Abstract
Cocaine addiction is characterized by a gradual loss of control over drug use, but molecular mechanisms regulating vulnerability to this process remain unclear. Here we report that microRNA-212 (miR-212) is upregulated in the dorsal striatum of rats with a history of extended access to cocaine. Striatal miR-212 decreases responsiveness to the motivational properties of cocaine by dramatically amplifying the stimulatory effects of the drug on CREB signaling. This action occurs through miR-212-enhanced Raf-1 activity, resulting in adenylyl cyclase sensitization and increased expression of the essential CREB co-activator TORC (Transducer of Regulated CREB; also known as CRTC). Our findings suggest that striatal miR-212 signaling plays a key role in determining vulnerability to cocaine addiction, reveal novel molecular regulators that control the complex actions of cocaine in brain reward circuitries, and provide an entirely new direction for the development of anti-addiction therapeutics based on modulation of noncoding RNAs.
Cocaine triggers a constellation of cellular and molecular alterations in brain reward systems, and cocaine addiction is commonly viewed as a disorder of neuroplasticity 1,2. Such long-lasting structural and functional modifications are thought to increase sensitivity to the motivational effects of cocaine and associated environmental stimuli, culminating in a loss of control over intake 3. However, recent findings support a more nuanced view in which cocaine also triggers adaptations in brain reward systems that decrease responsiveness to the drug 4,5. This notion of “yin-yang” neuroplastic responses to cocaine that can increase or decrease sensitivity to its motivational effects likely accounts for the fact that only a small fraction (~15%) of human cocaine users lose control over intake and develop compulsive drug seeking behaviors 6. Similar to human cocaine users, addiction-like drug seeking can be detected in rats that intravenously self-administer cocaine 7,8,9. In particular, rats with a history of extended cocaine access demonstrate increasing motivation to obtain the drug, reflected in escalating consumption 7, and higher “break-points” under progressive ratio reinforcement schedules 10. In a small percentage of rats (~15%) with extended cocaine access, motivation to obtain the drug increases to such a degree that their behavior can be considered “addiction-like” when assessed according to the same diagnostic criteria used for human drug users 9,11. Understanding factors that regulate the motivational properties of cocaine in rats under extended access conditions may therefore reveal important insights into the neurobiological basis of vulnerability to addiction.
Considering the complexity of cocaine-induced neuroadaptive responses in brain reward systems, it is likely that highly synchronized programs of gene regulation are involved. Particularly interesting in this regard are the microRNAs (miRNAs), which are small (~21–23 nucleotides) noncoding RNA transcripts that regulate gene expression at the post-transcriptional level. miRNAs control gene expression by binding to complementary sequences (miRNA response elements; MREs) in the 3′ untranslated region (3′ UTR) of target mRNA transcripts to facilitate their degradation and/or inhibit their translation 12. Because of their ability to coordinate the expression of networks of related genes responsible for brain structure and function 13,14, it has been proposed that miRNAs may play important roles in complex psychiatric disorders 15,16. Until now, little has been known about their potential involvement in addiction 17,18,19. Here, we identify a key role for miRNAs in regulating compulsive-like cocaine intake.
Cocaine increases miR-212 expression
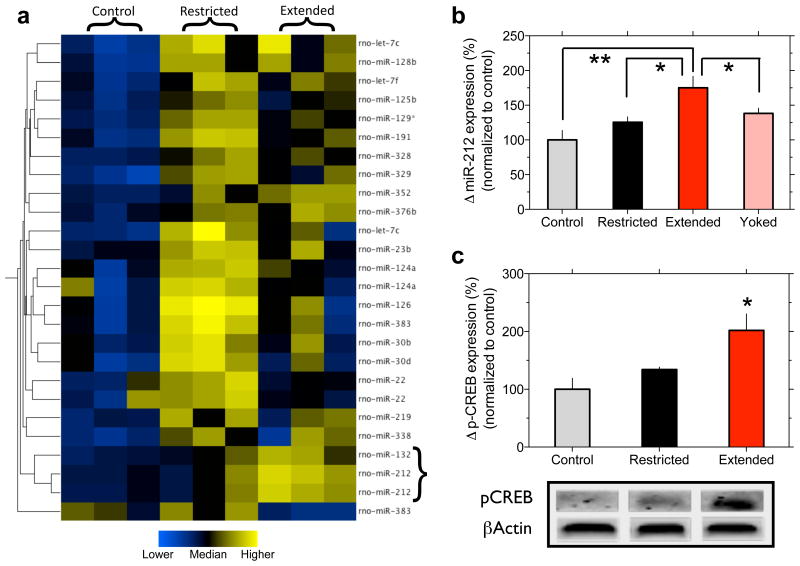
The dorsal striatum is a key brain region regulating the development of compulsive cocaine use 20,21,22,23. Expression profiling showed that miR-212, and the closely related miR-132 , was upregulated, 1.75-fold in the dorsal striatum of rats with extended (6 h) daily access to intravenous cocaine self-administration (0.5 mg kg -1 per injection; n=6) compared with cocaine-naive control rats (n=6) (Fig. 1a; Supplementary Figs. 1 and 2), an effect verified by Taqman assay (Fig. 1b; see Supplementary Fig. 3 for other brain regions). Striatal miR-212 expression was also increased compared to rats with restricted (1 h) access (n=6), and “yoked” rats (n=6) that received cocaine infusions at the same time points as extended access rats, but in a response-independent manner. miR-212 expression is highly responsive to cAMP response element binding protein (CREB) 14,24. Accordingly, striatal phospho-Ser133-CREB (p-CREB) (Fig. 1c) and total CREB (t-CREB; Supplementary Fig. 4) were increased in extended access rats (Fig. 1b).
Figure 1. Increased striatal miR-212 expression in extended access rats.
a, Expression analysis revealed that striatal miR-212 levels were increased in rats with extended access to cocaine self-administration. b, Taqman assay verified that striatal miR-212 levels were increased in extended access rats (F3,23=6.6, P<0.01; *P<0.05, **P<0.01, statistically significant compared with the extended access group). c, Relative amounts of p-CREB in the dorsal striatum were quantified by densitometry (F2,6=6.8, P<0.05; *P<0.05 compared with control). The lower panel is a representative immunoblot of the increased striatal p-CREB expression. Data are presented as mean ± SEM.
Effects of miR-212 oncocaine intake
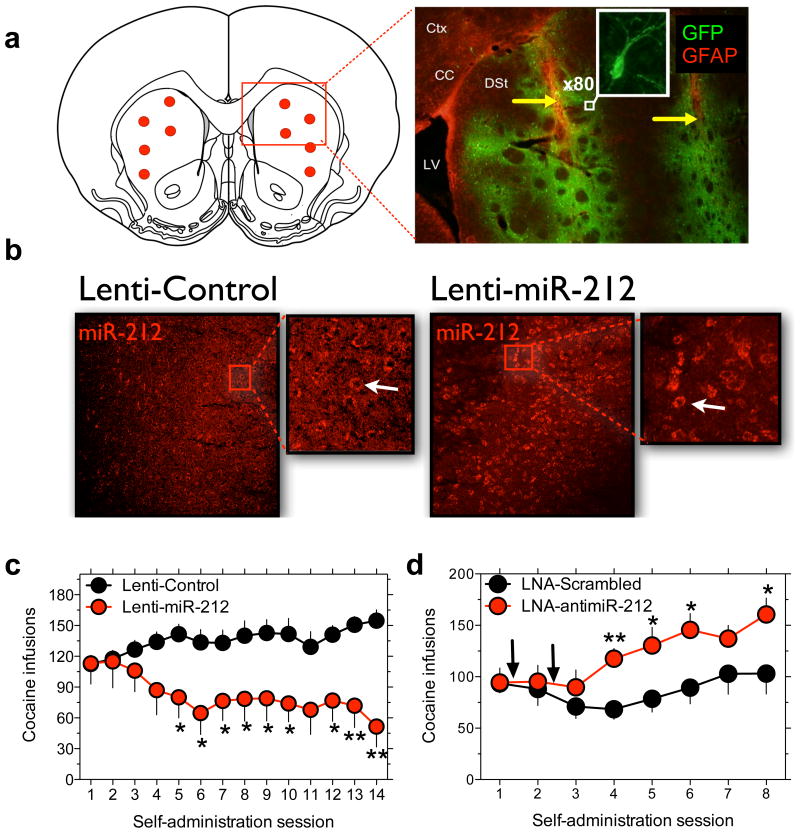
Next, we used a lentivirus vector to overexpress miR-212 (Lenti-miR-212) in dorsal striatum (Fig. 2a), and verified this effect using fluorescent in situ hybridization (FISH; Fig. 2b) and Taqman (Supplementary Fig. 5). Lenti-miR-212 rats, and rats treated with an empty vector (Lenti-Control rats), learned to respond for food reinforcement at similar rates (Supplementary Fig. 6), demonstrating that miR-212 overexpression did not alter operant performance. Cocaine intake did not differ between Lenti-miR-212 and Lenti-Control rats with restricted access (Supplementary Fig. 7). In contrast, strikingly different patterns of intake were revealed under extended access conditions (Fig. 2c). Consistent with previously reports 7, cocaine intake escalated in Lenti-Control rats with extended access (Fig. 2c). Conversely, Lenti-miR-212 initially consumed the same high levels of cocaine, but their intake became progressively lower as exposure to the drug increased across sessions (Fig. 2c; see Supplementary Fig. 8 for non-reinforced “inactive” lever responses). Striatal overexpression of miR-1 did not alter cocaine intake (Supplementary Fig. 9), confirming that these effects were specific to miR-212. When the unit dose of cocaine available for self-administration was varied, we found that the cocaine dose-response (D-R) curve was shifted upward in Lenti-Control rats with extended access compared to those with restricted access (Supplementary Fig. 10). Remarkably, the complete opposite effect occurred in Lenti-miR-212 rats, with the cocaine D-R curve shifting downward in those with extended access (Supplementary Fig.11). Upward or downward shifts in the D-R curve are interpreted as increased or decreased, respectively, in motivation to consume cocaine 7,25. Therefore, in contrast to the progressively increasing motivation to consume cocaine typically seen in rats with extended drug access 7,8,9, Lenti-miR-212 rats were progressively less motivated to consume cocaine across sessions, and cocaine may have become aversive reflected in low levels of intake across doses. The cocaine D-R curve was similar in Lenti-miR-212 and Lenti-Control rats with restricted access (Supplementary Fig. 12), demonstrating that striatal miR-212 regulates intake only under extended access conditions. Interestingly, non-reinforced responding during the 20-sec timeout period after each cocaine infusion, which may reflect compulsive drug-seeking behavior 9, increased over sessions in the extended access Lenti-Control rats (Supplementary Fig. 13), and striatal miR-212 overexpression abolished this effect.
Figure 2. Dissociable effects of striatal miR-212 oncocaine intake.
a, Red circles in left panel are locations at which viral infusions were targeted in dorsal striatum. The right panel is representative immunochemistry staining from a Lenti-miR-212 rat. Green is GFP from virus; red is the astrocyte marker glial fibrillary acidic protein (GFAP); Ctx, cortex; CC corpus callosum; LV, lateral ventricle; DSt, dorsal striatum; yellow arrows highlight the injector track used to deliver virus. Insert is an x80 confocal image of a virus-infected neuron. b, FISH was used to visualize striatal miR-212 expression (shown in red) in Lenti-Control and Lenti-miR-212 rats. c, Striatal miR-212 overexpression reverses the long-term trajectory of cocaine-taking behavior in rats with extended access (Virus x Session: F13,130=3.0, P<0.001; *P<0.05, **P<0.01 compared with intake on the same day in Lenti-Control rats). d, LNA-antimiR-212 delivered into dorsal striatum increases cocaine intake in extended access rats (LNA x Session: F4,36=5.3, P<0.005; *P<0.05, **P<0.01, compared with intake on the same day in the LNA-Scrambled rats). Data are presented as mean ± SEM.
Inhibition of striatal miR-212signaling, using a locked nucleic acid (LNA)-modified antisense oligonucleotide 26,27 (LNA-antimiR-212; Supplementary Figs. 14 and 15), increased cocaine intake in rats with extended (Fig. 2d) but not restricted (Supplementary Fig. 16) access, reflected in sharply increased consumption 48 h after the last LNA-antimiR-212 injection that persisted >4 days (Fig. 2d). Nonreinforced responding during timeout periods also increased in the LNA-antimiR-212-treated rats with extended but not restricted access (Supplementary Fig. 17). An antisense oligonucleotide against miR-1 (LNA-antimiR-1) did not alter cocaine intake (Supplementary Fig. 18), confirming that these effects were specific to miR-212. Disruption of striatal miR-212 signaling therefore precipitates compulsive-like responding for cocaine 9. Hence, cocaine-induced increases in striatal miR-212 may represent a novel anti-addiction counter-adaptive response in brain reward circuitries, and deficits in miR-212 signaling may increase vulnerability to addiction.
miR-212 amplifies CREB signaling
It has been proposed that gene regulatory networks in the brain, similar to electronic circuits, are comprised of feedforward and feedback loops, and that miRNAs can amplify or curtail activity in these networks 28. Cocaine-induced activation of CREB signaling is considered an important compensatory response that decreases the motivational properties of the drug 5,29,30, and miR-212 is highly CREB-responsive 14,24. We hypothesized that excessive cocaine intake may engage a novel feedforward circuit in the dorsal striatum in which activation of CREB signaling increases miR-212 expression, which in turn loops back to amplify the actions of this transcription factor and thereby decrease the motivation to consume cocaine.
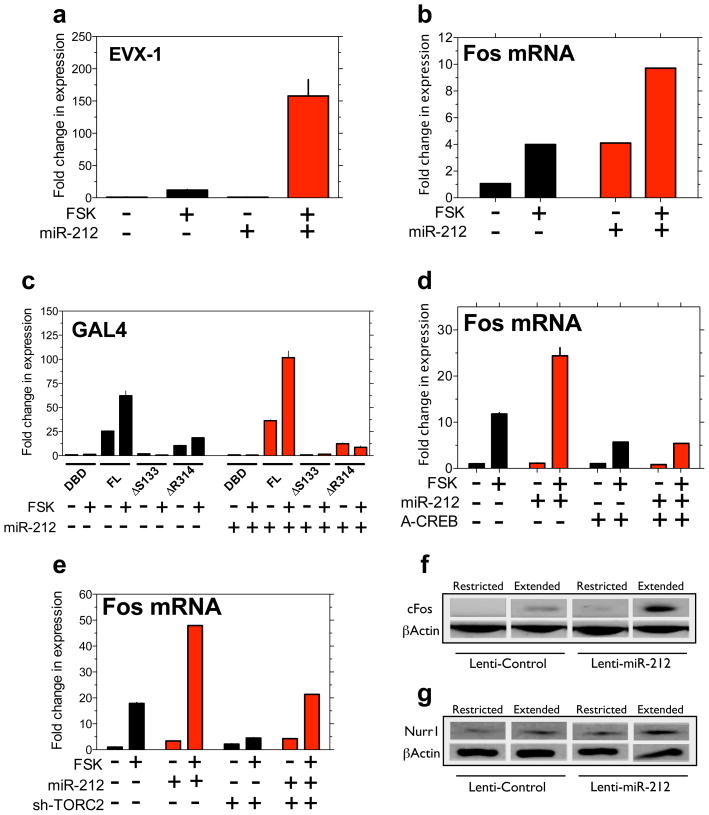
Consistent with this hypothesis, miR-212 markedly amplified CREB signalling evoked by a low concentration of forskolin (5 μM) compared with vector-transfected cells (Fig. 3a; Supplementary Fig. 19), measured using a luciferase-based CREB reporter construct (CRE-containing element from promoter of EVX-1) 31. CREB signaling was also amplified by the closely related miRNA, miR-132 (Supplementary Fig. 20), but not by unrelated miRNAs (miR-1 or miR-29b; Supplementary Fig. 21). Importantly, miR-212 potentiated increases in Fos mRNA (Fig. 3b), a known CREB-responsive gene, confirming that miR-212 acts on endogenous CREB-responsive genes and not just reporter constructs. Using a GAL4-luciferase reporter system in which the GAL4 DNA binding domain (DBD) is fused to the N-terminus of the full length CREB (FL), we found that a phosphorylation-defective CREB mutant (ΔS133) no longer responded to miR-212 (Fig. 3c). miR-212 also had no effects onGAL4-CREB with a mutation in the bZIP domain (ΔR314) that abolishes the recruitment of the essential CREB co-activator TORC (Fig. 3c). Similarly, a dominant-negative CREB polypeptide (A-CREB) 31 abolished the stimulatory effects of miR-212 on a CREB luciferase reporter (Supplementary Fig. 22), and on Fos gene expression (Fig. 3d). Knockdown of TORC2, the most abundant family member in HEK cells 31, also abolished the effects of miR-212 on the EVX-1 reporter (Supplementary Fig. 23) and on Fos expression (Fig. 3e). Finally, striatal CREB signaling was engaged in rats with extended but not restricted access to cocaine (Fig. 1c). Accordingly, striatal protein levels of the CREB-responsive genes Fos (Fig. 3f) and Nurr1 (Fig. 3g) were upregulated in Lenti-Control rats with extended access, and this effect was amplified in Lenti-miR-212 rats (Fig. 2c). Thus, miR-212 amplifies cAMP-responsive gene expression through the canonical CREB:TORC signaling cascade.
Figure 3. miR-212 amplifies CREB signaling.
a, miR-212 potentiated CREB signaling engaged by forskolin (FSK),measured using an EVX-1 luciferase reporter (miRNA x FSK: F1,8=99.2, P<0.001). b, miR-212 potentiated forskolin-induced increases in Fos mRNA in HEK cells(miRNA x FSK: F1,8>1000, p<0.001). c, The effects of miR-212 on CREB signaling were abolished in GAL4-luciferase reporters containing phosphorylation-defective (ΔSer133) or TORC recruitment-defective (ΔR314) CREB mutants. d, A-CREB polypeptide abolished the effects of miR-212 on Fos mRNA expression. e, RNAi-mediated knockdown of TORC2 also abolished the effects of miR-212 on Fos mRNA expression. f, Protein levels of Fos were enhanced in dorsal striatum from Lenti-miR-212 rats with extended cocaine access compared with other groups. g, Striatal levels of Nurr1, another CREB-responsive gene, were also enhanced inLenti-miR-212 rats with extended access. Data are presented as mean ± SEM.
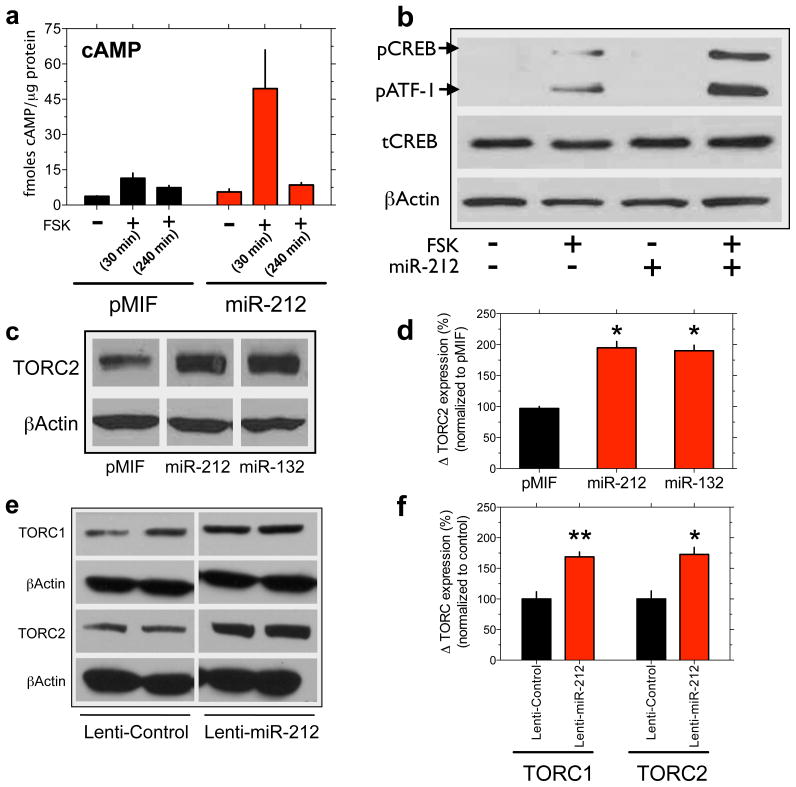
Next, we found that miR-212 potentiated forskolin-induced accumulation of cAMP in HEK cells with 30 min but not 240 min treatment (Fig. 4a). Type IV phosphodiesterases (PDE4) are major subtype of PDE responsible for cAMP degradation in HEK cells and striatum 32,33. The PDE4 inhibitor rolipram potentiated the effects of miR-212 on cAMP accumulation, and miR-212 did not alter PDE4D expression (Supplementary Fig. 24). Thus, miR-212 likely increases cAMP levels by sensitizing its production rather than inhibiting its breakdown. miR-212 also potentiated forskolin-stimulated increases of p-CREB (but not t-CREB; Fig. 4b), and increased expression of TORC2 (Fig. 4c and 4d; see Supplementary Fig. 25). cAMP can trigger CBP/p300-mediated acetylation of TORC, which protects against its degradation 34. miR-212 increased levels of total and acetylated TORC2 (Supplementary Fig. 26), suggesting that miR-212 increases TORC expression indirectly through increased cAMP levels. Finally, miR-212 also increased TORC1 and TORC2 expression in dorsal striatum (Fig. 4e and 4f). Together, these data reveal miR-212 as a novel regulator of CREB signaling that acts by sensitizing cAMP production and increasing expression of the TORC family of CREB coactivators.
Figure 4. miR-212 stimulates core CREB signaling components.
a, miR-212 potentiated cAMP accumulation in HEK cells (miRNA x FSK: F2,8=4.8, P<0.05). b, miR-212 also increased p-CREB and the related p-ATF-1, without altering total-CREB. c, Representative immunoblots of the increased TORC2 levels in HEK cells. d, Relative amounts of TORC2 in HEK cells expressing miR-212, miR-132 or vector (pMIF) were quantified by densitometry (*P<0.05 compared with pMIF). e, Representative immunoblots showing that miR-212 increased striatal levels of TORC1 and TORC2. f, The relative amounts of striatal TORC1 and TORC2 were quantified by densitometry (*P<0.05, **P<0.01, compared with Lenti-Control).Data are presented as mean ± SEM.
miR-212 amplifies CREB through Raf-1
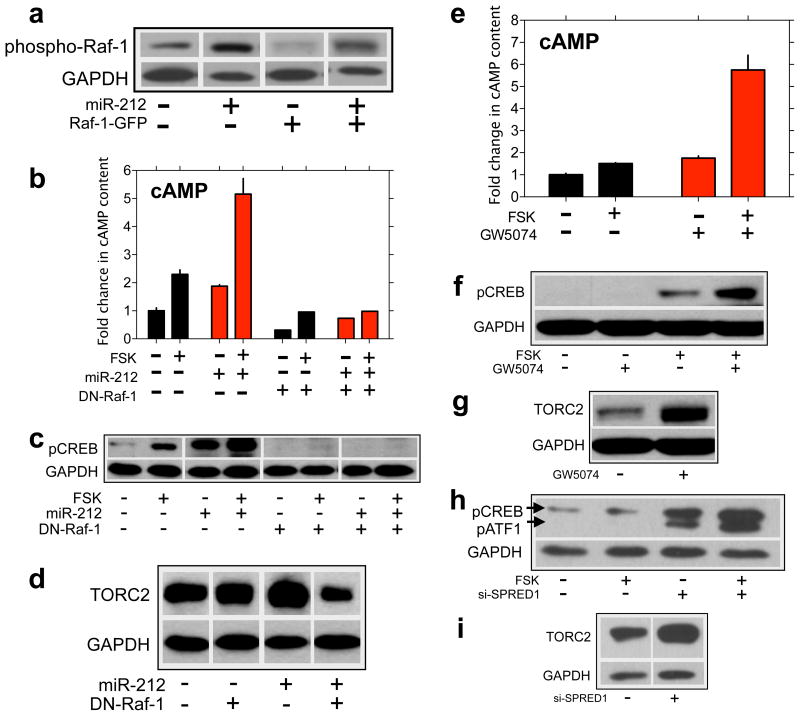
Raf-1 phosphorylates various isoforms of adenylyl cyclases and sensitizes their activity 35,36. Indeed, the characteristic increases in cAMP signaling associated with chronic opiate treatment 37 that are thought to contribute to the aversive opiate withdrawal syndrome occur through Raf-1-mediated sensitization of adenylyl cyclase activity 38. In many cases, phosphorylation of Raf-1 at Ser 338 is required for its activation 39, and we found that miR-212 increased phospho-Ser338-Raf-1 levels in HEK cells (Fig. 5a), without altering total Raf-1 levels (Supplementary Fig. 27). When we overexpressed exogenous GFP-tagged Raf-1 (to elevate basal levels of Raf-1), miR-212 also increased levels of the phosphorylated exogenous protein (Fig. 5a; for total Raf1 levels see Supplementary Fig. 27). Inhibition of Raf-1 function using a dominant-negative truncated version (amino acids 51-220) of the protein (DN-Raf-1), abolished the stimulatory effects of miR-212 on cAMP production (Fig. 5b), and lowered basal and forskolin-evoked increases in cAMP content in control cells (Fig. 5b). Disruption of Raf-1 also abolished miR-212-induced amplification of p-CREB levels in response to forskolin (Fig. 5c) and decreased basal and forskolin-enhanced p-CREB levels in control cells (Fig. 5c) without altering total CREB expression levels (Supplementary Fig. 28). Finally, disruption of Raf-1 signaling abolished miR-212-induced increases in TORC2 expression (Fig. 5d).
Figure 5. miR-212 amplifies CREB signaling through Raf-1.
a, miR-212 activates Raf-1 signaling, reflected in increased levels of phosphorylated endogenous and exogenous (Raf-1-GFP) Raf-1 protein. b, Dominant-negative Raf-1 (DN-Raf-1) abolished the stimulatory effects of miR-212 on cAMP accumulation. c, DN-Raf-1abolished miR-212-induced increases in p-CREB. d, DN-Raf-1 also abolished miR-212-induced increases in TORC2. e, Enhancing Raf-1 signaling by pulsing cells with the Raf-1 inhibitor GW5074 potentiated cAMP accumulation. f, Potentiation of Raf-1 signaling increased p-CREB levels. g, Potentiation of Raf-1 signaling increased TORC2 levels. h, Knockdown of the Raf-1 repressor SPRED1, a target for miR-212, increased p-CREB levels. i, SPRED1 knockdown also increased TORC2 levels. Data are presented as mean ± SEM.
A useful feature of pharmacological inhibitors of Raf is their bimodal action on the activity of the kinase in intact cells 40. Cells briefly exposed to a Raf inhibitor display a paradoxical increase in Raf activity when the inhibitor is washed away 40,41,42. We verified that a brief pulse (30 min) of the Raf-1 inhibitor GW5074 increased Raf-1 activity in HEK cells (Supplementary Fig. 29). Using this approach to upregulate Raf activity, we found that activation of Raf-1 signaling amplified forskolin-stimulated increases in cAMP content in cells (Fig. 5e), potentiated increases in p-CREB levels (Fig. 5f) and upregulated TORC2 levels (Fig. 5g), fully recapitulating the effects of miR-212.
miRNAs exert their actions by binding to the 3′ UTR of target mRNA transcripts to facilitate their degradation and/or inhibit their translation 12. To identify gene targets for miR-212 that may explain its actions we assessed gene expression profiles using microarrays in HEK cells transfected with miR-212 or vector 43,44. All predicted targets for miR-212 were collated (http://www.targetscan.org/) whose expression was reduced by ≥30% by miR-212. This analysis identified ~100 predicted targets for miR-212 whose expression was knocked down (Supplementary Tables1 and 2). Amongst these target transcripts were at least 7 previously shown to regulate Raf-1 signaling: SMAD4, SPRED1, DACH1, ERBB2IP (Erbin), APC, RASA1 (p120GAP), and SLK (STE20-like kinase; Fig. 6e). Most notable was SPRED1 (sprouty-related, EVH1 domain containing 1), a recently identified core repressor of Raf-1 function 45,46. We tested the hypothesis that miR-212-mediated repression of SPRED1 may contribute to its stimulatory effects on Raf-1 and CREB:TORC signaling. First, miR-212 knocked down protein levels of SPRED1 in HEK cells (Supplementary Fig. 30). SPRED1 was expressed in a punctuate fashion near the plasma membrane of dorsal striatal cells, and miR-212 knocked down striatal SPRED1 levels (Supplementary Fig. 31). Second, RNAi-mediated knockdown of SPRED1 increased levels of phospho-Ser338-Raf-1 in HEK cells (Supplementary Fig. 32), similar to the actions of miR-212. Moreover, SPRED1 knockdown potentiated increases in p-CREB in response to forskolin (Fig. 5h) without altering t-CREB (Supplementary Fig. 32) and increased TORC2 expression levels (Fig. 5i), further recapitulating the effects of miR-212. Third, expression of a clone containing only the coding sequence of the SPRED1 gene, but not the 3′UTR, attenuated but did not block the stimulatory effects of miR-212 on p-CREB levels (Supplementary Fig. 33). These data suggest that miR-212 activates Raf-1 and thereby amplifies the CREB:TORC cascade in part through repression of SPRED1.
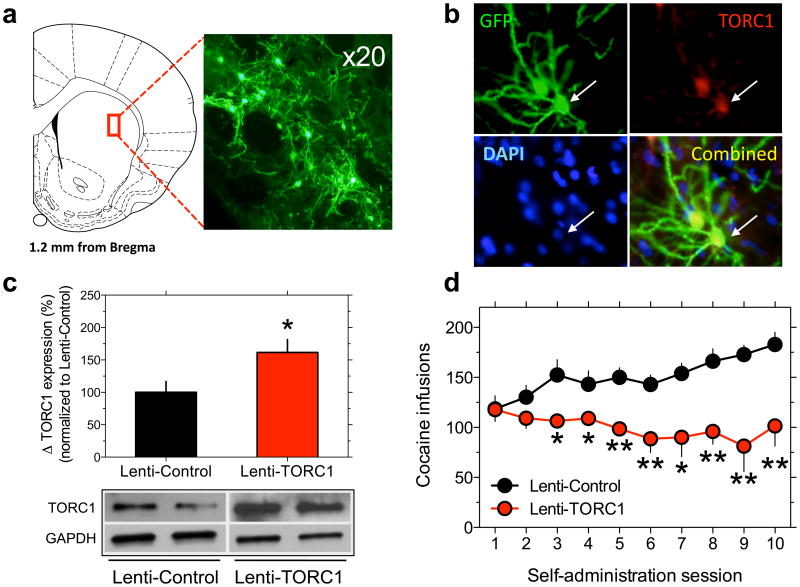
Figure 6. Striatal CREB:TORC signaling controls cocaine intake.
a, Representative image of Lenti-TORC1-infected neurons (green) in the dorsal striatum. b, Representative high-magnification images of Lenti-TORC1 infected cells (green), demonstrating the viral-driven upregulation of TORC1 expression (red). Cell nuclei are shown in blue using DAPI staining. c, Relative amounts of TORC1in dorsal striatum were quantified by densitometry (*P<0.05). The lower panel is a representative immunoblot. d, Cocaine intake under extended access conditions is far lower in Lenti-TORC1 rats compared with Lenti-Controls(Virus x Session: F9,.90=2.9, P<0.001; *P<0.05, **P<0.001 compared with intake on the same day in Lenti-Control rats).Data are presented as mean ± SEM.
Striatal CREB:TORC limits cocaine intake
A major consequence of striatal miR-212 overexpression was increased TORC expression. Knockdown of TORC1, the predominant family member in brain 47, abolishes CREB-regulated gene expression, dendritic growth and synaptic plasticity in neurons 48,49,50. Conversely, TORC1 overexpression enhances CREB-responsive gene expression 31. To directly investigate the functional relevance of miR-212-induced amplification of striatal CREB:TORC signaling, we next tested the effects of striatal TORC1 overexpression on cocaine intake.
Using a lentivirus vector to overexpress TORC1 (Lenti-TORC1) in dorsal striatum (Fig. 6a–c, and Supplementary Fig. 34), we found that Lenti-TORC1 and Lenti-Control rats acquired operant responding for food reinforcement at the same rate (Supplementary Fig. 35), and that their mean daily cocaine intake (Supplementary Fig. 36) and cocaine D-R curves (Supplementary Fig. 37) did not differ under restricted access conditions. Cocaine intake progressively escalated in the Lenti-Control rats under extended access conditions (Fig. 6d). In contrast, cocaine intake was significantly lower in the Lenti-TORC1 rats and did not escalate (Fig. 6d). Similarly, the cocaine D-R curve was shifted downward in the Lenti-TORC1 rats compared with the Lenti-controls (Supplementary Fig. 38). These data support a conceptual framework in which miR-212 amplification of striatal CREB:TORC signaling counters the motivational properties of the drug (see Supplementary Figure 39).
Conclusions
Repeated cocaine overconsumption is considered an important risk factor for drug-seeking behaviors to switch from controlled to uncontrolled 7,8,9. We show here that miR-212 is a cocaine-responsive noncoding RNA whose expression is increased in the dorsal striatum of rats with extended but not restricted cocaine access, and that striatal miR-212 signaling decreases responsiveness to the motivational properties of cocaine. miR-212 controls cocaine intake by dramatically amplifying the activity of CREB, a known negative regulator of cocaine reward. This action occurs through Raf-1-mediated sensitization of adenylyl cyclase activity and increased TORC expression (see Supplementary Fig. 39). Striatal miR-212 therefore protects against development of compulsive drug-taking, and factors that regulate miR-212 signaling may play key roles in determining vulnerability to cocaine addiction. Hence, miRNAs are revealed as important molecular regulators that control the complex actions of cocaine in brain reward circuitries and neuroadaptations associated with addiction.
Methods
Animals
Male Wistar rats (Charles River Laboratories, Raleigh, NC) weighing 300-320 g were housed in groups of 1-2 per cage in a temperature-controlled vivarium on a 12-h reverse light/dark cycle (lights off at 7:00 a.m.). Food and water were available ad libitum except when animals were trained to perform the operant response to receive food rewards, when animals were restricted to 20 g chow per day. All procedures were conducted in strict adherence with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee of The Scripps Research Institute.
Cocaine self-administration
Rats were anesthetized by inhalation of 1-3% isoflurane in oxygen, and surgically prepared with silastic catheters in the jugular vein 51. The catheter was passed subcutaneously to a polyethylene assembly mounted on the animal’s back. Rats were permitted at least 7 days recovery before behavioral training commenced. Animals were food restricted (20 g per day; 3-4 days) then trained to respond on an “active” lever for food pellets (45 mg; 60 min sessions) under a FR5TO20 reinforcement schedule 52. Rats were also presented with an “inactive” lever during training and testing sessions, responses on which were recorded but were without scheduled consequence. Rats responded for food until criteria was reached (>80 pellets per 1 h session). Rats then responded for cocaine on the FR5TO20 sec reinforcement schedule during 1 h daily testing sessions for at least 7 consecutive days. Cocaine hydrochloride was dissolved in sterile saline solution (0.9% w/v). Each cocaine infusion earned resulted in the delivery of 0.5 mg/kg/infusion cocaine (0.1 ml injection volume delivered over 4-sec), and initiated a 20-sec time-out period signaled by a light cue located above the active lever during which responding on the lever was recorded but was without consequence. After training in the cocaine self-administration procedure as described above, two balanced groups of rats were formed so that they consumed similar amounts of cocaine. One group of rats continued to respond for cocaine infusions during 1-h daily testing session, or responded for cocaine during 6-h daily sessions. Additionally, a separate control group of rats were matched to individual animals in the extended access group to receive the same number and frequency of food and cocaine rewards in a response-independent manner (yoked rats). To determine the cocaine dose-response (D-R) curve, the unit dose of cocaine available for self-administration was adjusted upward or downward during 3-hour testing sessions every other day between regular 6-h self-administration sessions. When comparing the D-R curves between restricted and extended access groups, only intake during the first 60 min of the 3 h session were included in statistical analyses, see 53. Doses of cocaine were tested once, and in the following order: 0.5, 0.0625, 0.25, 0.125, and 0 mg/kg/infusion.
Intracerebral injection procedures
For intra-striatal administration of lentivirus vectors, rats were first anaesthetized by inhalation of 1-3% isoflurane in oxygen and positioned in a stereotaxic frame (Kopf Instruments, Tujunga, CA). A total of 5 viral supernatant injections (1 μl per injection; viral supernatant concentrations ranged from 3.6 x 107 – 4.5 x 109 infection units/ml) were delivered into each side of the striatum, for a total of 10 striatal injections per rat. The viruses were directed toward medial and lateral portions of the dorsal striatum. Medial injection sites were according to the following stereotaxic coordinates 54: anterior/posterior (AP): 1.20 mm from Bregma; medial/lateral (ML): ±2.00 mm from midline; dorsal/ventral (DV): −5.0 and −3.8 mm below dura. Lateral injection sites were according to the following stereotaxic coordinates 54: AP: 1.20 mm from Bregma; ML: ±3.25 mm from midline; DV: −6.5, −5.5 and −4.5 mm below dura. To deliver the virus, a small hole was drilled through the skull at the ML coordinate, and a stainless steel injector (32 gauge, 14 mm in length) was lowered to the most ventral injection site. The viral supernatant injection was delivered over 60 sec. After the infusion, the injector was left in place for an additional 60 sec. The injector was then raised to the next more dorsal injection site, and the injection procedure repeated. After the final virus supernatant injection, the drill holes in the skull were filled with dental acrylic, the scalp sutured, and the incision site treated with antibiotic ointment.
For intra-striatal administration of LNA oligonucleotides, chronic indwelling intracerebral cannula were first implanted above the dorsal striatum. Briefly, rats were anaesthetized by inhalation of 1–3% isoflurane in oxygen and positioned in a stereotaxic frame (Kopf Instruments, Tujunga, CA). Bilateral stainless steel guide cannulae (23 gauge, 12 mm in length) were implanted 2.0 mm above the most dorsal injection site in the dorsal striatum according to the following stereotaxic coordinates 54: AP: 1.20 mm from bregma; ML: ±3.25 mm from midline; DV: −2.40 mm from dura. Four stainless steel skull screws and dental acrylic held the cannulae in place. Cannulae were kept patent using 12 mm long stainless steel stylets. The oligonucleotides were delivered on two consecutive days. On each day, animals were anesthetized after their daily cocaine self-administration session, and received a total of 3 oligonucleotide injections (1 μl per injection; 25 μM concentration) into each side of the dorsal striatum (a total of 6 striatal injections per rat per day). A stainless steel injector (32 gauge, 16 mm in length) was lowered into the most ventral injection site. The molecule was delivered over 60 sec. After the infusion, the injector was left in place for an additional 60 sec. The injector was then raised 1 mm to the next more dorsal injection site, and the injection procedure repeated. After the final LNA injection, the 12 mm long stylet was re-inserted into the cannula.
Statistical Analyses
Cocaine intake data were analyzed by one- or two-way repeated-measures analysis of variance (ANOVA). Significant main or interaction effects were followed by Bonforroni post-tests or Newman-Keuls post-hoc tests as appropriate. All statistical analyses were performed using Graphpad Prism software. The level of significance was set at 0.05.
RNA isolation -Cells
Total RNA was extracted from HEK 293 cells using RNASTAT60 (Tel-Test Company) kit according to manufacturer’s instructions. cDNA was prepared by reverse transcription of 500 ng total RNA using Superscript II enzyme and oligo dT primer. The resulting cDNAs were amplified using the Taqman kits (System Biosciences) and an ABIPRISM 7700 sequence detector (Perkin Elmer). All mRNA expression data was normalized to GAPDH expression in the corresponding sample.
RNA isolation -Tissues
Rats were euthanized and brains rapidly removed and frozen in chilled isopentane previously stored at −80°C and kept on dry ice. Brains were sliced on a cryostat (HM 505 E, Microm, Walldorf, Germany) and kept at −20°C until each region of interest comes into the cutting plane. Bilateral 1 mm2 punches of the PFC, dorsal striatum, NAcc, hippocampus, VTA and cerebellum were collected into Eppendorf tubes on dry ice for RNA isolation. Total RNA for miRNA and mRNA expression analyses was isolated from tissue samples using phenol and chloroform extractions. ~30 mg of tissue was homogenized in 1 ml of RNASTAT60 (Tel-Test Company) using 26 or 27 gauge needles, followed by addition of 250 μl of chloroform and vortexed for 1 min. After centrifugation of the samples for 15 min at 12,000 x g (at 4°C), the upper aqueous RNA containing layer was removed for an additional RNASTAT60/chloroform extraction. The RNA was precipitated with 2 X volumes of isopropanol overnight at -20°C and centrifuged for 30 min at 12000 X g. The RNA pellets were washed twice with 70% ethanol (made using RNAase-free water) and subsequently re-suspended in RNAsecure (Ambion). ~10 μg of RNA from each sample was treated with Turbo DNase (Ambion) for 60 min at 37°C to degrade genomic DNA.
miRNA Expression Profiling and data analysis
Samples of small RNA (≤200 nucleotides) from the isolated RNA samples generated from rat dorsal striatum tissues were obtained by size fractionation on YM-100 ultrafiltration columns (Millipore). The small RNAs were 3′-end labeled with Oyster-550 fluorescent dye using the Flash-Tag kit (Genisphere). The labeled RNA was hybridized overnight to epoxide glass slides double-spotted with the NCode version 2.0 microRNA oligonucleotide probe set (Invitrogen). There was a total of n=6 rats for each access group (extended, restricted and naïve). Three microarrays were analyzed for each access condition, with each microarray containing pooled RNA from two animals per group. Hence, a total of 9 microarrays were analyzed, three per cocaine access level. Microarrays were scanned on an Axon Genepix 4000B scanner, and data was extracted from images using GenePix V4.1 software. After scanning and feature extraction, background-subtracted data were log base 2-transformed, then normalized and scaled based on the median rat miRNA probe intensity for each probe. The normalized data (transformed from log to linear) was imported into Agilent GeneSpring GX 7.3. Experiment interoperation mode was set to Log of ratio. Only probes with signal higher than the array median (721) at least in one array were selected for further analysis (141 probes out of 474). Each probe signal was normalized to the median signal value of that probe across all nine arrays. Differentially expressed probes (2 fold up or down-regulated, p<0.05) were identified using GX Volcano Plot feature (26 probes). In order to visualize the results, a heatmap was generated using GX Gene Tree (hierarchical clustering) feature (shown in Fig. 1a). Pearson Correlation was used as the Similarity measure between probes and Clustering Algorithm was set to Average Linkage.
Gene Expression Profiling and data analysis
Total RNA was isolated from HEK cells as described above. Samples were quantified using the NanoDrop ND-1000 spectrophotometer. Double-stranded cDNA was prepared from 1 μg of total RNA using the Affymetrix cDNA synthesis kit and was then in vitro-transcribed using an IVT labeling kit (Affymetrix), with the cRNA product purified using a GeneChip Sample Cleanup Module (Affymetrix). Biotin-labeled cRNA (20 μg) was fragmented and hybridized to an Affymetrix Rat Genome 230 2.0 microarray overnight in the Affymetrix 640 hybridization oven with a speed of 60 rpm for 16 h. Microarrays were washed and stained using an Affymetrix Fluidics Station FS400. GeneChip arrays were scanned using a GeneChip Scanner 3000 (Affymetrix). The probe set intensities were quantified using the GeneChip Operating Software and analyzed with GeneChip Robust Multichip Average (GC-RMA) normalization using Array Assist Software (Stratagene, La Jolla, CA). All hybridized chips met standard quality control criteria, and mean fluorescence values of each array were scaled to a mean intensity of 500. Only probe sets with intensities greater than 100 were considered reliably detected.
Taqman real-time PCR
Taqman analyses of miRNA expression levels were performed using miRNA assays and other stock primers commercially available from Applied Biosystems (ABI). For all reactions, 1-2 μg of total RNA were reverse-transcribed using miRNA-specific primers (ABI), and primers to small nuclear RNA (snoRNA) as endogenous controls for miRNA (ABI), or GAPDH for Fos. The protocol followed the manufacturer’s specifications with the exception of using 1.5 μl for each primer in a 15 μl total reaction volume. The cDNA was amplified by real-time PCR using Universal Taqman Mix (with no Amperase Ung) and miRNA-specific primers (ABI) according to manufacturer protocol. Comparison between groups was made using the method of 2−ΔΔCt.
In Situ Hybridization
To detect miR212 in frozen rat brain tissue, a 3′ digitonin (DIG)-labeled LNA probe was purchased from Exiqon. Rat brain sections (12-18 μm) in the coronal plane that incorporated the dorsal striatum on a cryostat (HM 505 E, Microm, Walldorf, Germany) kept at −80°C. miR-212 expression was detected exactly as described in ref. 55.
Immunochemistry
Rats were anesthetized and perfused transcardially with cold 4% paraformaldehyde (PFA) in phosphate buffered saline (PBS) PBS, pH 7.4. Brains were post-fixed in 4% PFA overnight and stored in 30% sucrose in PBS. Brains were sectioned (30 μm) in the coronal plane that incorporated the dorsal striatum on a cryostat (HM 505 E, Microm, Walldorf, Germany) kept at −20°C. Immunohistochemistry was performed on free-floating sections using the following primary antibodies: anti-copGFP (Axxora); anti-GFAP (Covance) and/or TORC1 (Santa Cruz). Primary antibodies were incubated overnight at 4°C in 1% bovine serum albumin (BSA) in PBS at 1:1000 (copGFP), 1:1000 (GFAP), and 1:50 (DARPP-32) dilutions. This was followed by incubation for 1-h with the following fluorescent-tagged secondary antibodies: Alexa Fluor 488 goat anti-rabbit (Invitrogen) or Alexa Fluor 660 donkey anti-goat IgG (Invitrogen), both at a 1:100 dilution. Sections were mounted on Superfrost Plus slides (Fischer Scientific, Pittsburg, PA), dehydrated, and coverslipped. Sections were visualized by using a BX61 (Olympus) fluorescence microscope at 20x and 40x.
Immunoblotting
For protein analyses, brain sections were collected on a frozen cryostat as described above, and sonicated in 1X RIPA buffer. Protein content was determined using the Bio-Rad DC Protein Assay kit, and the concentration of each sample was adjusted to 2 mg/ml protein or 4-6 mg/ml for less abundant proteins. Protein samples were separation by gel electrophoresis, and proteins transferred to Nitrocellulose (NC membrane, Invitrogen iBlot system). HiMark high molecular weight pre-stained standards (BIO-RAD) were also run on each gel. Nonspecific binding sites on the membranes was blocked by 5% nonfat dry milk in TBS and 0.1% Tween 20 (TBS-T). Blots were incubated in primary antibody in PBS-T, washed, then incubated in secondary antibody. Blots were washed and immunological detection was carried out using SuperSignal Chemiluminescent Substrate (Thermo Scientific). Antibodies were stripped from the blots, and the blots probed for anti-β-actin or GAPDH (Sigma or Cell Signaling). Primary antibodies used were: anti-CREB (Millipore); S133-p-CREB (Santa Cruz or Millipore); Fos (Santa Cruz); Nurr1 (NR4A) (Santa Cruz); TORC1 (Cell Signaling); phospho-TORC1 (Cell Signaling); acetylated lysine (Cell Signaling), phospho-Raf-1 (Cell Signaling), Raf-1 (Santa Cruz), SPRED1 (Santa Cruz), ERK (Cell Signaling) and phospho-ERK (Santa Cruz). Antiserum against TORC2 was generated using peptide [H]-CAETDKTLSKQSWDSKKAG-[NH2] at Covance as previously describe in Amelio et al. 56. Secondary antibodies used were: Amersham ECL anti-rabbit IgG, horseradish peroxidase linked species-specific whole antibody from donkey (GE Healthcare).
Clones, miRNA precursors and lentiviral constructs
The Raf-1-GFP and dominant negative Raf-151-220-GFP constructs were obtained from Dr. Tamas Balla 57. The SPRED1 vector expressing only the coding sequence was purchased from OriGene Technologies (Rockville, MD); catalog number SC101247. The pMIF-copGFP, and expression vectors for immature microRNAs (rat in each case) were purchased from System Biosciences Inc, and were feline immunodeficiency virus lentiviral vectors that contain the miRNA precursor and a copGFP reporter under the control of a cytomegalovirus (CMV) promoter. TORC1 was cloned into the pCDF1 lentiviral expression vector from System Biosciences, Inc. The pCDF1 vector is a feline immunodeficiency virus that contained the cloned TORC1 gene under the control of a CMV promoter and a copGFP reporter under the control of an EF1 promoter. Correct insertion of the TORC1 gene was verified in functional assays (potentiation of CREB signaling) and via Western blotting.
Generation of lentivirus
To generate lentivirus supernatant, HEK-293FT packaging cells (3.75 x 106 293TN cells per 10 cm plate) were transfected with the vectors (pMIF or pCDF1, respectively), along with the pPACKF1TM Lentiviral Packaging Kit (http://www.systembio.com/pPACK.htm) using Lipofectamine Reagent and Plus Reagent (Invitrogen) according to the manufacturer’s instructions. Medium containing virus particles (~10 ml) was harvested 48-60 hr post-transfection by centrifuged at 3000 rpm at room temperature for 5 min to pellet cell debris, and filtered through 0.45 mm PVDF filers (Millex-HV). To concentrate the viral supernatant for intra-striatal administration, supernatants were centrifuged at 32,000 x g for 90 min at 4°C, and the precipitate re-suspended in 100 μl cold PBS. Supernatants were aliquoted into 10 ml volumes, and stored at −80°C until use.
Estimation of lentivirus titer
Viral supernatant titers were determined using the Lentivector Rapid Titer Kit from System Biosciences Inc, according to the manufacturer’s instructions. The number of infectious units per ml of supernatant (IFU/ml) was calculated as follows: (MOI of the sample) X (The number of cells in the well upon infection) X 1000/(μl of viral supernatant used).
Oligonucleotides and short-hairpin RNAs
The SPRED1 siRNA was purchased from Invitrogen. The LNA-antimiR molecules were purchased from Exiqon Inc. The shRNA targeting TORC2 (GGTCTCTGCCCAATGTTAACCA) was created by subcloning annealed oligos into the pBS/U65 expression vectors 58, as previously described 59.
3′UTR Analyses
Bioinformatic identification of 3′UTRs MREs was carried using TargetScan 5.1. The HMGA2 3′UTR renilla luciferase reporter construct (Addgene) was previously described 60. To assess the effects of miRNA overexpression on HMGA2 3′UTR luciferase expression, HEK-293 cells, (3 × 105 cells/well) were co-transfected with the immature microRNA constructs or the empty control vector (pMIF) (500 ng), the HMGA2 3′UTR renilla luciferase reporter construct (100 ng), the RSV b-galactosidase control vector (50 ng), and where indicated with LNA-antimiR-212 or LNA-Scrambled (40 nM) in lipofectamine according to the manufacturer’s instructions, as described in 60,61. Renilla luciferase assays were performed (Renilla Lucieferase Assay system, Promega) and normalized to β-galactosidase activity.
CREB Reporter assays
CRE-containing luciferase reporter plasmids were used as described in detail previously 59. To assess the effects of miRNA overexpression on CREB signaling, HEK-293 cells (3 × 105 cells/well/96-well plates) were reverse transfected with the EVX-1 CRE-containing luciferase reporter plasmid (80 ng), the pre-miR-212, pre-miR-1, pre-miR-132 or pre-miR-29b expression vector (40 ng), and Rous sarcoma virus-β-galactosidase (RSV-β-galactosidase) expression plasmid such that the total amount of transfected DNA was held constant at (150 ng/well). 24-48 h post-transfection, cells were treated for 30 min or 4 h with 5, 10 or 15 μM forskolin or vehicle (0.75% DMSO) as indicated. The cells were then lysed and assayed for luciferase activity using the Bright Glo luciferase assay system (Promega) as described previously 59, normalizing to activity from co-transfected RSV-β-galactosidase expression plasmid.
cAMP assay
cAMP accumulation was measured using a competitive immunoassay (BioVision). Briefly, HEK-293 cells were transiently transfected with the miR-212 or control pMIF expression vectors (2.5 μg). 48-h after transfection cells were treated with 10 μM forskolin or vehicle (0.1% DMSO) for 30-min or 240-min, and the cell lysed in 0.1 N HCl. Levels of cAMP were then assayed according to the manufacture’s instructions.
Supplementary Material
Acknowledgments
Supported by a grant from the National Institute on Drug Abuse (NIDA) to P.J.K.; and NIDA post-doctoral awards to J.A.H. and H.-I.I. We thank Mohammad Fallahi-Sichani for help with miRNA expression analysis and Dr. Tamas Balla for the Raf-1 expression constructs. We also thank Drs. Christie Fowler and Patrick Griffin for helpful comments on the manuscript, and NIDA for supplying the cocaine used in these studies. This is manuscript 19873 from Scripps Florida.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions J.A.H., H-.I.I, A.L.A, J.K., P.B. and Q.L. performed all experiments; D.W. performed microarray analysis; M.D.C. and C.W. provided essential reagents and helpful advice; P.J.K. designed the molecular experiments; J.A.H. and P.J.K. designed the behavioral experiments, performed the statistical analyses, and wrote the manuscript.
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
References
- 1.McClung CA, Nestler EJ. Neuroplasticity mediated by altered gene expression. Neuropsychopharmacology. 2008;33:3–17. doi: 10.1038/sj.npp.1301544. [DOI] [PubMed] [Google Scholar]
- 2.Kalivas PW, O’Brien CP. Drug addiction as a pathology of staged neuroplasticity. Neuropsychopharmacology. 2008;33:166–180. doi: 10.1038/sj.npp.1301564. [DOI] [PubMed] [Google Scholar]
- 3.Thomas MJ, Kalivas PW, Shaham Y. Neuroplasticity in the mesolimbic dopamine system and cocaine addiction. Br J Pharmacol. 2008;154:327–342. doi: 10.1038/bjp.2008.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pulipparacharuvil S, et al. Cocaine regulates MEF2 to control synaptic and behavioral plasticity. Neuron. 2008;59:621–633. doi: 10.1016/j.neuron.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlezon WAJ, et al. Regulation of cocaine reward by CREB. Science. 1998;282:2272–2275. doi: 10.1126/science.282.5397.2272. [DOI] [PubMed] [Google Scholar]
- 6.Anthony JC, Warner LA, Kessler RC. Comparative epidemiology of dependence on tobacco, alcohol, controlled substances, and inhalants: Basic findings from the National Comorbidity Survey. Experimental and Clinical Psychopharmacology. 2004;2:244–268. [Google Scholar]
- 7.Ahmed SH, Koob GF. Transition from moderate to excessive drug intake: change in hedonic set point. Science. 1998;282:298–300. doi: 10.1126/science.282.5387.298. [DOI] [PubMed] [Google Scholar]
- 8.Vanderschuren LJ, Everitt BJ. Drug seeking becomes compulsive after prolonged cocaine self-administration. Science. 2004;305:1017–1019. doi: 10.1126/science.1098975. [DOI] [PubMed] [Google Scholar]
- 9.Deroche-Gamonet V, Belin D, Piazza PV. Evidence for addiction-like behavior in the rat. Science. 2004;305:1014–1017. doi: 10.1126/science.1099020. [DOI] [PubMed] [Google Scholar]
- 10.Paterson NE, Markou A. Increased motivation for self-administered cocaine after escalated cocaine intake. Neuroreport. 2003;14(17):2229–32. doi: 10.1097/00001756-200312020-00019. [DOI] [PubMed] [Google Scholar]
- 11.Belin D, Balado E, Piazza PV, Deroche-Gamonet V. Pattern of intake and drug craving predict the development of cocaine addiction-like behavior in rats. Biol Psychiatry. 2009;65:863–868. doi: 10.1016/j.biopsych.2008.05.031. [DOI] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 13.Schratt GM, et al. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 14.Wayman GA, et al. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc Natl Acad Sci U S A. 2008;105:9093–9098. doi: 10.1073/pnas.0803072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perkins DO, Jeffries C, Sullivan P. Expanding the ‘central dogma’: the regulatory role of nonprotein coding genes and implications for the genetic liability to schizophrenia. Mol Psychiatry. 2005;10:69–78. doi: 10.1038/sj.mp.4001577. [DOI] [PubMed] [Google Scholar]
- 16.Rogaev EI. Small RNAs in human brain development and disorders. Biochemistry (Mosc) 2005;70:1404–1407. doi: 10.1007/s10541-005-0276-z. [DOI] [PubMed] [Google Scholar]
- 17.Pietrzykowski AZ, et al. Posttranscriptional regulation of BK channel splice variant stability by miR-9 underlies neuroadaptation to alcohol. Neuron. 2008;59:274–287. doi: 10.1016/j.neuron.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chandrasekar V, Dreyer JL. microRNAs miR-124, let-7d and miR-181a regulate cocaine-induced plasticity. Mol Cell Neurosci. 2009;42:350–362. doi: 10.1016/j.mcn.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 19.Nudelman AS, et al. Neuronal activity rapidly induces transcription of the CREB-regulated microRNA-132, in vivo. Hippocampus. 20:492–498. doi: 10.1002/hipo.20646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Everitt BJ, Robbins TW. Neural systems of reinforcement for drug addiction: from actions to habits to compulsion. Nat Neurosci. 2005;8:1481–1489. doi: 10.1038/nn1579. [DOI] [PubMed] [Google Scholar]
- 21.Belin D, Everitt BJ. Cocaine seeking habits depend upon dopamine-dependent serial connectivity linking the ventral with the dorsal striatum. Neuron. 2008;57:432–441. doi: 10.1016/j.neuron.2007.12.019. [DOI] [PubMed] [Google Scholar]
- 22.Ito R, Dalley JW, Robbins TW, Everitt BJ. Dopamine release in the dorsal striatum during cocaine-seeking behavior under the control of a drug-associated cue. J Neurosci. 2002;22:6247–6253. doi: 10.1523/JNEUROSCI.22-14-06247.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vanderschuren LJ, Di Ciano P, Everitt BJ. Involvement of the dorsal striatum in cue-controlled cocaine seeking. J Neurosci. 2005;25:8665–8670. doi: 10.1523/JNEUROSCI.0925-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vo N, et al. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci U S A. 2005;102:16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piazza PV, Deroche-Gamonent V, Rouge-Pont F, Le Moal M. Vertical shifts in self-administration dose-response functions predict a drug-vulnerable phenotype predisposed to addiction. J Neurosci. 2000;20:4226–4232. doi: 10.1523/JNEUROSCI.20-11-04226.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kocerha J, et al. MicroRNA-219 modulates NMDA receptor-mediated neurobehavioral dysfunction. Proc Natl Acad Sci U S A. 2009;106:3507–3512. doi: 10.1073/pnas.0805854106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elmen J, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 28.Tsang J, Zhu J, van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell. 2007;26:753–767. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McClung CA, Nestler EJ. Regulation of gene expression and cocaine reward by CREB and DeltaFosB. Nat Neurosci. 2003;6:1208–1215. doi: 10.1038/nn1143. [DOI] [PubMed] [Google Scholar]
- 30.Dinieri JA, et al. Altered sensitivity to rewarding and aversive drugs in mice with inducible disruption of cAMP response element-binding protein function within the nucleus accumbens. J Neurosci. 2009;29:1855–1859. doi: 10.1523/JNEUROSCI.5104-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conkright MD, et al. TORCs: transducers of regulated CREB activity. Mol Cell. 2003;12:413–423. doi: 10.1016/j.molcel.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 32.Perry SJ, et al. Targeting of cyclic AMP degradation to beta 2-adrenergic receptors by beta-arrestins. Science. 2002;298:834–836. doi: 10.1126/science.1074683. [DOI] [PubMed] [Google Scholar]
- 33.Nishi A, et al. Distinct roles of PDE4 and PDE10A in the regulation of cAMP/PKA signaling in the striatum. J Neurosci. 2008;28:10460–10471. doi: 10.1523/JNEUROSCI.2518-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y, et al. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature. 2008;456:269–273. doi: 10.1038/nature07349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ding Q, et al. Raf kinase activation of adenylyl cyclases: isoform-selective regulation. Mol Pharmacol. 2004;66:921–928. [PubMed] [Google Scholar]
- 36.Beazely MA, Alan JK, Watts VJ. Protein kinase C and epidermal growth factor stimulation of Raf1 potentiates adenylyl cyclase type 6 activation in intact cells. Mol Pharmacol. 2005;67:250–259. doi: 10.1124/mol.104.001370. [DOI] [PubMed] [Google Scholar]
- 37.Duman RS, Tallman JF, Nestler EJ. Acute and chronic opiate-regulation of adenylate cyclase in brain: specific effects in locus coeruleus. J Pharmacol Exp Ther. 1988;246:1033–1039. [PubMed] [Google Scholar]
- 38.Varga EV, et al. Involvement of Raf-1 in chronic delta-opioid receptor agonist-mediated adenylyl cyclase superactivation. Eur J Pharmacol. 2002;451:101–102. doi: 10.1016/s0014-2999(02)02220-3. [DOI] [PubMed] [Google Scholar]
- 39.King AJ, et al. The protein kinase Pak3 positively regulates Raf-1 activity through phosphorylation of serine 338. Nature. 1998;396:180–183. doi: 10.1038/24184. [DOI] [PubMed] [Google Scholar]
- 40.Hatzivassiliou G, et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010 doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 41.Hall-Jackson CA, et al. Paradoxical activation of Raf by a novel Raf inhibitor. Chem Biol. 1999;6:559–568. doi: 10.1016/s1074-5521(99)80088-x. [DOI] [PubMed] [Google Scholar]
- 42.Chin PC, et al. The c-Raf inhibitor GW5074 provides neuroprotection in vitro and in an animal model of neurodegeneration through a MEK-ERK and Akt-independent mechanism. J Neurochem. 2004;90:595–608. doi: 10.1111/j.1471-4159.2004.02530.x. [DOI] [PubMed] [Google Scholar]
- 43.Selbach M, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 44.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wakioka T, et al. Spred is a Sprouty-related suppressor of Ras signalling. Nature. 2001;412:647–651. doi: 10.1038/35088082. [DOI] [PubMed] [Google Scholar]
- 46.Brems H, et al. Germline loss-of-function mutations in SPRED1 cause a neurofibromatosis 1-like phenotype. Nat Genet. 2007;39:1120–1126. doi: 10.1038/ng2113. [DOI] [PubMed] [Google Scholar]
- 47.Altarejos JY, et al. The Creb1 coactivator Crtc1 is required for energy balance and fertility. Nat Med. 2008;14:1112–1117. doi: 10.1038/nm.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kovacs KA, et al. TORC1 is a calcium- and cAMP-sensitive coincidence detector involved in hippocampal long-term synaptic plasticity. Proc Natl Acad Sci U S A. 2007;104:4700–4705. doi: 10.1073/pnas.0607524104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou Y, et al. Requirement of TORC1 for late-phase long-term potentiation in the hippocampus. PLoS ONE. 2006;1:e16. doi: 10.1371/journal.pone.0000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li S, Zhang C, Takemori H, Zhou Y, Xiong ZQ. TORC1 regulates activity-dependent CREB-target gene transcription and dendritic growth of developing cortical neurons. J Neurosci. 2009;29:2334–2343. doi: 10.1523/JNEUROSCI.2296-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caine B, Koob GF. In: Behavioural Neuroscience: A Practical Approach. Sahgal A, editor. Vol. 2. IRL Press; 1993. pp. 117–143. [Google Scholar]
- 52.Kenny PJ, Markou A. Nicotine self-administration acutely activates brain reward systems and induces a long-lasting increase in reward sensitivity. Neuropsychopharmacology. 2006;31:1203–1211. doi: 10.1038/sj.npp.1300905. [DOI] [PubMed] [Google Scholar]
- 53.Ahmed SH, Koob GF. Transition from moderate to excessive drug intake: change in hedonic set point. Science. 1998;282:298–300. doi: 10.1126/science.282.5387.298. [DOI] [PubMed] [Google Scholar]
- 54.Paxinos G, Watson C. The rat brain in stereotaxic coordinates. 3. Academic Press; 1997. [DOI] [PubMed] [Google Scholar]
- 55.Silahtaroglu AN, et al. Detection of microRNAs in frozen tissue sections by fluorescence insitu hybridization using locked nucleic acid probes and tyramide signal amplification. Nat Protoc. 2007;2:2520–2528. doi: 10.1038/nprot.2007.313. [DOI] [PubMed] [Google Scholar]
- 56.Amelio AL, et al. A coactivator trap identifies NONO (p54nrb) as a component of the cAMP-signaling pathway. Proc Natl Acad Sci U SA. 2007;104:20314–20319. doi: 10.1073/pnas.0707999105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bondeva T, Balla A, Varnai P, Balla T. Structural determinants of Ras-Raf interaction analyzed in live cells. Mol Biol Cell. 2002;13:2323–2333. doi: 10.1091/mbc.E02-01-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sui G, et al. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc Natl Acad Sci U S A. 2002;99:5515–5520. doi: 10.1073/pnas.082117599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Conkright MD, et al. TORCs: transducers of regulated CREB activity. Mol Cell. 2003;12:413–423. doi: 10.1016/j.molcel.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 60.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.