive cell proliferation and morphogenetic changes, and develop into the neural retina, as well as non-neural tissues, such as the retinal pigmented epithelium (RPE), ciliary body, and iris (Chow and Lang, 2001; Davis-Silberman and Ashery-Padan, 2008). The ciliary body and iris are part of the anterior segment structures of the eye and the ciliary body is required for accommodation, the process by which the lens changes shape, allowing for changes in focal length. The ciliary body and iris comprise the inner bilayered epithelia and the overlying stroma. The inner two layers originate from the inner and outer layers of the optic cup, which is derived from the ventral forebrain. The inner non-pigmented layer of the ciliary epithelium is continuous with the inner pigmented iris epithelium and the retina. The outer pigmented layer of the ciliary epithelium is continuous with the outer iris epithelium and RPE (Beebe, 1986). The third layer derives primarily from the neural crest, which contributes melanocytes and connective tissue cells to the iris, as well as connective tissue and muscle to the ciliary body. However, this layer is a mixture of different cells, some of which are derived from the mesoderm (Gage et al., 2005).
The primary role of the ciliary body is the production and secretion of the aqueous humor of the posterior chamber, which then flows to the anterior chamber where it maintains the intraocular pressure (IOP) of the eye. Maintenance of the proper IOP is important both for the normal functioning of the adult eye (Gould et al., 2004) and the continued growth of the developing embryonic eye (Coulombre and Coulombre, 1957). In addition to its important role in IOP establishment and maintenance, the ciliary body also is responsible for synthesizing many proteins of the inner limiting membrane, a structure that is crucial for the survival of retinal ganglion cells (Halfter et al., 2005). Intriguingly, recent studies suggest that the ciliary body and iris harbor cells with stem cell/progenitor characteristics (Ahmad et al., 1999; Tropepe et al., 2000; Sun et al., 2006).
The iris is responsible for controlling the amount of light that passes through the lens and strikes the retina, operating as a shutter, analogous to the role of the shutter in a camera. It is also involved in the circulation of the aqueous humor that is secreted by the ciliary body (Davis-Silberman and Ashery-Padan, 2008). The iris is composed of several cellular layers. The pigmented epithelium of the iris is on lens/retinal side of the tissue, abutting the outer layers, which are composed of the iris muscles and stroma. The iris root is attached directly to the ciliary body. An intriguing and unique developmental plasticity is exhibited by the iris. The pigmented, postmitotic cells of the immature iris transdifferentiate into the sphincter and dilator muscles, the muscles that open and close the iris (Volpe et al., 1993). During development, the iris cells lose their pigment, enter mitosis, and become these peripheral muscles. These iris-derived muscles are the only known ectodermally derived muscles of the body (Imaizumi and Kuwabara, 1971). Developmental abnormalities of the anterior segment structures are readily observed in humans (Idrees et al., 2006). Notably, for many of these disorders involving the anterior segment, there is a high occurrence of associated glaucoma (Gould et al., 2004).
Glaucoma, a disease characterized by the degeneration of the optic nerve, is usually linked to an increase in IOP in the anterior chamber, possibly controlled by aqueous humor secretion of the ciliary body (Gould et al., 2004). Another disorder, aniridiae, is commonly caused by the mutations in the Pax6 gene and is characterized by a partial or complete loss of the iris (Davis-Silberman and Ashery-Padan, 2008). While the disorders described above are the most common, there exist other ocular diseases that have been linked to developmental defects in the peripheral tissues (Gould et al., 2004; Idrees et al., 2006; Davis-Silberman and Ashery-Padan, 2008), although the precise mutations causing these disorders have yet to be identified.
Although the ciliary body and iris play critical roles in vision and might harbor the source of retinal stem cells, our understanding of the molecular mechanisms governing the development of these tissues remains limited. It has been shown that the perturbation of several distinct signaling pathways can disrupt the formation of the ciliary body and iris. In particular, BMP signaling was the first to be demonstrated to have an important role in this process (Zhao et al., 2002). The ciliary body was completely absent in transgenic mice engineered to overexpress Noggin, a BMP antagonist, using a lens specific promoter (Zhao et al., 2002). BMP signaling appears to involve FGFs as forced expression of Fgf4 from the RPE showed that there is an interplay between FGF and BMP in the generation of the ciliary body domain (Dias da Silva et al., 2007). Finally, the canonical Wnt signaling pathway has also been shown to play a crucial role in the determination of peripheral ciliary/iris epithelial fates during early optic vesicle/cup development (Liu et al., 2003; Cho and Cepko, 2006; Liu et al., 2007).
In addition to the links between ciliary body development and several signaling pathways, there are a handful of transcription factors that have been shown to be necessary for formation of the anterior segment structures of the eye (Napier and Kidson, 2007). Mice that lack Otx1, for instance, have smaller irises than their wildtype counterparts and are devoid of ciliary processes (Acampora et al., 1996). Similarly, mice mutant for Lmx1b, a LIM homeodomain transcription factor, also display iris and ciliary body hypoplasia (Pressman et al., 2000). Interestingly, in humans, haploinsufficiency at the LMX1B locus results in nail-patella syndrome, an inherited condition characterized by developmental defects in the nails, kneecaps and elbows (McIntosh et al., 1998; Vollrath et al., 1998). In many of these patients there is also an associated rise in IOP and an increased risk for developing open angle glaucoma (Dreyer et al., 1998; McIntosh et al., 1998; Vollrath et al., 1998).
To identify candidate genes expressed in a variety of ocular tissues during development, we have profiled single cells from the developing eye. A post hoc classification scheme was used to identify the single cell profiles based upon clusters of gene expression (Trimarchi et al., 2008). During this analysis, it became apparent that two of the single cells expressed sets of genes that distinguished them from the other profiled cells. By comparing the expression profiles of these cells to many other retinal cell types, candidate genes to determine the identity of these cells were determined. Section in situ hybridization (ISH) both in the mouse and the chick was used to demonstrate the expression of these candidate genes in the developing ciliary body/iris. Therefore, these cells most likely originate in the developing ciliary body/iris and provide sets of marker genes expressed during the development of this tissue. In addition, we examined the expression of these genes in a chick retina overexpressing a constitutively activated β-catenin (CA-β-catenin) (Capdevila et al., 1998). Previously, CA-β-catenin was shown to convert retinal progenitor cells to more peripheral fates (Cho and Cepko, 2006; Liu et al., 2007). Therefore, ectopic expression of the newly identified genes after overexpression of CA-β-catenin confirmed that these genes were useful markers of peripheral tissues and revealed those genes that could potentially act downstream of Wnt signaling.
RESULTS AND DISCUSSION
Single Cell Isolation and Retrospective Identification
Retinas were isolated from several stages of mouse development, ranging from embryonic day 12.5 (E12.5) to postnatal day 0 (P0) (Trimarchi et al., 2007). These time points were chosen to maximize the number of retinal progenitor cells (RPCs) harvested and profiled (Trimarchi et al., 2008), as well as to provide additional cells such as newborn neurons for comparison. Subsequently, the retinas were dissociated into single cells and individual cells were collected and lysed. The mRNAs from these lysed cells were reverse transcribed, and the resulting cDNAs were PCR amplified (Brady and Iscove, 1993; Tietjen et al., 2003; Trimarchi et al., 2007). The cDNA quality was assessed through visual inspection on an agarose gel and those cDNA preparations that displayed a smear centered at 1kb were chosen for further analysis (data not shown). Ten micrograms of cDNA from each single cell were biotinylated and hybridized to an Affymetrix mouse 430 2.0 oligonucleotide array using standard Affymetrix protocols (Trimarchi et al., 2007). These arrays allowed for the sampling of the expression of over 40,000 probesets corresponding to approximately 20,000 genes (www.netaffx.com), or almost the entire mouse transcriptome. The Affymetrix signal data were collected for each single cell and normalized using the Affymetrix MAS 5.0 software.
Since the retinal cells isolated during these developmental times had not yet adopted their mature morphologies, a post hoc strategy was employed to classify each cell based upon the expression of clusters of genes (Trimarchi et al., 2008). These gene clusters were generated using a Fisher’s exact test to identify genes that are associated with previously characterized marker genes of the different retinal cell types. This classification method allowed for the identification of 21 single cells as developing retinal ganglion (RGC), amacrine (AC) or photoreceptor cells (PR) (Trimarchi et al., 2007) and an additional 42 cells as cycling RPCs (Trimarchi et al., 2008). However, a number of the single cells did not score significantly for any of the retinal cell types. Also, in the course of classifying these single cell profiles, one of the additional methods used to evaluate the data involved a visual inspection in Microsoft Excel. During this examination of the single cell expression data, it became apparent that two cells (E12 cell A6 and P0 cell E8) expressed numerous transcripts that were not detected in the majority of the other single cell profiles (Figure 1). Given these readily apparent differences in gene expression, the profiles of these two cells were subjected to further scrutiny and validation to assess precisely which type of cell(s) produced these profiles.
Figure 1.
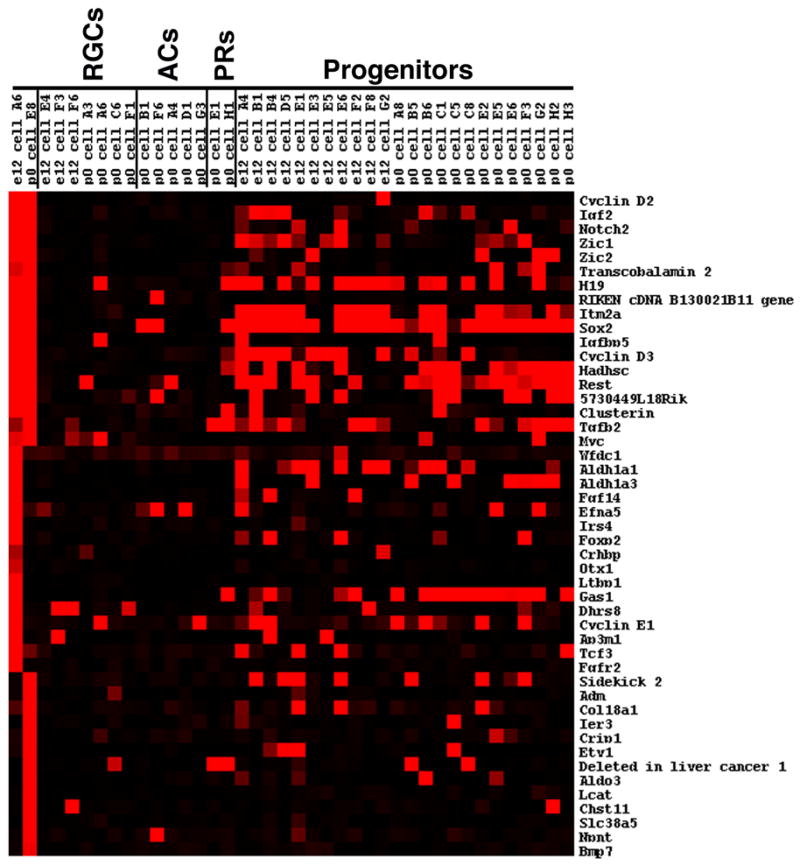
Single cell transcriptional profiles of selected genes expressed in cells of the peripheral retina. The heatmap shown was generated using Treeview software and displays the expression of genes identified by visual inspection of the single cell microarray data. The expression is shown for the two new single cells that are putative ciliary body cells compared with individual developing ganglion cells (RGCs), developing amacrine cells (ACs), developing photoreceptors (PR), and cycling progenitor cells. The intensities of the Affymetrix signals have been scaled in the Treeview software such that signals of 10,000 and above are colored bright red, signals below 1,000 (designated as absent by the Affymetrix software) are black, and signals in between 10,000 and 1,000 are graded according to their signal value.
Analysis of transcripts expressed in the mouse peripheral margin of the optic cup and developing ciliary body/iris
Visual inspection of the microarray data revealed that several genes were present in both E12 cell A6 and P0 cell E8 and largely absent from the previously examined developing RGCs and ACs (Figure 1). However, a further examination of the single cell profiles from these two cells revealed that they shared some expression in common with RPCs (Figure 1 and data not shown). This encompassed a range of expression levels such that some genes were broadly expressed in RPCs (for example, see Sox2), some were only expressed in a small subset of RPCs (for example, see Igf2) and some were not found in the RPC profiles (for example see RIKEN cDNA B130021B11) (Figure 1). Despite sharing some gene expression in common with RPCs, these two cells did not score significantly for RPC character using our previously developed classification scheme (data not shown). Additionally, for many of the RPC expressed genes, the expression levels found on the Affymetrix microarrays were lower in these two cells than in the cells that scored highly as RPCs in the classification scheme (data not shown).
Included among the genes whose expression was enriched in these two cells were cell cycle regulators (Cyclin D2), signaling molecules (Igf2 and Notch2), and transcription factors (Zic1 and Zic2). To identify additional genes that were associated with these markers, hierarchical clustering was performed using E12 cell A6, P0 cell E8 and a set of 21 previously characterized developing RGCs, ACs and PRs (Trimarchi et al., 2007). An examination of the genes that clustered with cyclin D2 with a correlation coefficient of >0.75 revealed several of the genes (Igf2, Notch2 and Zic1) that had been identified by visual inspection (Figure 1 and Supplemental Figure S1A). Many genes that were not readily apparent by visual inspection were also identified through clustering. These genes included, for example, a cell surface molecule (CD38 antigen) that has been shown to be present in both the pigmented and non-pigmented ciliary body (Khoo and Chang, 1999) and many uncharacterized RIKEN cDNAs (Supplemental Figure S1A).
To more fully explore the identity of these single cells and to validate the microarray findings, ISH was performed on cryosections of E12.5, E16.5 and P0 mouse eyes. Prominent expression was detected either in the peripheral margin of the optic cup or in the developing ciliary body for each of the genes from this list that was tested (Figure 2 and Supplemental Figure S2). It was somewhat surprising that cells from the developing ciliary body/iris would have been isolated given that they represent a small percentage of the total cells at any given stage. However, in situ hybridizations performed on dissociated cells have shown that cells from the ciliary body/iris are present in these preparations from all three stages examined (data not shown). Also, previous single cell profiling analyses from these timepoints examined developing ganglion and amacrine cells, which would also have been present at a low percentage in the developing retina at these timepoints (Trimarchi et al., 2007).
Figure 2.
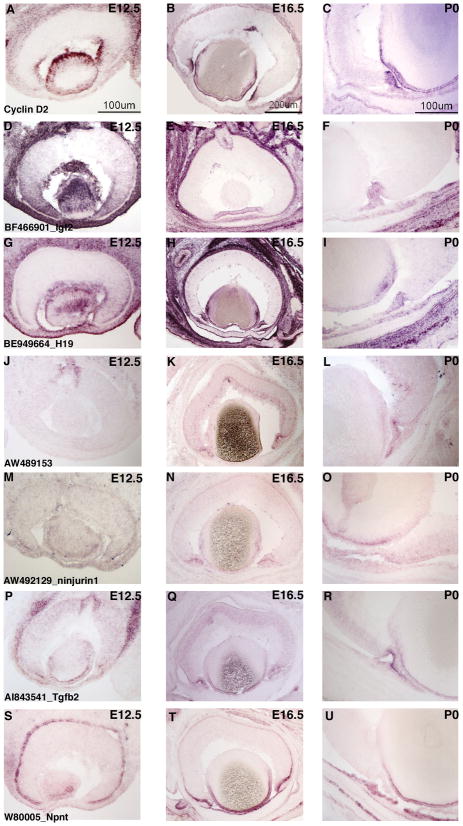
ISH analysis of candidate genes for the peripheral mouse eye. ISH on retinal cryosections was performed at three stages: E12.5 (A, D, G, J, M, P, S), E16.5 (B, E, H, K, N, Q, T) and P0 (C, F, I, L, O, R, U). The following probes were used: Cyclin D2 (AC), BF466901 [Igf2] (D–F), BE949664 [H19] (G–I), AW489153 (J–L), AW492129 [ninjurin1] (M–O) AI843541 [Tgfb2] (P–R), and W8005 [Npnt] (S–U). Representative scale bars are shown.
At all three stages, Cyclin D2 expression was observed in the lens and in the developing ciliary body (Figure 2A–C). No significant expression was detected in the central portion of the retina at E12.5 and E16.5, while some faint staining was observed in the outer neuroblastic layer (ONBL), where the cycling progenitor cells are located, at P0 (Figure 2C). Likewise, the transcript for the growth factor Igf2 was found enriched at the peripheral margin of the optic cup at E12.5 and then in the developing ciliary body at E16.5 and P0 (Figure 2D–F). Staining was also detected for Igf2 in the lens (E12.5, Figure 2D) and the optic nerve head (E16.5, Figure 2E). H19, a non-coding RNA that is located in the same chromosomal region as Igf2 and is reciprocally imprinted with Igf2 (Zemel et al., 1992), was similarly found in the lens, the developing periphery of the optic cup (Figure 2G) and developing ciliary body (Figure 2H and I). However, unlike Igf2, robust expression of H19 also was observed in a subset of cells in the retinal ONBL at E16.5 and P0 (Figure 2H and I, Supplemental Figure S3A, and B). These cells were predicted to be RPCs based upon the presence of H19 transcripts in the profiles of many RPCs at E12.5 and P0 (Figure 1). To fully characterize the identity of these additional H19+ cells, double DISH was performed for H19 and Fgf15, a previously characterized marker of RPCs (Blackshaw et al., 2004; Trimarchi et al., 2008), on dissociated P0 retinas that had been labeled with [3H]-thymidine for 1 hour. There was no overlap observed between H19+ cells and [3H]-thymidine+ cells (Supplemental Figure S3C and D), while a robust overlap was seen between Fgf15 and [3H]-thymidine (Supplemental Figure S3F). These data indicate that the H19+ cells in the ONBL were likely not progenitor cells in the S phase of the cell cycle. Intriguingly, many of the H19+ cells were also Fgf15+ (Supplemental Figure S3E) suggesting that these cells may be RPCs in either the G1 or G2 phase of the cell cycle.
Transcription factors represented another category of genes detected in these two single cell profiles. Zic1, for example, was observed in both E12 cell A6 and P0 cell E8, not in any of the developing RGCs or ACs, and in a subset of RPCs (Figure 1). ISH experiments on retinal cryosections confirmed that Zic1 mRNA was found in the peripheral margin of the optic cup at E12.5 (Supplemental Figure S2A) and then in the developing ciliary body at E16.5 and P0 (Supplemental Figure S2B and C). Additionally, transcripts for the related family member, Zic2, were also enriched in the developing ciliary body/iris, but unlike Zic1, staining for Zic2 was also detected throughout the ONBL (Supplemental Figure S2D–F). These data are consistent with previous reports detailing the retinal expression of these zinc finger transcription factors (Nagai et al., 1997).
Uncharacterized transcripts also were observed in these two single cell profiles (Figure 1 and Supplemental Figure S1). ISH for one of these (RIKEN cDNA 5730449L18/riboprobe AW489153) showed expression in the optic nerve head at E12.5 (Figure 2J) and subsequent expression in the developing ciliary body/iris at E16.5 (Figure 2K) and P0 (Figure 2L). Additional expression was noted in a subset of cells on the vitreal side of the developing inner neuroblastic layer (INBL), consistent with expression in developing RGCs (Figure 2K and L).
Hierarchical clustering produced a group of genes that associated significantly with cyclin D2 (Supplemental Figure S1A). To validate this clustering and more fully examine the expression patterns of some of the cyclin D2 associated genes, ISH were performed for ninjurin1 and rhomboid family 1 (Rhbdf1). Ninjurin1 is an adhesion molecule originally identified through its role in nerve regeneration (Araki and Milbrandt, 1996) that could have a similar adhesive role in the developing ciliary body. The expression of ninjurin1 was very weak at E12.5 in the peripheral margin of the optic cup, but was not found in the retina itself (Figure 2M). At E16.5 and P0, ninjurin1 transcripts were detected in the developing ciliary body/iris and in the ONBL (Figure 2N and O). Rhbdf1 is an atypical member of a family of serine proteases, the Drosophila homolog of which has been demonstrated to activate to growth factors (Urban, 2006). However, while Rhbdf1 has been shown to associate with growth factors, it lacks key residues responsible to protease activity and, therefore, its function remains to be fully determined (Nakagawa et al., 2005). Similar to ninjurin1, Rhbdf1 was detected weakly in the peripheral margin of the optic cup at E12.5 (Supplemental Figure 2G) and then stronger signal was found in the developing ciliary body/iris at both E16.5 and P0 with weaker staining in the ONBL at E16.5 (Supplemental Figure 2H and I). These ISH experiments validate that the cluster of genes that associate with cyclin D2 are expressed in the developing ciliary body/iris and provide new markers for this tissue.
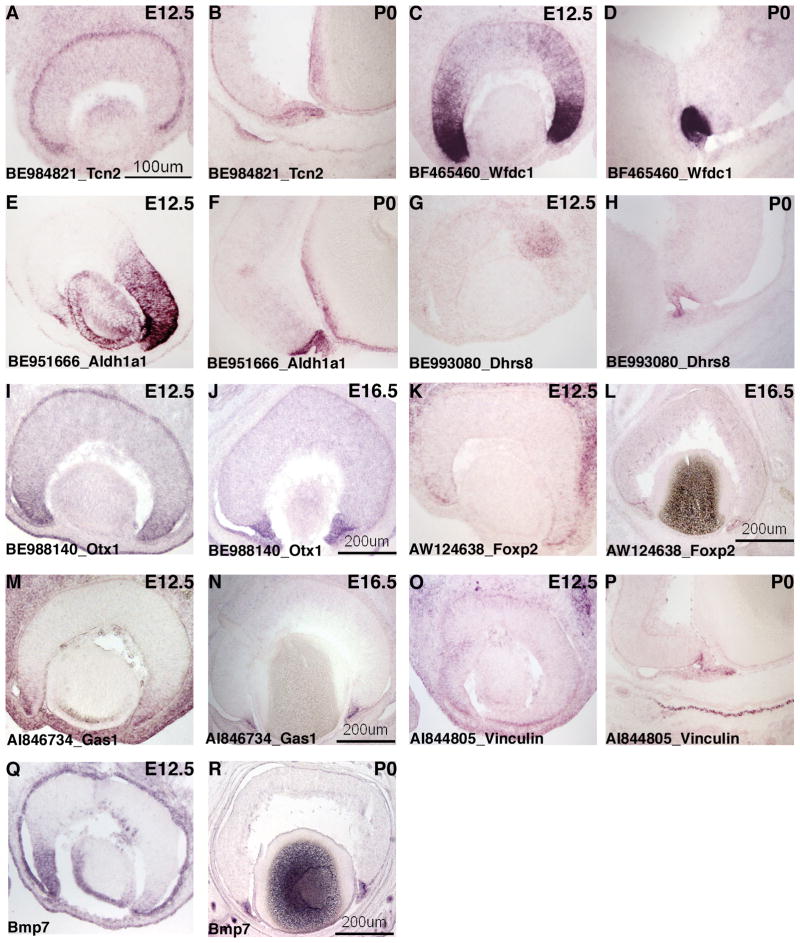
There were transcripts that were enriched in both E12 cell A6 and P0 cell E8 that also were present in subsets of the developing RGCs and ACs (Figure 1) or in subsets of RPCs (data not shown). These genes included a cell cycle regulator (cyclin D3), signaling molecules (Tgfb2), an IGF binding protein (Igfbp5) and transcription factors (Sox2, Myc). ISH performed for several of these transcripts confirmed that they were expressed in the developing ciliary body/iris (Figure 2 and data not shown), while also having other domains of expression. Tgfb2 mRNA, for instance, was detected in both the ONBL and the developing ciliary body/iris at E12.5, E16.5 and P0 (Figure P–R). The staining in the ciliary body/iris was consistently more intense than that in the ONBL at E16.5 and P0 (Figure 2P and R). Additionally, at P0 the staining for Tgfb2 mRNA was more pronounced in the outer layer of the developing ciliary body/iris than the inner layer (Figure 2R). Cyclin D3 displayed a very similar staining pattern as Tgfb2 in that it was also significantly enriched in the ciliary body/iris at E16.5 and P0 while being present in the ONBL as well (data not shown). Myc, however, was expressed in the peripheral margin of the optic cup at E12.5 and then at E16.5 and P0 was more strongly expressed in subsets of cells in the most vitreal portion of the INBL, consistent with expression in developing RGCs (data not shown). Transcobalamin 2 (Tcn2) was found in the profiles for both of the developing ciliary body single cells and not in those for the developing RGCs and ACs (Figure 1). However, Tcn2 was detected in a subset of the RPC single cell profiles (Figure 1). ISH performed on retinal cryosections showed intense staining for Tcn2 in the retinal pigment epithelium (RPE) at E12.5 and P0 and weaker staining in the ONBL (Figure 3A and B). The ISH also revealed Tcn2 expression in the developing ciliary body/iris, which was especially pronounced at E16.5 and P0 (Figure 3B and data not shown).
Figure 3.
ISH analysis of candidate genes for the peripheral mouse eye. ISH on retinal cryosections was performed at: E12.5 (A, C, E, G, I, K, M, O, Q), E16.5 (J, L, N) and P0 (B, D, F, H, P, R). The following probes were used: BE984821 [Tcn2] (A,B), BF465460 [Wfdc1] (C,D), BE951666 [Aldh1a1] (E,F), BE993080 [Dhrs8] (G,H), BE988140 [Otx1] (I,J), AW124638 [Foxp2] (K,L), AI846734 [Gas1] (M,N), AI844805 [Vinculin] (O,P) and Bmp7 (Q,R). Representative scale bars are shown.
Additionally, transcripts were observed that were present in only one or the other of the putative developing ciliary body single cells (Figure 1). As observed with the sets of genes characterized above, the categories of these transcripts included transcription factors (Foxp2, Otx1, Tcf3, Etv1), cell cycle regulators (Wfdc1, cyclin E1), and signaling molecules (Fgf14, Bmp7, Ltbp1). Finding associated genes with a significant correlation coefficient through hierarchical clustering was more difficult for these genes since they were only expressed in one of the single cell profiles. Nonetheless, one cluster did emerge around the genes Foxp2 and Aldh1a3 (Supplemental Figure S1B). As was the case with the clusters of genes observed above, many of the genes associated with Foxp2 were uncharacterized RIKEN cDNAs and other expressed sequence tags (Supplemental Figure S1B).
ISH on retinal cryosections from three developmental stages confirmed the expression of the genes that clustered with Foxp2 and Aldh1a3 in the peripheral margin of the optic cup at E12.5 and in the developing ciliary body/iris at E16.5 and P0 (Figure 3 and data not shown). A riboprobe for the growth inhibitor Wfdc1 (Larsen et al., 1998) showed strong staining in the peripheral margin of the optic cup at E12.5, and while additional staining was detected in cells located in the ONBL, the most central part of the retina was not stained (Figure 3C). At subsequent developmental stages, Wfdc1 mRNA became localized only to the developing ciliary body (Figure 3D and data not shown). An additional growth inhibitory gene, Gas1 (Del Sal et al., 1992), was also found in the single cell profile for E12 cell A6 (Figure 1). ISH confirmed that Gas1 was expressed in the peripheral margin of the optic cup at E12.5 (Figure 3M) and then became restricted to the ciliary body/iris at E16.5 (Figure 3N). It is curious that several growth inhibitory genes (Gas1, Wfdc1, and p57) are expressed in the same cell as genes that stimulate cell growth (several cyclin genes, Myc, and Notch2) [see figure 1 and (Rowan et al., 2004)]. While it is possible that the explanation is simply a lack of correlation between mRNA expression and protein expression, this observation warrants further study to understand the interplay between genes that drive the cell cycle and those that impede it in the developing ciliary body/iris.
Aldh1a1, which codes for an enzyme that catalyzes the second step of the conversion of retinoic acid to retinol (Duester, 2000), was found to be strongly expressed in the lens and in the ONBL of the dorsal retina at E12.5 and E16.5 (Figure 3E and data not shown), as previously reported (Fan et al., 2003). Additionally, this gene was observed in both the dorsal and ventral developing ciliary body at all three stages (Figure 3E and F). Aldh1a3, a related member of the aldehyde dehydrogenase family, was observed in E12 cell A6, much like Aldh1a1 (Figure 1). ISH on retinal cryosections for Aldh1a3 showed that while there was robust expression in the peripheral margin of the optic cup at E12.5, no expression was detected in the developing ciliary body/iris at P0 (data not shown), unlike in the case of Aldh1a1. Another enzyme, Dhrs8, a member of the alcohol dehydrogenase family, was initially observed in a subset of cells in the dorsal portion of the ONBL at E12.5 (Figure 3G), but then became expressed in the ciliary body at E16.5 and P0 (Figure 3H and data not shown).
Several transcription factors were found in the single cell profile of E12 cell A6, including Foxp2, Otx1 and Tcf3 (Figure 1). Otx1 has been reported previously to be expressed in the developing ciliary body (Martinez-Morales et al., 2001) and ISH on retinal cryosections demonstrated robust expression of this transcript in both the E12.5 peripheral margin of the optic cup and the E16.5 ciliary body (Figure 3I and J). Foxp2 also was detected in the peripheral margin of the optic cup at E12.5 and the developing ciliary body at E16.5, but this gene also displayed additional staining in a subset of cells in the INBL (Figure 3K and L). The location of these cells to the scleral side of the INBL indicated that they might be developing ACs (Figure 3L). Uncovering the distinct transcription factors expressed in the developing ciliary body/iris will enable future studies dissecting the specific transcription factor networks driving the differentiation of this region.
Several additional genes were found to be expressed in one of the putative two ciliary body cells. Insulin receptor substrate proteins (Irs1–4) dock with the insulin receptor upon its activation and phosphorylation and act as bridges molecules with the downstream signaling pathway (Giovannone et al., 2000). While the other members of this family play roles in growth and glucose metabolism, the function of Irs4 is not well understood (Fantin et al., 2000). Irs4 was found in the transcriptional profile for E12 cell A6 (Figure 1) and ISH at E12.5 showed robust expression in the peripheral margin of the optic cup (Supplemental Figure S2J) and the developing ciliary body/iris at E16.5 (Supplemental Figure S2K). The expression of Irs4 was absent from both the retina and the developing ciliary body at P0, suggesting this gene may play a role in early ciliary body development. The transcript for Ltbp1, which encodes a protein that is part of a larger complex that binds Tgfβ family members and keeps them inactive while assisting in their assembly and targeting (Oklu and Hesketh, 2000), was also found in the profile for E12 cell A6 (Figure 1). ISH for Ltbp1 revealed that this gene was expressed in the peripheral margin of the optic cup at E12.5 and then in the developing ciliary body/iris at E16.5 and P0 (Supplemental Figure S2M–O). Vinculin, which encodes an actin binding protein that plays a role in cell/cell and cell/extracellular matrix adhesion (Ziegler et al., 2006), also was observed in E12 cell A6 (data not shown). ISH further demonstrated that this gene was expressed in a subset of cells in the ONBL at E12.5 (Figure 3O), but then was detected in the outer layer of the developing ciliary body at E16.5 and P0 (Figure 3P and data not shown). Additional strong staining was observed in the developing iris tip, both in the mouse at P0 and in the chick at E6 and E8 (Figure 3P, Supplemental Figure S2P and R).
Nephronectin (Npnt) is an EGF-repeat containing protein that has been identified in tissues undergoing an epithelial-mesenchymal transition during development (Brandenberger et al., 2001; Morimura et al., 2001). Transcripts for Npnt were found in the profile for P0 cell E8, but not E12 cell A6 (Figure 1). ISH on E12.5 retinal cryosections revealed strong staining for Npnt in the RPE (Figure 2S). At E16.5, the RPE staining decreased considerably and the most prominent signal was found in the tip of the developing ciliary body and in the developing iris (Figure 2T). This pattern continued at P0 with weak staining for Npnt in the ciliary body and more pronounced signal in the developing iris (Figure 2U). The expression pattern for Npnt suggests that P0 cell E8 may originate from a different portion of the developing ciliary body/iris as E12 cell A6 and, therefore, provides distinct marker genes for this region.
BMP7, a potent signaling molecule that has been shown to play an important role in the development of the mouse ciliary body (Zhao et al., 2002), was found in the single cell profile for P0 cell E8 (Figure 1). ISH on retinal cryosections showed that BMP7 was expressed in the peripheral margin of the optic cup at E12.5 (Figure 3Q). In addition, BMP7 was found in the lens and in a subset of cells in the central retina, located in a position consistent with newborn RGCs at this stage (Figure 3Q). At E16.5, BMP7 was expressed robustly in the ciliary body/iris, but now was also found in a subset of cells in the ONBL (data not shown). By P0, BMP7 retinal expression was confined to the developing ciliary body/iris (Figure 3R). These microarray profiling experiments coupled with the ISH validation demonstrate the dynamic nature of gene expression in the developing mouse peripheral margin of the optic cup and ciliary body/iris and the extensive diversity of the classes of genes that are expressed in this region of the developing eye.
Analysis of transcripts expressed in the developing chick peripheral margin of the optic cup
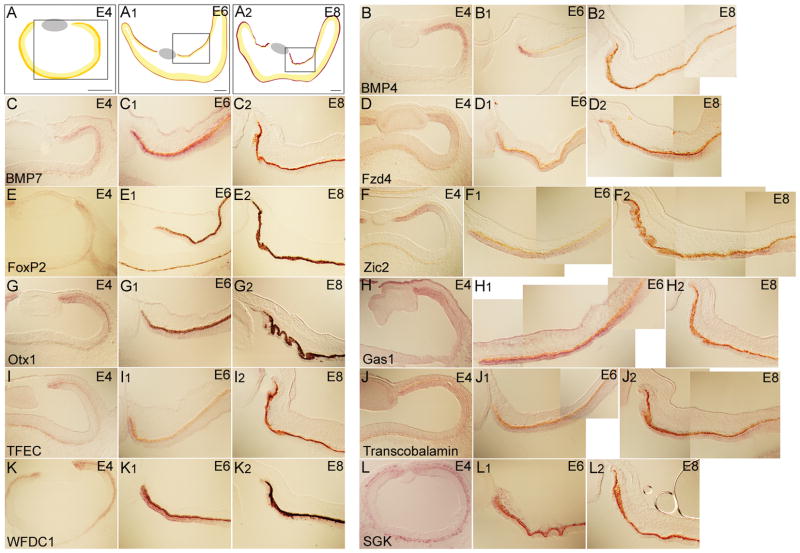
It has been suggested that the intricate morphogenetic events that occur during ciliary body morphogenesis may be quite different between the mouse and the chick (Napier and Kidson, 2007). Given this potential for both similarities and differences in how the anterior segment structures form in these organisms, a comparison in gene expression between mouse and chick peripheral retina might be informative. To identify the genes that also were expressed in the periphery of the developing chick eye, ISH was performed on cryosections from E4, E6 and E8 (Figure 4A). These stages were chosen to most closely reflect E12.5, E16.5 and P0 stages of mouse development. The genes examined included those identified from the single cell mouse microarrays (Figure 1), and genes previously shown to be expressed in the developing peripheral mouse eye (Rowan et al., 2004). Among the genes identified from mouse, the transcripts of the signaling molecules BMP4 and BMP7 (Figure 4B and C), the Wnt receptor, Frizzled 4 (Figure 4D), transcription factors such as FoxP2, Zic1, Otx1, and TFEC (Figure 4E 4G, 4I), cell cycle regulators such as GAS1, WFDC1, SGK, c-Myc, and Cyclin E1 (Figure 4H, 4K–4L, and data not shown) and other genes such as Transcobalamin 2, Clusterin, and Collagen IX (Figure 4J and data not shown) were enriched in the periphery of the optic cup between E4 to E8. In general, ISH signals specific to the periphery of the retina were detected with two different probes corresponding to different parts of the gene (Supplemental Table 1 and data not shown). The expression was highest at the tip of the optic cup and decreased toward the center of the retina. In addition, the intensity of the signal decreased over time, so that the expression at E8 was very low in most cases. It is not clear whether this was due to the profound thinning of the ciliary and iris epithelia at this time, or a reduction in transcript levels.
Figure 4.
ISH analysis for the expression of eleven candidate genes for the peripheral chick eye. The scheme in A, A1 and A2 shows the area of peripheral eye represented in this figure for chick E4, E6 and E8, respectively. The probes used were BMP4 (B-B2), BMP7 (C-C2), Fzd4 (D-D2), FoxP2 (E-E2), Zic2 (F-F2), Otx1 (G-G2), Gas1 (H-H2), TFEC (I-I2), Transcobalamin 2 (J-J2), WFDC1 (K-K2), and SGK (L-L2). Scale bars: 150μm.
The kinetics of the expression was variable during these developmental stages as well. For some genes, the ISH signal intensified over time, whereas in most cases, the expression at E4 and E6 was stronger than that of E8. The extent of expression in the periphery varied at a given stage. For example, TFEC and SGK (Figure 4I, 4I1, 4I2 and 4L, 4L1, 4L2) displayed a narrow expression domain located at the tip of the optic cup, which is thought to develop into the iris, while most other genes showed a much broader expression pattern in the periphery of the optic cup at E4.
Although the majority of the genes tested were expressed in both the mouse and chick, some of the genes (cyclin D2, Igf2, cyclin D3, IGFBP5, Crhbp, REST, FGFR2, and CD63) expressed in the mouse were either weakly expressed, not expressed, or expressed in different parts of the developing optic tissues of the chick embryo (Supplemental Table 1). Because several genes of this class showed expression in other tissues surrounding the developing optic cup, the absence of expression is likely to be the result of species differences, rather than technical issues. For example, cyclin D2, CD63 and FGFR2 showed strong expression in the lens and periocular mesenchyme (data not shown), whereas IGF2 showed broad expression in the ventral optic cup (data not shown). This may indicate that there are molecular distinctions in the ciliary body and iris development between mouse and chick. For example, it is known that the mechanism of periocular mesenchyme development is significantly different between mouse and chick. In this tissue, the neural crest- and mesoderm- derived cells differentially contribute to the periocular mesenchymal cells surrounding the eye in mouse and chick (Gage et al., 2005).
Ectopic expression of ciliary body markers upon induction of the Wnt signaling pathway
The canonical Wnt signaling pathway plays an important role during early specification of the ciliary body and iris in the periphery of the developing optic vesicle/cup (Liu et al., 2003; Cho and Cepko, 2006; Liu et al., 2007). In order to investigate whether the peripheral genes might be downstream of Wnt signaling, the expression of these genes was examined in the retina following overexpression of a constitutively active form of beta-catenin (CA-β-catenin) (Capdevila et al., 1998; Cho and Cepko, 2006). In previous experiments, an avian replication competent retroviral vector encoding an N-terminal deletion mutant of β-catenin (RCAS:CA-β-catenin), that rendered it constitutively active, was introduced into the chick retina. Infection with this virus led to retinal progenitor cells adopting a more peripheral retinal fate. Additionally, downregulation of retinal genes and upregulation of several previously known markers of the ciliary body/iris was observed (Cho and Cepko, 2006; Fu et al., 2006).
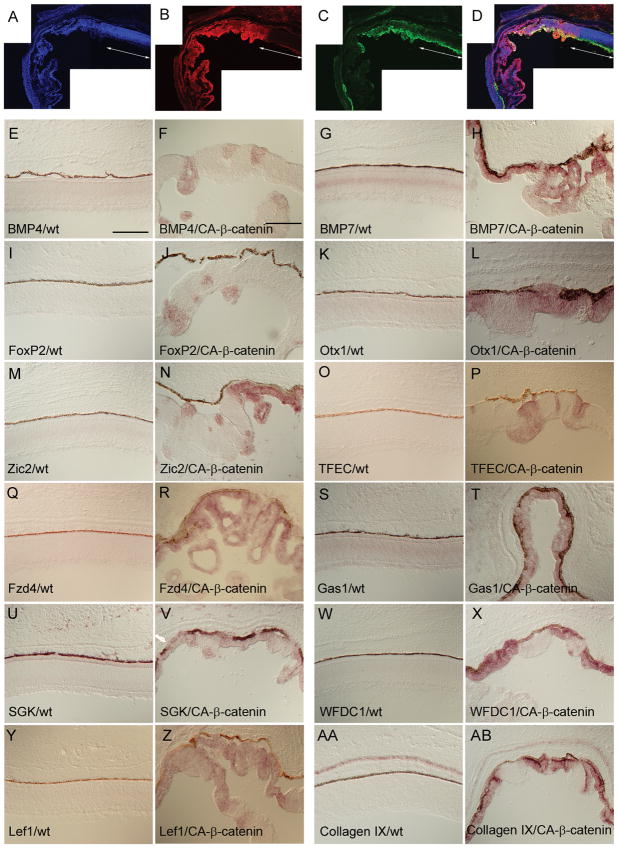
To examine which of the newly identified ciliary body markers was potentially downstream of Wnt signaling, RCAS:CA-β-catenin was used to infect the optic vesicle at stage 10. The infected retinas were harvested at E9.5, sectioned and stained with an anti-gag antibody (3C2) to determine the extent of the infected region that also expressed CA-β-catenin (Figure 5A–D). Retinal epithelia without 3C2 staining displayed both a normal thickness and a normal pattern of neurogenesis as determined by anti-β-tubulin staining (Figure 5C), whereas 3C2+ areas showed a gradual diminution of β-tubulin staining and a thinning of the retina (Figure 5A–D). Since the correlation between the retinal thinning and the induction of ciliary body/iris fate was highly reproducible, these sections were examined for the expression of the newly identified ciliary body/iris markers.
Figure 5.
ISH analysis for the induction of peripheral marker genes upon canonical Wnt signaling activation. Optic vesicles were infected with a RCAS:CA-β-catenin at HH stage 10 and the analysis was performed on embryos harvested at E9.5. Using a representative section from the central part of the retina, the overlap between the virally infected region (B) and the area showing retinal thinning and folding [shown as the absence of β-tubulin antibody staining in (C)] is visualized in merged image (D). A DAPI-stained retinal section is shown in (A). The white arrowed line demarcates the normal uninfected portion of the retina. Probes used were BMP4 (E and F), BMP7 (G and H), FoxP2 (I and J), Otx1 (K and L), Zic2 (M and N), TFEC (O and P), Fzd4 (Q and R), Gas1 (S and T), SGK (U and V), WFDC1 (W and X), Lef1 (Y and Z), and Collagen IX (AA and AB). Images shown on the left are from the central retinal of uninfected chicks. The images on the right are also from the central retina of chicks infected with the RCAS:CA-β-catenin. The ventral side is down. Scale bars: 150μm.
To insure that the viral overexpression assay was functioning properly, the ectopic induction in the retina of a few previously identified iris/ciliary body marker genes was examined. These genes included the secreted signaling molecule (BMP7, Figure 5G, H), the cell cycle regulator (WFDC1, Figure 5W, X), and the matrix protein (Collagen IX, Figure 5AA, AB). The expression of a Wnt receptor (Fzd4) and a Wnt signal transducer (Lef1) have been characterized previously in the peripheral retina (Jasoni et al., 1999; Kubo et al., 2003). To determine whether the expression of Fzd4 and Lef1 was potentially downstream of Wnt signaling, Stage 10 optic vesicles were infected with RCAS:CA-β-catenin and eyes were harvested at E9.5. Both Fzd4 (Figure 5Q, R) and Lef1 (Figure 5Y, Z) were ectopically upregulated in the central retina upon activation of the Wnt pathway. This upregulation of Lef1 and Fzd4 may be caused by a positive feedback mechanism of β-catenin mediated Wnt signaling (Kubo et al., 2003; Liu et al., 2003).
The newly characterized genes identified from the combined microarray analysis/section ISH from the mouse, described above, and additional periphery-enriched genes reported previously (Rowan et al., 2004) also were examined for their responsiveness to Wnt signaling. All of the results were observed in ISH on sections from multiple RCAS:CA-β-catenin infected retinas (minimum of two). In a similar fashion as Fzd4 and Lef1, infection with RCAS:CA-β-catenin led to an upregulation of the secreted signaling molecule BMP4 (compare Figure 5E to 5F), the transcription factors FoxP2, Otx1, Zic2 and TFEC (compare Figure 5I to 5J, 5K to 5L, 5M to 5N, and 5O to 5P, respectively), and the cell cycle regulators Gas1 and SGK (compare Figure 5S and 5U to 5T and 5V, respectively). In general the induction of these markers was present across the entire range of the thinner retina except in a few cases where the upregulation was confined to only a small area of the thinner epithelium (see Figure 5F, J and N). Additionally, in any given retinal section, a similar upregulation was observed in any part of the epithelium where thinning had occurred. The absence of a larger induction in these cases may be due to a requirement of higher levels of Wnt signaling to attain full transcriptional upregulation. Ectopic induction of negative cell cycle regulators, such as SGK, Gas1 and WFDC1 suggests that canonical Wnt signaling in the developing periphery of the optic cup may participate in the suppression of cell proliferation. This interpretation is consistent with previous results demonstrating that the cell proliferation index of the peripheral retinal is lower than that of central retina, and is lower in the central retina following infection with the RCAS:CA-β-catenin (Cho and Cepko, 2006).
Since infection with the RCAS:CA-β-catenin virus leads to a repatterning of the eye, such that the central retina takes on the peripheral fate (Cho and Cepko, 2006), the observed ectopic expression in the central retina of these candidate genes may be either downstream of Wnt signaling directly, or simply be a consequence of the fate change. Either way, these experiments provide further validation of these newly identified as bona fide peripheral markers. They have now also been shown to be potential downstream targets of the Wnt signaling pathway. Their exact position in the hierarchy of gene expression downstream of Wnt signaling will need to be established by further analysis.
Comparison of ciliary body marker studies
Several strategies have been employed to identify the genes specifically expressed in the peripheral structures of the eyes from various animals. These approaches have included ISH screens, subtractive hybridization strategies and microarrays using mouse, human and chick anterior segment structures (Thut et al., 2001; Escribano and Coca-Prados, 2002; Kubota et al., 2004; Diehn et al., 2005). In the current study, a combination of single cell transcriptional profiling and ISH was used to identify markers of the ciliary body in the developing mouse and chick retina. A previous high throughput ISH screen in the developing mouse eye identified 6 genes as having specific expression in the optic cup margin, the location of the developing ciliary body (Thut et al., 2001). Three of these markers (Tgfb2, Col9a1 and Slc16a1) were expressed in both CB single cell profiles and observed in very few single RGC, AC or PR profiles (Supplemental Figure S4A). Tac1 was not observed in either putative single CB cell and Ptmb4 could not be located on the Affymetrix microarray (Supplemental Figure S4A). The strongest marker for the developing ciliary body found in the previous study of Thut et al. was Tgfb1i4 (Thut et al., 2001). Another group further characterized the expression of this marker in more mature peripheral structures (Napier and Kidson, 2007). Tgfb1i4 was not identified as a ciliary body marker in our screen since robust expression of this gene was also detected in all of the single RGCs and ACs profiled between E12.5 and P0 (Supplemental Figure S4A). To examine whether Tgfb1i4 was expressed outside of the developing ciliary body, ISH were performed on E12.5, E14.5, E16.5 and P0 retinas. Strong staining was observed at each of these stages in the developing ciliary body/iris (Supplemental Figure S4B–E). However, at E14.5, E16.5 and P0, significant signal also was present in the INBL (Supplemental Figure S4C–E), where the developing RGCs and ACs reside, consistent with the single cell microarray results. While Tgfb1i4 is strongly expressed in the developing ciliary body, this gene does not mark this area specifically and is expressed in other nearby retinal cells as well.
Additional studies searching for markers of human ciliary body structures have been performed. These analyses utilized either a microarray based approach (Diehn et al., 2005) or EST database mining (Escribano and Coca-Prados, 2002) to produce lists of candidate ciliary body specific genes. To examine the expression of these sets of markers gene in our single cell transcriptional profiles, heatmaps were generated with the genes identified by EST mining (Supplemental Figure S5A) and by microarray analysis (Supplemental Figure S5B). In general, the markers found by EST mining were expressed in many more single cells than merely the putative CB cells (Supplemental Figure S4A). Additionally, not all of these markers were expressed in either E12 cell A6 or P0 cell E8. Conversely, the majority of the markers identified in the microarray screen were not expressed in either of the two single cells from this study that may have originated from the developing ciliary body or in any of the other single cell profiles (Supplemental Figure S5B). The two exceptions were Tpm2 and Bmp7. The principal reason behind these differences is most likely that these other studies were conducted using adult human tissue, whereas the current study focused solely on the developing mouse and chick ciliary body.
Finally, one study was conducted to find genes specifically expressed in the chick ciliary epithelium. A combination of subtractive hybridization and cDNA microarrays were used to identify candidate genes and ISH was employed subsequently to validate the specificity of expression of these candidate markers. The majority of the genes identified as specific markers of the ciliary epithelium by Kubota et al. (Kubota et al., 2004) did not overlap with those from our study. Only three genes (Zic2, Tgfβ2 and Collagen IX) were commonly isolated by the two approaches. This is most likely due to the differences in stages of the tissue samples taken for the comparison. In our experiments, embryonic and early postnatal cells were used for the analysis in mice and E4 to E8 eyes were used for the chick analysis, whereas Kubota et al. used posthatch day 1 chick eyes (Kubota et al., 2004). This stage is after most of development of the eye structures is complete. Alternatively, the failure to identify known markers of the ciliary body could be due to some technical shortcomings in the protocols used. For example, the Msh homeobox-containing genes Msx1 and Msx2 are expressed in the developing ciliary body at multiple developmental timepoints (Monaghan et al., 1991). However, these genes were not detected in the transcriptional profiles of the two presumptive ciliary body cells in this study. In fact, these two genes were never detected in any of the single cells profiled to date, suggesting that either they are expressed at levels which are too low to be sampled by the single cell profiling method or that the target sequences on the Affymetrix microarray fail to hybridize with the cDNAs for these genes.
EXPERIMENTAL PROCEDURES
Single cell harvesting and cDNA generation
Single cells were isolated from CD-1 mice (Charles River Laboratories) exactly as described in (Trimarchi et al., 2007). Briefly, retinas were dissected, dissociated in a papain solution, and plated in PBS (pH 7.4) containing 0.1% BSA. Each individual cell was harvested into a drawn glass pipette using capillary action, washed, and collected into lysis buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 1.5 mM MgCl2, 5 mM DTT, 0.5% NP-40, 200 ng/ml modified Oligo dT primer [TATAGAATTCGCGGCCGCTCGCGATTTTTTTTTTTTTTTTTTTTTTTT] [Oligos etc.], 250 U/ml RNAguard [Amersham], 15 U/ml Prime RNase inhibitor [Eppendorf], and 40 μM each of dATP, dGTP, dCTP and dTTP). The mRNAs were reverse transcribed with Superscript II (Invitrogen) for 30 min. at 37°C and a poly(A) tract was added to the first strand synthesis using TdT (Roche). The second strand was generated and the subsequent mixture PCR amplified using the following program: 95°C for 2 min., 37°C for 5 min., 72°C for 16 min., 93°C for 40 sec., 67°C for 1 min., 72°C for 6 min with a 6 sec extension each cycle for 35 cycles.
Affymetrix microarray hybridizations
Ten micrograms of single cell cDNA was RNase treated (Roche) for exactly 13 minutes at 37°C. The samples were heated to 99°C for 15 minutes and end labeled with 25 μM Biotin N6 ddATP (Enzo Biosciences). Mouse Genome 430 2.0 Genechip® oligonucleotide arrays were hybridized with the labeled single cell cDNAs using standard Affymetrix protocols as detailed in (Trimarchi et al., 2007). Global scaling of each array dataset was performed using the Affymetrix Microarray software (MAS 5.0) and the target intensity was set to 500. The resulting signal data for each probe set was exported as a tab delimited text file and subsequent analyses were performed using Microsoft Excel. The raw and processed Affymetrix data files have been deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under the accession number GSE15566.
In situ hybridizations and immunohistochemistry
In the mouse, both ISH on retinal cryosections and dissociated cell ISH were performed exactly as detailed in (Trimarchi et al., 2007; Trimarchi et al., 2008).
For the chick ISH, freshly fertilized chicken eggs were purchased from SPAFAS (CT) and incubated at 37°C for four, six and eight days to embryonic day (E) 4, 6 and 8 for section ISH. Twenty-micron frozen cryosections were generated from either whole heads or eyes that had been fixed in 4% paraformaldehyde. ISH were performed as previously described (Murtaugh et al., 1999).
Immunohistochemical detection of RCAS:CA-β-catenin infected cells and β-tubulin was done as described previously (Cho and Cepko, 2006). Primary antibodies (α-β-Tubulin III (Upstate Biotechnology) and 3C2 mAB (Developmental Studies Hybridoma Bank; DSHB) were used at 1: 250 and at 1:50, respectively. Secondary antibodies were Cy2- and Cy3- conjugated anti-rabbit antibodies or anti-mouse (Jackson Immunoresearch Laboratories) used at 1:300.
Chicken embryos and in ovo injection
For the in ovo injections, concentrated viral particles encoding CA-β-catenin (RCAS:CA-β-catenin) (Capdevila et al., 1998) were injected into the right optic vesicle of the Hamilton-Hamburger (HH) stage 10 (Hamburger and Hamilton, 1992) chicken embryo as described previously (Cho and Cepko, 2006) and eggs were further incubated to E9.5. All experimental procedures with embryos were approved by the Institutional Animal Care and Use Committee at Harvard University.
Supplementary Material
Clusters of genes expressed in the developing ciliary body. Hierarchical clustering of 44 microarrays including developing RGCs, ACs, PRs, cycling RPCs, and the two putative ciliary body cells. (A) Genes that clustered with Cyclin D2 with a correlation coefficient of >0.7 are shown. (B) Genes that clustered with Foxp2 with a correlation coefficient of >0.7 are shown.
ISH analysis of candidate genes for the peripheral mouse and chick eye. ISH on retinal cryosections was performed at three stages of mouse development: E12.5 (A, D, G, J), E16.5 (B, E, H, K) and P0 (C, F, I, L) and two stages of chick development: E6 (M) and E8 (N). The following probes were used: AI848240 [Zic1] (A–C), BE988982 [Zic2] (D–F), AI842763 [Rhbdf1] (G–I), AW045815 [Irs4] (J–L), and vinculin (M and N).
Further characterization of H19 expression. Close-ups of in situ hybridizations using an H19 riboprobe (BE949664) and E16.5 mouse retinas [A, B]. The ONBL is denoted by a red bar. P0 retinas were explanted, labeled with [3H]-thymidine for 1 hour, dissociated and probed for H19 alone [C, D] or for H19 (red) and Fgf15 (green) [E, F]. Images are shown with [C, E] and without [D, F] the development of the [3H]-thymidine. The arrows in E indicate cells that are positive for both H19 and Fgf15.
A comparison of ciliary body/iris marker genes from this study and others. (A) A Treeview generated heatmap showing the expression of 6 previously identified ciliary body marker genes (Thut et al., 2001) in E12 cell A6, P0 cell E8 along with the single RGCs, ACs, and PRs. ISH for Tgfb1i4 was performed on retinal cryosections at E12.5 (B), E14.5 (C), E16.5 (D) and P0 (E). Arrows indicate the location of the INBL staining. Scale bars are shown.
A comparison of ciliary body/iris marker genes from this study and others. (A) A Treeview generated heatmap showing the expression of a set of previously identified adult human ciliary body marker genes (Diehn et al., 2005) in E12 cell A6, P0 cell E8 along with the single RGCs, ACs, and PRs. (B) A Treeview generated heatmap showing the expression of a second set of previously identified adult human ciliary body marker genes (Escribano and Coca-Prados, 2002) in E12 cell A6, P0 cell E8 along with the single RGCs, ACs, and PRs.
Summary of gene expression in the mouse and chicken embryos using ISH and the probes used in this study. For each gene the corresponding cDNA used to generate the ISH riboprobe in both the chick and mouse is listed. A summary of the expression pattern observed at each stage is included. Terms used to describe the in situ patterns: Outer neuroblastic layer/ventricular zone (ONBL), inner neuroblastic layer (INBL), retinal pigment epithelium (RPE), not detectable (ND), (NT) not tested, and (NA) no probes available.
Acknowledgments
This work was supported by the Howard Hughes Medical Institute (C.L.C.).
References
- Acampora D, Mazan S, Avantaggiato V, Barone P, Tuorto F, Lallemand Y, Brulet P, Simeone A. Epilepsy and brain abnormalities in mice lacking the Otx1 gene. Nat Genet. 1996;14:218–222. doi: 10.1038/ng1096-218. [DOI] [PubMed] [Google Scholar]
- Ahmad I, Dooley CM, Thoreson WB, Rogers JA, Afiat S. In vitro analysis of a mammalian retinal progenitor that gives rise to neurons and glia. Brain Res. 1999;831:1–10. doi: 10.1016/s0006-8993(99)01376-1. [DOI] [PubMed] [Google Scholar]
- Araki T, Milbrandt J. Ninjurin, a novel adhesion molecule, is induced by nerve injury and promotes axonal growth. Neuron. 1996;17:353–361. doi: 10.1016/s0896-6273(00)80166-x. [DOI] [PubMed] [Google Scholar]
- Beebe DC. Development of the ciliary body: a brief review. Trans Ophthalmol Soc U K. 1986;105 (Pt 2):123–130. [PubMed] [Google Scholar]
- Blackshaw S, Harpavat S, Trimarchi J, Cai L, Huang H, Kuo WP, Weber G, Lee K, Fraioli RE, Cho SH, Yung R, Asch E, Ohno-Machado L, Wong WH, Cepko CL. Genomic analysis of mouse retinal development. PLoS Biol. 2004;2:E247. doi: 10.1371/journal.pbio.0020247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady G, Iscove NN. Construction of cDNA libraries from single cells. Methods Enzymol. 1993;225:611–623. doi: 10.1016/0076-6879(93)25039-5. [DOI] [PubMed] [Google Scholar]
- Brandenberger R, Schmidt A, Linton J, Wang D, Backus C, Denda S, Muller U, Reichardt LF. Identification and characterization of a novel extracellular matrix protein nephronectin that is associated with integrin alpha8beta1 in the embryonic kidney. J Cell Biol. 2001;154:447–458. doi: 10.1083/jcb.200103069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capdevila J, Tabin C, Johnson RL. Control of dorsoventral somite patterning by Wnt-1 and beta-catenin. Dev Biol. 1998;193:182–194. doi: 10.1006/dbio.1997.8806. [DOI] [PubMed] [Google Scholar]
- Cho SH, Cepko CL. Wnt2b/beta-catenin-mediated canonical Wnt signaling determines the peripheral fates of the chick eye. Development. 2006;133:3167–3177. doi: 10.1242/dev.02474. [DOI] [PubMed] [Google Scholar]
- Chow RL, Lang RA. Early eye development in vertebrates. Annu Rev Cell Dev Biol. 2001;17:255–296. doi: 10.1146/annurev.cellbio.17.1.255. [DOI] [PubMed] [Google Scholar]
- Coulombre AJ, Coulombre JL. The role of intraocular pressure in the development of the chick eye: III. Ciliary body. Am J Ophthalmol. 1957;44:85–93. doi: 10.1016/0002-9394(57)90435-x. [DOI] [PubMed] [Google Scholar]
- Davis-Silberman N, Ashery-Padan R. Iris development in vertebrates; genetic and molecular considerations. Brain Res. 2008;1192:17–28. doi: 10.1016/j.brainres.2007.03.043. [DOI] [PubMed] [Google Scholar]
- Del Sal G, Ruaro ME, Philipson L, Schneider C. The growth arrest-specific gene, gas1, is involved in growth suppression. Cell. 1992;70:595–607. doi: 10.1016/0092-8674(92)90429-g. [DOI] [PubMed] [Google Scholar]
- Dias da Silva MR, Tiffin N, Mima T, Mikawa T, Hyer J. FGF-mediated induction of ciliary body tissue in the chick eye. Dev Biol. 2007;304:272–285. doi: 10.1016/j.ydbio.2006.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehn JJ, Diehn M, Marmor MF, Brown PO. Differential gene expression in anatomical compartments of the human eye. Genome Biol. 2005;6:R74. doi: 10.1186/gb-2005-6-9-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyer SD, Zhou G, Baldini A, Winterpacht A, Zabel B, Cole W, Johnson RL, Lee B. Mutations in LMX1B cause abnormal skeletal patterning and renal dysplasia in nail patella syndrome. Nat Genet. 1998;19:47–50. doi: 10.1038/ng0598-47. [DOI] [PubMed] [Google Scholar]
- Duester G. Families of retinoid dehydrogenases regulating vitamin A function: production of visual pigment and retinoic acid. Eur J Biochem. 2000;267:4315–4324. doi: 10.1046/j.1432-1327.2000.01497.x. [DOI] [PubMed] [Google Scholar]
- Escribano J, Coca-Prados M. Bioinformatics and reanalysis of subtracted expressed sequence tags from the human ciliary body: Identification of novel biological functions. Mol Vis. 2002;8:315–332. [PubMed] [Google Scholar]
- Fan X, Molotkov A, Manabe S, Donmoyer CM, Deltour L, Foglio MH, Cuenca AE, Blaner WS, Lipton SA, Duester G. Targeted disruption of Aldh1a1 (Raldh1) provides evidence for a complex mechanism of retinoic acid synthesis in the developing retina. Mol Cell Biol. 2003;23:4637–4648. doi: 10.1128/MCB.23.13.4637-4648.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantin VR, Wang Q, Lienhard GE, Keller SR. Mice lacking insulin receptor substrate 4 exhibit mild defects in growth, reproduction, and glucose homeostasis. Am J Physiol Endocrinol Metab. 2000;278:E127–133. doi: 10.1152/ajpendo.2000.278.1.E127. [DOI] [PubMed] [Google Scholar]
- Fu X, Sun H, Klein WH, Mu X. Beta-catenin is essential for lamination but not neurogenesis in mouse retinal development. Dev Biol. 2006;299:424–437. doi: 10.1016/j.ydbio.2006.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage PJ, Rhoades W, Prucka SK, Hjalt T. Fate maps of neural crest and mesoderm in the mammalian eye. Invest Ophthalmol Vis Sci. 2005;46:4200–4208. doi: 10.1167/iovs.05-0691. [DOI] [PubMed] [Google Scholar]
- Giovannone B, Scaldaferri ML, Federici M, Porzio O, Lauro D, Fusco A, Sbraccia P, Borboni P, Lauro R, Sesti G. Insulin receptor substrate (IRS) transduction system: distinct and overlapping signaling potential. Diabetes Metab Res Rev. 2000;16:434–441. doi: 10.1002/1520-7560(2000)9999:9999<::aid-dmrr159>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Gould DB, Smith RS, John SW. Anterior segment development relevant to glaucoma. Int J Dev Biol. 2004;48:1015–1029. doi: 10.1387/ijdb.041865dg. [DOI] [PubMed] [Google Scholar]
- Halfter W, Willem M, Mayer U. Basement membrane-dependent survival of retinal ganglion cells. Invest Ophthalmol Vis Sci. 2005;46:1000–1009. doi: 10.1167/iovs.04-1185. [DOI] [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. 1951. Dev Dyn. 1992;195:231–272. doi: 10.1002/aja.1001950404. [DOI] [PubMed] [Google Scholar]
- Idrees F, Vaideanu D, Fraser SG, Sowden JC, Khaw PT. A review of anterior segment dysgeneses. Surv Ophthalmol. 2006;51:213–231. doi: 10.1016/j.survophthal.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Imaizumi M, Kuwabara T. Development of the rat iris. Invest Ophthalmol. 1971;10:733–744. [PubMed] [Google Scholar]
- Jasoni C, Hendrickson A, Roelink H. Analysis of chicken Wnt-13 expression demonstrates coincidence with cell division in the developing eye and is consistent with a role in induction. Dev Dyn. 1999;215:215–224. doi: 10.1002/(SICI)1097-0177(199907)215:3<215::AID-AJA4>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Khoo KM, Chang CF. Characterization and localization of CD38 in the vertebrate eye. Brain Res. 1999;821:17–25. doi: 10.1016/s0006-8993(98)01347-x. [DOI] [PubMed] [Google Scholar]
- Kubo F, Takeichi M, Nakagawa S. Wnt2b controls retinal cell differentiation at the ciliary marginal zone. Development. 2003;130:587–598. doi: 10.1242/dev.00244. [DOI] [PubMed] [Google Scholar]
- Kubota R, McGuire C, Dierks B, Reh TA. Identification of ciliary epithelial-specific genes using subtractive libraries and cDNA arrays in the avian eye. Dev Dyn. 2004;229:529–540. doi: 10.1002/dvdy.20000. [DOI] [PubMed] [Google Scholar]
- Larsen M, Ressler SJ, Lu B, Gerdes MJ, McBride L, Dang TD, Rowley DR. Molecular cloning and expression of ps20 growth inhibitor. A novel WAP-type “four-disulfide core” domain protein expressed in smooth muscle. J Biol Chem. 1998;273:4574–4584. doi: 10.1074/jbc.273.8.4574. [DOI] [PubMed] [Google Scholar]
- Liu H, Mohamed O, Dufort D, Wallace VA. Characterization of Wnt signaling components and activation of the Wnt canonical pathway in the murine retina. Dev Dyn. 2003;227:323–334. doi: 10.1002/dvdy.10315. [DOI] [PubMed] [Google Scholar]
- Liu H, Xu S, Wang Y, Mazerolle C, Thurig S, Coles BL, Ren JC, Taketo MM, van der Kooy D, Wallace VA. Ciliary margin transdifferentiation from neural retina is controlled by canonical Wnt signaling. Dev Biol. 2007;308:54–67. doi: 10.1016/j.ydbio.2007.04.052. [DOI] [PubMed] [Google Scholar]
- Martinez-Morales JR, Signore M, Acampora D, Simeone A, Bovolenta P. Otx genes are required for tissue specification in the developing eye. Development. 2001;128:2019–2030. doi: 10.1242/dev.128.11.2019. [DOI] [PubMed] [Google Scholar]
- McIntosh I, Dreyer SD, Clough MV, Dunston JA, Eyaid W, Roig CM, Montgomery T, Ala-Mello S, Kaitila I, Winterpacht A, Zabel B, Frydman M, Cole WG, Francomano CA, Lee B. Mutation analysis of LMX1B gene in nail-patella syndrome patients. Am J Hum Genet. 1998;63:1651–1658. doi: 10.1086/302165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan AP, Davidson DR, Sime C, Graham E, Baldock R, Bhattacharya SS, Hill RE. The Msh-like homeobox genes define domains in the developing vertebrate eye. Development. 1991;112:1053–1061. doi: 10.1242/dev.112.4.1053. [DOI] [PubMed] [Google Scholar]
- Morimura N, Tezuka Y, Watanabe N, Yasuda M, Miyatani S, Hozumi N, Tezuka Ki K. Molecular cloning of POEM: a novel adhesion molecule that interacts with alpha8beta1 integrin. J Biol Chem. 2001;276:42172–42181. doi: 10.1074/jbc.M103216200. [DOI] [PubMed] [Google Scholar]
- Murtaugh LC, Chyung JH, Lassar AB. Sonic hedgehog promotes somitic chondrogenesis by altering the cellular response to BMP signaling. Genes Dev. 1999;13:225–237. doi: 10.1101/gad.13.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai T, Aruga J, Takada S, Gunther T, Sporle R, Schughart K, Mikoshiba K. The expression of the mouse Zic1, Zic2, and Zic3 gene suggests an essential role for Zic genes in body pattern formation. Dev Biol. 1997;182:299–313. doi: 10.1006/dbio.1996.8449. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Guichard A, Castro CP, Xiao Y, Rizen M, Zhang HZ, Hu D, Bang A, Helms J, Bier E, Derynck R. Characterization of a human rhomboid homolog, p100hRho/RHBDF1, which interacts with TGF-alpha family ligands. Dev Dyn. 2005;233:1315–1331. doi: 10.1002/dvdy.20450. [DOI] [PubMed] [Google Scholar]
- Napier HR, Kidson SH. Molecular events in early development of the ciliary body: a question of folding. Exp Eye Res. 2007;84:615–625. doi: 10.1016/j.exer.2006.07.012. [DOI] [PubMed] [Google Scholar]
- Oklu R, Hesketh R. The latent transforming growth factor beta binding protein (LTBP) family. Biochem J. 2000;352(Pt 3):601–610. [PMC free article] [PubMed] [Google Scholar]
- Pressman CL, Chen H, Johnson RL. LMX1B, a LIM homeodomain class transcription factor, is necessary for normal development of multiple tissues in the anterior segment of the murine eye. Genesis. 2000;26:15–25. [PubMed] [Google Scholar]
- Rowan S, Chen CM, Young TL, Fisher DE, Cepko CL. Transdifferentiation of the retina into pigmented cells in ocular retardation mice defines a new function of the homeodomain gene Chx10. Development. 2004;131:5139–5152. doi: 10.1242/dev.01300. [DOI] [PubMed] [Google Scholar]
- Sun G, Asami M, Ohta H, Kosaka J, Kosaka M. Retinal stem/progenitor properties of iris pigment epithelial cells. Dev Biol. 2006;289:243–252. doi: 10.1016/j.ydbio.2005.10.035. [DOI] [PubMed] [Google Scholar]
- Thut CJ, Rountree RB, Hwa M, Kingsley DM. A large-scale in situ screen provides molecular evidence for the induction of eye anterior segment structures by the developing lens. Dev Biol. 2001;231:63–76. doi: 10.1006/dbio.2000.0140. [DOI] [PubMed] [Google Scholar]
- Tietjen I, Rihel JM, Cao Y, Koentges G, Zakhary L, Dulac C. Single-cell transcriptional analysis of neuronal progenitors. Neuron. 2003;38:161–175. doi: 10.1016/s0896-6273(03)00229-0. [DOI] [PubMed] [Google Scholar]
- Trimarchi JM, Stadler MB, Cepko CL. Individual retinal progenitor cells display extensive heterogeneity of gene expression. PLoS ONE. 2008;3:e1588. doi: 10.1371/journal.pone.0001588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi JM, Stadler MB, Roska B, Billings N, Sun B, Bartch B, Cepko CL. Molecular heterogeneity of developing retinal ganglion and amacrine cells revealed through single cell gene expression profiling. J Comp Neurol. 2007;502:1047–1065. doi: 10.1002/cne.21368. [DOI] [PubMed] [Google Scholar]
- Tropepe V, Coles BL, Chiasson BJ, Horsford DJ, Elia AJ, McInnes RR, van der Kooy D. Retinal stem cells in the adult mammalian eye. Science. 2000;287:2032–2036. doi: 10.1126/science.287.5460.2032. [DOI] [PubMed] [Google Scholar]
- Urban S. Rhomboid proteins: conserved membrane proteases with divergent biological functions. Genes Dev. 2006;20:3054–3068. doi: 10.1101/gad.1488606. [DOI] [PubMed] [Google Scholar]
- Vollrath D, Jaramillo-Babb VL, Clough MV, McIntosh I, Scott KM, Lichter PR, Richards JE. Loss-of-function mutations in the LIM-homeodomain gene, LMX1B, in nail-patella syndrome. Hum Mol Genet. 1998;7:1091–1098. doi: 10.1093/hmg/7.7.1091. [DOI] [PubMed] [Google Scholar]
- Volpe P, Biral D, Pizzo P, Salviati G, Margreth A. Ontogenesis of chick iris intrinsic muscles: evidence for a smooth-to-striated muscle transition. Dev Biol. 1993;159:441–449. doi: 10.1006/dbio.1993.1254. [DOI] [PubMed] [Google Scholar]
- Zemel S, Bartolomei MS, Tilghman SM. Physical linkage of two mammalian imprinted genes, H19 and insulin-like growth factor 2. Nat Genet. 1992;2:61–65. doi: 10.1038/ng0992-61. [DOI] [PubMed] [Google Scholar]
- Zhao S, Chen Q, Hung FC, Overbeek PA. BMP signaling is required for development of the ciliary body. Development. 2002;129:4435–4442. doi: 10.1242/dev.129.19.4435. [DOI] [PubMed] [Google Scholar]
- Ziegler WH, Liddington RC, Critchley DR. The structure and regulation of vinculin. Trends Cell Biol. 2006;16:453–460. doi: 10.1016/j.tcb.2006.07.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clusters of genes expressed in the developing ciliary body. Hierarchical clustering of 44 microarrays including developing RGCs, ACs, PRs, cycling RPCs, and the two putative ciliary body cells. (A) Genes that clustered with Cyclin D2 with a correlation coefficient of >0.7 are shown. (B) Genes that clustered with Foxp2 with a correlation coefficient of >0.7 are shown.
ISH analysis of candidate genes for the peripheral mouse and chick eye. ISH on retinal cryosections was performed at three stages of mouse development: E12.5 (A, D, G, J), E16.5 (B, E, H, K) and P0 (C, F, I, L) and two stages of chick development: E6 (M) and E8 (N). The following probes were used: AI848240 [Zic1] (A–C), BE988982 [Zic2] (D–F), AI842763 [Rhbdf1] (G–I), AW045815 [Irs4] (J–L), and vinculin (M and N).
Further characterization of H19 expression. Close-ups of in situ hybridizations using an H19 riboprobe (BE949664) and E16.5 mouse retinas [A, B]. The ONBL is denoted by a red bar. P0 retinas were explanted, labeled with [3H]-thymidine for 1 hour, dissociated and probed for H19 alone [C, D] or for H19 (red) and Fgf15 (green) [E, F]. Images are shown with [C, E] and without [D, F] the development of the [3H]-thymidine. The arrows in E indicate cells that are positive for both H19 and Fgf15.
A comparison of ciliary body/iris marker genes from this study and others. (A) A Treeview generated heatmap showing the expression of 6 previously identified ciliary body marker genes (Thut et al., 2001) in E12 cell A6, P0 cell E8 along with the single RGCs, ACs, and PRs. ISH for Tgfb1i4 was performed on retinal cryosections at E12.5 (B), E14.5 (C), E16.5 (D) and P0 (E). Arrows indicate the location of the INBL staining. Scale bars are shown.
A comparison of ciliary body/iris marker genes from this study and others. (A) A Treeview generated heatmap showing the expression of a set of previously identified adult human ciliary body marker genes (Diehn et al., 2005) in E12 cell A6, P0 cell E8 along with the single RGCs, ACs, and PRs. (B) A Treeview generated heatmap showing the expression of a second set of previously identified adult human ciliary body marker genes (Escribano and Coca-Prados, 2002) in E12 cell A6, P0 cell E8 along with the single RGCs, ACs, and PRs.
Summary of gene expression in the mouse and chicken embryos using ISH and the probes used in this study. For each gene the corresponding cDNA used to generate the ISH riboprobe in both the chick and mouse is listed. A summary of the expression pattern observed at each stage is included. Terms used to describe the in situ patterns: Outer neuroblastic layer/ventricular zone (ONBL), inner neuroblastic layer (INBL), retinal pigment epithelium (RPE), not detectable (ND), (NT) not tested, and (NA) no probes available.