Abstract
Fungi produce an impressive array of secondary metabolites (SMs) including mycotoxins, antibiotics and pharmaceuticals. The genes responsible for their biosynthesis, export, and transcriptional regulation are often found in contiguous gene clusters. To facilitate annotation of these clusters in sequenced fungal genomes, we developed the web-based software SMURF (www.jcvi.org/smurf/) to systematically predict clustered SM genes based on their genomic context and domain content. We applied SMURF to catalog putative clusters in 27 publicly available fungal genomes. Comparison with genetically characterized clusters from six fungal species showed that SMURF accurately recovered all clusters and detected additional potential clusters. Subsequent comparative analysis revealed the striking biosynthetic capacity and variability of the fungal SM pathways and the correlation between unicellularity and the absence of SMs. Further genetics studies are needed to experimentally confirm these clusters.
Index descriptors: NRPS, PKS, prenyltransferases, polyketides, antibiotics, secondary metabolism, filamentous fungi, Aspergillus, genome annotation
Introduction
Secondary metabolites (SMs) are small bioactive molecules produced by many organisms including bacteria, plants and fungi. These compounds are particularly abundant in soil-dwelling filamentous fungi, which exist as multicellular communities competing with each other for nutrients, minerals and water (Keller et al., 2005). Unlike primary metabolites, most SMs – as their name suggests – are not essential for fungal growth, development, or reproduction under in vitro conditions. They can however provide protection against various environmental stresses and during antagonistic interactions with other soil inhabitants or a eukaryotic host. Scientific appreciation of the importance of fungal SMs grew in the 1940s as the massive impact of penicillin on human health began to be seen. Since then, many other beneficial SM compounds have been discovered including immunosuppressants, cholesterol-lowering drugs, antiviral drugs, and anti-tumor drugs (for a recent review see Hoffmeister and Keller, 2007). At the same time, fungi are also known to produce numerous mycotoxins such as aflatoxin, fumonisin, trichothecene, and zearalone.
The first committed step in biosynthesis of an SM is catalyzed by one of five proteins, which we refer to here as “backbone” enzymes. They include nonribosomal peptide synthases (NRPSs), polyketide synthases (PKSs), hybrid NRPS-PKS enzymes, prenyltransferases (DMATSs), and terpene cyclases (TCs). These multidomain enzymes are associated, respectively, with production of the five classes of SM: nonribosomal peptides, polyketides, NRPS-PKS hybrids, indole alkaloids, and terpenes (Hoffmeister and Keller, 2007). Terpenes, which are composed of isoprene units, are not considered further in our analysis, because terpene cyclases are highly variable in sequence and difficult to detect by bioinformatic methods (Keller et al., 2005; Townsend, 1997). Intermediate products formed by the backbone enzymes can undergo further modifications catalyzed by “decorating” enzymes. The final product is then often steered by a transporter outside the fungal cell wall or sometimes remains within the cell. All these genes tend to be found in contiguous gene clusters, which are coordinately regulated by a specific Zn2Cys6 transcription factor and/or by the global regulator of secondary metabolism, putative methyltransferase LaeA (Keller and Hohn, 1997; Keller et al., 2005).
The availability of data from fungal genome sequencing projects has facilitated the discovery and characterization of new compounds and their biosynthetic pathways. Thus within months after completion of the first A. fumigatus genome (Nierman et al., 2005), several secondary metabolite clusters were characterized at the molecular level including the gliotoxin (Gardiner and Howlett, 2005), fumigaclavines (Coyle and Panaccione, 2005; Unsold and Li, 2005; Unsold and Li, 2006), fumitremorgin (Maiya et al., 2006), and siderophores (Reiber et al., 2005) biosynthesis clusters. Genome sequencing also revealed that the number of secondary metabolites characterized from a given species falls far behind the numbers of clusters that can be predicted based on its genomic sequence (Bok et al., 2006; Chiang et al., 2008). This has been attributed to the fact that not all clusters may be expressed under normal laboratory conditions.
Despite the medical and agricultural importance of fungal SMs, most putative SM clusters in fungal genomes have been predicted by ad hoc strategies based on manual reviews of BLAST searches generated for backbone genes and their neighbors (e.g. (Nierman et al., 2005)). Manual annotation of SM clusters, however, is time-consuming and may result in inconsistent annotation.
To facilitate systematic mapping of SM clusters in fungal genomes, we developed a web-based software tool, Secondary Metabolite Unknown Regions Finder (SMURF; www.jcvi.org/smurf/). It is based on three hallmarks of fungal SM biosynthetic pathways: (i) the presence of backbone genes, (ii) clustering, and (iii) characteristic protein domain content. Subsequent analyses of the predicted clusters present in 27 sequenced fungal genomes (Supplementary Table 1) shows SM gene enrichment in the genus Aspergillus, the absence of the clusters in unicellular fungi, and unexpected abundance and variability of the fungal clusters. Our results are also consistent with the view that SM profiles can be used as means of differentiating species and strains in filamentous fungi (Frisvad et al., 2008), and show that gene duplication plays an essential role in the creation and expansion of the SM repertoires of fungi.
Methods
Identification of putative backbone enzymes
SMURF relies on hidden Markov model (HMM) searches to detect backbone genes in sequenced fungal genomes. The HMMER program (http://hmmer.janelia.org) was used to search for conserved Pfam and TIGRFAM domains of backbone enzymes in the protein set of each sequenced species. Trusted threshold bit score cutoffs (predefined in HMMER) were used for each HMM search. NRPS enzymes were identified as enzymes with at least one module composed of an amino acid adenylation domain (A), a thiolation domain (PCP) and a condensation domain (C). PKS enzymes were identified as enzymes with at least one acyl transferase domain (AT), a beta-ketoacyl synthase C-terminal domain (BKS-C), and a beta-ketoacyl synthase N-terminal domain (BKS-N). Hybrid PKS-NRPS enzymes were identified as enzymes with at least one instance from each set of three domains listed above.
NRPS-like enzymes were identified with a combination of at least two domains from any of those in the NRPS enzyme module; or a combination of an A domain and a NAD_binding_4 domain; or a combination of an A domain and short chain dehydrogenase domain. PKS-like enzymes were identified with a combination of at least two domains from any of those in the PKS enzyme module. To eliminate false positives among PKS-like enzymes, they were defined as proteins with AT, BKS-C and BKS-N domains that scored below a trusted HMM cut-off. In contrast, to eliminate false positives such as alpha-aminoadipate reductase among NRPSs, we required the score of the C-terminal domain of L-aminoadipate-semialdehyde dehydrogenase alpha subunit to be above the cut-off.
Prenyltransferase enzymes were identified as enzymes with at least one DMATS-type prenyltransferase domain (DMATS). The corresponding de novo HMM model for this domain (TIGR03429) was created in this study from the seed alignment generated using the A. fumigatus dimethylallyl tryptophan synthase FtmPT2 as a seed sequence as previously described (Sonnhammer et al., 1998). Characterized or partially characterized seed members include several dimethylallyltryptophan synthases, a brevianamide F prenyltransferase, the LtxC enzyme involved in lyngbyatoxin biosynthesis, and a probable dimethylallyl tyrosine synthase.
Identification of putative decorating enzymes
To define protein domains commonly present in SM decorating enzymes, transporter, and transcriptional regulators; we examined the domains detected in the 22 A. fumigatus clusters we used as a training set. The list of clusters included two genetically characterized A. fumigatus clusters involved in biosynthesis of fumitremorgin (Grundmann et al., 2008; Kato et al., 2009; Maiya et al., 2006) and melanin (Fujii et al., 2004; Tsai et al., 1999) and 10 clusters predicted based on expression data: A. fumigatus clusters Pes1, siderophore, fumigaclavine, pseurotin, the gliotoxin-like polyketide (McDonagh et al., 2008; Perrin et al., 2007), and gliotoxin (Gardiner and Howlett, 2005). The rest of the 22 clusters were predicted manually based on genes' name and their proximity to the adjacent backbone gene (Perrin et al., 2007). Some domains were present almost exclusively in clusters, while others were evenly distributed throughout the entire genome (Supplementary Table 2). The final 27 SM-defining domains were selected as domains most likely to be found in a cluster based on their distribution.
Identification of putative SM clusters
Once all putative backbone genes are identified in a genome, the SMURF algorithm then evaluates their adjacent genes to test whether they are part of an SM gene cluster (Supplementary Figure 1). A window of ±20 genes on each side of a backbone gene is scanned for the 27 SM-defining domains using HMMer. The number 20 was established empirically based on the training set of 22 A. fumigatus clusters. Genes in the window are tagged as “SM domain positive” if they contain at least one of these domains, or “SM domain negative” if they do not. Then the boundaries of any putative cluster are defined by the algorithm that evaluates each gene by walking rightwards from the backbone gene until it reaches as a stop signal, which is defined below. The last gene on the rightwards walk before the stop signal is given the label alpha. After that SMURF carries out an identical walk leftwards from the backbone gene, until a stop signal is encountered defining a left-limit gene beta. The interval between alpha and beta is the preliminary extent of the cluster.
The algorithm requires two key parameters: d, the maximum intergenic distance (in base pairs) permitted between two adjacent genes in the same cluster; and y, the maximum number of SM domain negative genes, which is allowed within a cluster. By a trial-and-error process, we identified the parameters d = 3,814 bp and y = 10 genes as optimal based on the training set of 22 clusters. A stop signal is defined as either an intergenic distance that is larger than the limit d, or a cumulative number of negative genes between the backbone gene and the current position that is larger than y (Supplementary Figure 1).
To take into account the intergenic distances, the SMURF algorithm trims each cluster to ensure that the interval between alpha and beta is less than y. Then, additional genes are trimmed at both ends of the cluster until the algorithm reaches the first backbone or SM domain positive gene on each side. In some instances, SMURF predicts overlapping clusters, in which case the two clusters are merged into one.
Results
Parameter optimization
SMURF predicts putative secondary metabolism clusters by using an algorithm that takes into account the domain content of putative “backbone” genes and adjacent “decorating” genes. One of the key challenges in developing this tool was identification of the adjacent genes. In choosing parameters for SMURF we were confronted with the dilemma of striking a balance between levels of under-prediction and over-prediction. We chose to favor the latter, because over-prediction is easier to address in the future once a more comprehensive training set becomes available.
Our underlying hypothesis was that some domains may be disproportionately present in SM clusters. To select these domains, we considered clusters that have been identified either by standard genetic methods or by transcriptional profiling of the A. fumigatus ΔlaeA strain (Perrin et al., 2007). We thus identified 27 SM-defining domains over-represented in clusters (Supplementary Table 2). For most of the domains, one or more corresponding domain models already existed in the PFAM (Mistry and Finn, 2007) or TIGRFAM (Selengut et al., 2007) databases; and we built a new model for the N-methyltransferase domain (TIGR03439). Genes containing at least one of these 27 domains were called SM domain positive.
Specificity and sensitivity
After parameter optimization with the training set, we compared SMURF output against eight A. fumigatus Af293 clusters and ten clusters from other species that were all experimentally linked to a secondary metabolite product (Supplementary Table 3). The ten clusters from other species encoded the following metabolites: Aspergillus nidulans sterigmatocystin (Brown et al., 1996), penicillin (reviewed in (Brakhage et al., 2005)), asperfuranone (Chiang et al., 2009), asperthecin (Szewczyk et al., 2008), and terrequinone (Bouhired et al., 2007); Aspergillus flavus aflatoxin (Yu et al., 2007); and aflatrem (Zhang et al., 2004); Penicillin chrysogenum penicillin (Smith et al., 1990); Fusarium graminearum zearalenone (Kim et al., 2005), and aurofusarin (Malz et al., 2005); Fusarium verticillioides fumonisin (Proctor et al., 2003).
The algorithm was able to recover all the backbone genes in the clusters. We further evaluated the algorithm's performance by counting the number of over-predicted and under-predicted genes. An over-predicted gene is defined here as a gene detected by SMURF, but not by the previous annotations, and an under-predicted gene as the opposite. Assuming previous annotations are correct, over- and under-predictions correspond to false-positive and false-negative calls, respectively (Supplementary Table 3). Note that it is possible for SMURF to simultaneously over-predict some genes and under-predict other genes for the same cluster.
Among the eight A. fumigatus clusters, we found only one predicted cluster (Pes1) that was under-predicted by SMURF. The cluster was previously annotated as containing only two genes based on expression studies (Perrin et al., 2007). SMURF omitted one of these genes, because the intergenic distance between them was unusually long and, simultaneously, identified six additional genes in the cluster. The siderophore, epipolythiodioxopiperazine type toxin (ETP), and pseurotin clusters were considerably over-predicted as compared to the experimentally annotated clusters. The mean for over-prediction (7.0) was largely appreciably affected by the over-prediction of these three clusters. Optimizing SMURF to detect the three clusters decreased the accuracy for the remaining A. fumigatus clusters.
Notably, the algorithm performed better for non-A. fumigatus species with the mean for over-prediction being 3.9. This was unexpected considering that parameter optimization was done using only A. fumigatus clusters (Supplementary Table 3). Only two clusters, terrequinone and asperthecin, were notably over-predicted. SMURF under-predicted 4 clusters (again mostly due to unusually large intergenic distances) with a mean of -0.5 per cluster. For all species, SMURF-predicted clusters are larger than those annotated experimentally with the median number of over- and under- predicted genes being 5.0 and 0.0 per cluster, respectively.
Uneven taxonomic distribution of backbone enzymes
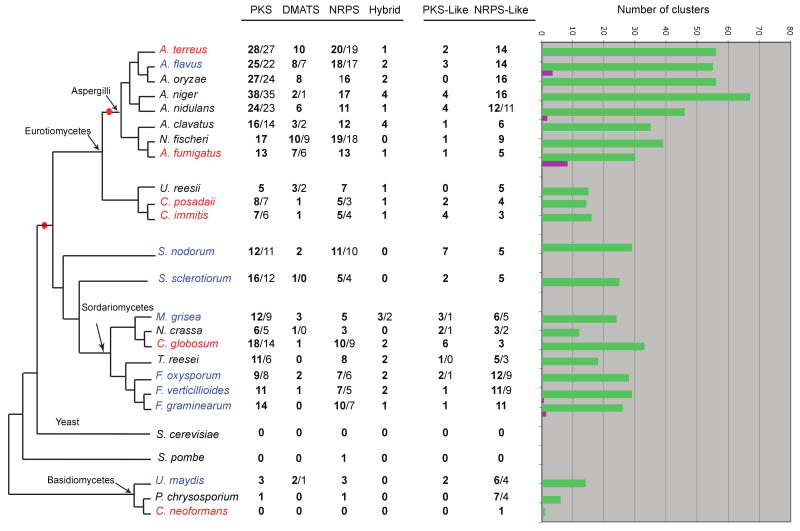
Having validated SMURF, we then systematically searched the genome sequences of 27 fungal species (24 Ascomycota and three Basidiomycota; Supplementary Table 1) for the presence of putative backbone genes and clusters. As expected, the search revealed that the numbers of backbone genes varies greatly (from 0-61) from one fungal taxon to another (Fig. 1). We found no backbone genes in two of the three unicellular species examined here: the ascomycete yeast Saccharomyces cerevisiae and the basidiomycete yeast Cryptococcus neoformans (though the latter species does have one NRPS-like gene). Similarly, there is only one backbone gene (Schwecke et al., 2006) in the genome of the third unicellular fungus, the archiascomycete yeast Schizosaccharomyces pombe.
Figure 1.
Numbers of backbone genes and SM clusters in the 27 sequenced fungal genomes we analyzed. The central columns show the numbers of backbone genes of each type in a species. Each column contains two numbers separated by a slash; the first (in bold) is the number of backbone genes, and the second is the number of putative SM clusters predicted by SMURF. If both numbers are identical, only one (in bold) is shown. The tree topology is based on the phylogenetic tree by Fitzpatrick and colleagues (Figure 2 in (Fitzpatrick et al., 2006)). Species named in red are human pathogens (some are also animals and/or plant pathogens), blue are plant pathogens, and black are non-pathogenic fungi. Red bullets mark two internal branches on which enrichment in backbone genes occurred during evolution. In the histograms on the right, green bars show the total numbers of SM clusters predicted by SMURF in each genome (excluding SM clusters containing only PKS-like and NRPS-like genes), and purple bars show the numbers of SM clusters that have been characterized experimentally. PKS, polyketide synthase; DMATS, prenyltransferase; NRPS, nonribosomal peptide synthase.
In addition to the canonical NRPS and PKS genes, we also catalogued NRPS-like and PKS-like enzymes, because some SM clusters such as the fumonisin cluster in Fusarium species include backbone enzymes with atypical domain composition (Song et al., 2004; Zaleta-Rivera et al., 2006). In addition, our estimate is that most fungal backbone genes in public databases have incorrect gene structures including split gene models (Fedorova, unpublished), which also may result in atypical domain composition. Our analysis shows that the numbers of NRPS-like and PKS-like genes fluctuate in correlation with their counterparts, the canonical NRPS and PKS genes (Fig. 1).
Fig. 1 also shows an expansion of backbone genes in Pezizomycotina, especially in Eurotiomycetes, as compared to Basidiomycetes and Sordariomycetes. However, within the Eurotiomycetes, there are notably fewer backbone genes in the genomes of the human pathogens Coccidioides immitis and Coccidioides posadasii than in the section Aspergillus. This difference is probably more due to the phylogenetic distance between Coccidioides and Aspergillus than to lifestyle differences. Among the Pezizomycota, Neurospora crassa has a significantly reduced number of backbone genes (10), even when compared to Fusarium oxysporum which has the second lowest number of backbone genes in the Pezizomycota (P < 10-16, Chi-square test). This difference is presumably attributable to the presence of the repeat-induced point mutation (RIP) process in N. crassa, which has dramatically reduced the rate of formation of new gene duplications in that species (Galagan et al., 2003).
PKSs and NRPSs are found in significantly higher numbers than DMATSs and hybrid enzymes in almost all species. We also observed that the number of backbone genes in aspergilli is significantly higher than in Sordariomycetes (P = 0.001, Wilcoxon test). This difference is due to increases in the numbers of NRPS (P = 7 × 10-4), PKS (P = 0.001), and DMATS (P = 0.002) enzymes in the aspergilli, but not hybrid enzymes (P = 0.9).
Species specificity of SM clusters
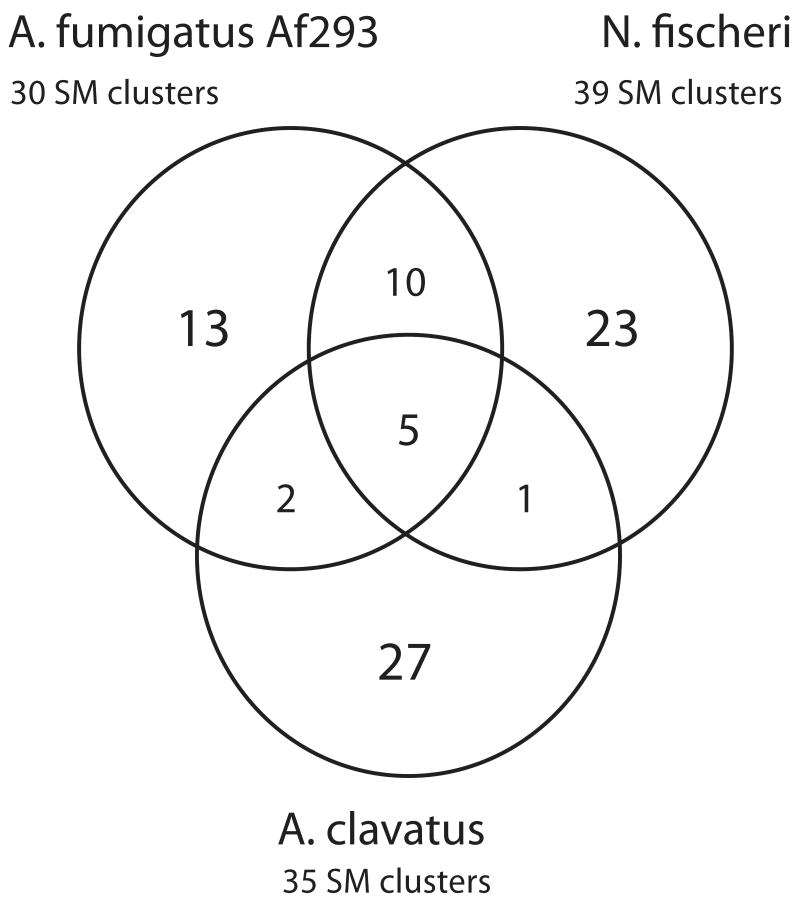
The large numbers of putative SM gene clusters identified by SMURF (Fig. 1) emphasizes the unusual diversity of the SM repertoires in fungal species. To what extent are these metabolites and their biosynthetic pathways species-specific? To answer this question, we further analyzed the genomes of the three closely related species A. fumigatus Af293, A. clavatus, and Neosartorya fischeri (Fedorova et al., 2008; Nierman et al., 2005). For accessory genes in each of these species, we assumed their reciprocal best BLASTP hits to be putative orthologs. For backbone genes, we defined orthology based on the sequence identity, alignment length, and domain content. We then defined two SM clusters as orthologous if at least 80% of their genes were orthologous. This approach sometimes yielded hidden paralogs, which were excluded from further analysis based on manual examination.
The comparative analysis (Fig. 2) shows that only five SM gene clusters are common to all three genomes, while most other clusters are species-specific and appear relatively young in evolutionary terms. The core set includes clusters, such as Pes1, siderophores, and melanin biosynthesis clusters. Their orthologs can be found in all other aspergilli and many distantly related fungi such as Penicillium marneffei and Talaromyces stipitatus. Interestingly most of the “core” clusters are involved in protection against oxidative stress (Eisendle et al., 2003; Reeves et al., 2006; Schrettl et al., 2004), while the species-specific clusters either have been linked to antifungal or antibacterial compounds.
Figure 2. Core orthologous and species-specific SM clusters in A. fumigatus, A. clavatus and N. fischeri.
This Venn diagram shows relationships between putative SM clusters that were identified by SMURF in these three species. Non-overlapping areas represent the number of clusters unique to each species. Overlapping areas represent the number of orthologous clusters shared by two or three species. The total number of clusters is shown under the species name. The figure is not drawn to scale.
Interspecies comparison of the clusters present in the genomes of two strains (Af293 and A1163) of A. fumigatus also confirmed the prominent role of gene loss in the evolution of SM gene clusters. This search showed that two putative SM clusters present in Af293 are absent from A1163. One of them (AFUA_1G17710–AFUA1G17740) has an orthologous cluster in A. clavatus (ACLA_098870–ACLA_098920) as shown in Fig. 1. The other Af293-specific cluster has no orthologs in any other species.
Discussion
Validations and Limitations
SMURF is the first web-based tool that can systematically predict putative backbone genes in fungal genomes with high accuracy. Currently, there are only two publicly available software programs (Starcevic et al., 2008; Weber et al., 2009) designed to annotate PKS, NRPS and hybrid genes and both have been tailored to bacterial genomes. In addition to the backbone genes, SMURF can also generate rule-based sets of clusters, which can be used as a first approximation in comparative genomics and genetic studies. Notably, the algorithm predicts clusters that can be overlooked by an expert eye. For example, it identified eight additional clusters in A. fumigatus Af293 that had not been found in previous annotations (data not shown). Since none of these new clusters have been characterized experimentally, more studies are needed to estimate the true accuracy of the algorithm at predicting novel clusters.
When it comes to predicting boundaries, SMURF tends to inflate the number of decorating genes within a cluster by 4.0 on average. We chose not to adjust the d and y parameters and to err on the side of keeping false positives, as these can later be rejected based on experimental data or manual review. This relatively high false positive rate can be explained by the low number of clusters available for parameter optimization. Unexpectedly, SMURF performed better on non-A. fumigatus genomes, although only A. fumigatus clusters were used for parameter optimization. Again this can be related to the limited set of experimentally characterized clusters.
Since so few SM clusters have been experimentally characterized, we used previously described A. fumigatus SM clusters to find SM-defining domains over-represented in decorating proteins and to optimize parameters d and y used by the SMURF algorithm. This approach allowed us to validate prediction made by SMURF by comparing them to experimentally characterized clusters in other fungal genomes. The potential limitation of this approach is that this training set may be biased towards A. fumigatus type clusters. As more fungal clusters become characterized, this limitation will be addressed in future iterations of the algorithm. This can be achieved by including new SM-defining domains, changing the weights assigned to particular domains, or limiting the searches to specific pathways or taxonomic groups.
Most likely additional information about clusters boundaries will come from expression profiling, which appears to be the most expeditious approach to defining the boundaries. Future expression studies involving putative methyltransferase LaeA (Bok et al., 2006), the histone deacetylase HdaA (Shwab et al., 2007; Williams et al., 2008), and other chromatin modifiers and pathway-specific transcriptional regulators (Brakhage et al., 2008) can also facilitate the discovery of new clusters. Not all clusters, however, can be expressed under in vitro conditions. Likewise, experimental conditions can affect the number of differentially expressed genes in a cluster as have been shown for the A. fumigatus gliotoxin cluster (McDonagh et al., 2008; Perrin et al., 2007). Identification of putative “boundary” DNA motifs that get recognized by transcription factors and epigenetic regulators could further improve the algorithm accuracy. Ultimately, however, gene knock-out experiments followed by biochemical characterization of the enzymes are required to validate a cluster and to demarcate its ends.
Research application
Our preliminary analysis of the putative SM clusters predicted by SMURF in 27 fungal genomes showed that the numbers of potential SMs produced by fungi appears to be much higher than previously anticipated. This apparent discrepancy between the encoded and observed secondary metabolite repertoire can be explained by the presence of silent or “orphan” gene clusters, which do not get expressed under common in vitro conditions. Based on SMURF predictions, nonribosomal peptides and polyketides are the most abundant secondary metabolites produced by fungi. Among the taxa studied by genome sequencing, the aspergilli and sordariomycete genomes encode the largest numbers of these metabolites. Since over 50% of all SM compounds are estimated to have antibacterial, antifungal, or antitumor activity as revealed by a recent study (Palaez, 2005), these hidden clusters may represent a large unexplored reservoir of natural products of medical, agricultural, or industrial importance.
SM clusters are very unevenly distributed among fungal taxa consistent with the view that they can be used as species or diagnostic markers at either an inter-species or an inter-strain level (Frisvad et al., 2008). Cross-species comparison of SM clusters shows that very few of them are shared even among very closely related fungi (Fig. 2). This suggests that, with the exception of the small number of conserved “core” clusters, most SM clusters are relatively young in evolutionary terms and have been subject to rapid gene gain and loss.
What kind of selective pressures could have created this chemical diversity of fungal natural products? This has been attributed to diversifying selection (also known as positive Darwinian selection) driven by a chemical arms race between fungi and their predators, competitors, and hosts (Magan, 2006). Our results show that most conserved core clusters in the aspergilli have been linked to protection against oxidative stress (Eisendle et al., 2003; Reeves et al., 2006; Schrettl et al., 2004). In contrast, many lineage specific clusters are involved in biosynthesis of mycotoxins and antimicrobial compounds (e.g. gliotoxin, aflatoxin, penicillin). The observed lineage-specific expansions of SM genes in aspergilli and other soil fungi may be responsible for adaptation to the ever changing soil microbiome.
Our results indicate a correlation between the presence of SM pathways and competence for filamentous growth form among fungal taxa. The species phylogeny shows that numerous SM backbone genes were lost, on three separate occasions, on branches leading to species with primarily unicellular growth habits. Although the number of secondary metabolite clusters in the ancestor of all fungi is unknown, this ancestor is thought to be a multicellular organism (Liu and Hall, 2004). This suggests that losses of secondary metabolism genes may have coincided with transitions to a unicellular lifestyle during the evolution of each of these lineages (Fig. 1). In contrast to filamentous fungi that live in soil, the unicellular fungi analyzed here have adapted to highly specialized niches like decaying fruit or a eukaryotic host, so they may not need SMs as defense mechanisms. Similarly, we found fewer backbone genes in plant pathogens than in other fungi that have to thrive in a wide range of conditions. Finally the SM pathways expansions are found in aspergilli and other filamentous fungi that are characterized by ubiquity, metabolic versatility and opportunism.
Supplementary Material
Supplementary Figure 1. The key steps in the SMURF algorithm. The vertical arrows represent each subsequent step in the algorithm. The horizontal strings of squares and circles show intermediate outputs of these steps. Backbone genes (NRPSs, PKSs, Hybrids, and DMATSs) are shown as black squares; SM positive decorating genes as shown as black circles; and SM negative genes are shown as gray circles.
Acknowledgments
This work was supported by Science Foundation Ireland [07/IN1/B911 to KHW], NIAID [N01-AI30071, R21-AI052236, U01 AI48830], and USDA [2004-35600-14172].
Footnotes
Supplementary data are available online.
Conflict of Interest: None declared.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bok JW, et al. Secondary metabolic gene cluster silencing in Aspergillus nidulans. Mol Microbiol. 2006;61:1636–45. doi: 10.1111/j.1365-2958.2006.05330.x. [DOI] [PubMed] [Google Scholar]
- Bouhired S, et al. Accurate prediction of the Aspergillus nidulans terrequinone gene cluster boundaries using the transcriptional regulator. LaeA Fungal Genet Biol. 2007;44:1134–45. doi: 10.1016/j.fgb.2006.12.010. [DOI] [PubMed] [Google Scholar]
- Brakhage AA, et al. Evolution of beta-lactam biosynthesis genes and recruitment of trans-acting factors. Phytochemistry. 2005;66:1200–10. doi: 10.1016/j.phytochem.2005.02.030. [DOI] [PubMed] [Google Scholar]
- Brakhage AA, et al. Activation of fungal silent gene clusters: a new avenue to drug discovery. Prog Drug Res. 2008;66(1):3–12. doi: 10.1007/978-3-7643-8595-8_1. [DOI] [PubMed] [Google Scholar]
- Brown DW, et al. Twenty-five coregulated transcripts define a sterigmatocystin gene cluster in Aspergillus nidulans. Proc Natl Acad Sci U S A. 1996;93:1418–22. doi: 10.1073/pnas.93.4.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang YM, et al. A Gene Cluster Containing Two Fungal Polyketide Synthases Encodes the Biosynthetic Pathway for a Polyketide, Asperfuranone, in Aspergillus nidulans. J Am Chem Soc. 2009;131:2965–2970. doi: 10.1021/ja8088185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyle CM, Panaccione DG. An ergot alkaloid biosynthesis gene and clustered hypothetical genes from Aspergillus fumigatus. Appl Environ Microbiol. 2005;71:3112–8. doi: 10.1128/AEM.71.6.3112-3118.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisendle M, et al. The siderophore system is essential for viability of Aspergillus nidulans: functional analysis of two genes encoding l-ornithine N 5-monooxygenase (sidA) and a non-ribosomal peptide synthetase (sidC) Mol Microbiol. 2003;49:359–75. doi: 10.1046/j.1365-2958.2003.03586.x. [DOI] [PubMed] [Google Scholar]
- Fedorova ND, et al. Genomic islands in the pathogenic filamentous fungus Aspergillus fumigatus. PLoS Genet. 2008;4:e1000046. doi: 10.1371/journal.pgen.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisvad JC, et al. The use of secondary metabolite profiling in chemotaxonomy of filamentous fungi. Mycol Res. 2008;112:231–40. doi: 10.1016/j.mycres.2007.08.018. [DOI] [PubMed] [Google Scholar]
- Fujii I, et al. Hydrolytic polyketide shortening by ayg1p, a novel enzyme involved in fungal melanin biosynthesis. J Biol Chem. 2004;279:44613–20. doi: 10.1074/jbc.M406758200. [DOI] [PubMed] [Google Scholar]
- Galagan JE, et al. The genome sequence of the filamentous fungus Neurospora crassa. Nature. 2003;422:859–68. doi: 10.1038/nature01554. [DOI] [PubMed] [Google Scholar]
- Gardiner DM, Howlett BJ. Bioinformatic and expression analysis of the putative gliotoxin biosynthetic gene cluster of Aspergillus fumigatus. FEMS Microbiol Lett. 2005;248:241–8. doi: 10.1016/j.femsle.2005.05.046. [DOI] [PubMed] [Google Scholar]
- Grundmann A, et al. FtmPT2, an N-prenyltransferase from Aspergillus fumigatus, catalyses the last step in the biosynthesis of fumitremorgin B. Chembiochem. 2008;9:2059–63. doi: 10.1002/cbic.200800240. [DOI] [PubMed] [Google Scholar]
- Hoffmeister D, Keller NP. Natural products of filamentous fungi: enzymes, genes, and their regulation. Nat Prod Rep. 2007;24:393–416. doi: 10.1039/b603084j. [DOI] [PubMed] [Google Scholar]
- Kato N, et al. Identification of cytochrome P450s required for fumitremorgin biosynthesis in Aspergillus fumigatus. Chembiochem. 2009;10:920–8. doi: 10.1002/cbic.200800787. [DOI] [PubMed] [Google Scholar]
- Keller NP, Hohn TM. Metabolic Pathway Gene Clusters in Filamentous Fungi. Fungal Genet Biol. 1997;21:17–29. [PubMed] [Google Scholar]
- Keller NP, et al. Fungal secondary metabolism - from biochemistry to genomics. Nat Rev Microbiol. 2005;3:937–47. doi: 10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- Kim YT, et al. Two different polyketide synthase genes are required for synthesis of zearalenone in Gibberella zeae. Mol Microbiol. 2005;58:1102–13. doi: 10.1111/j.1365-2958.2005.04884.x. [DOI] [PubMed] [Google Scholar]
- Liu YJ, Hall BD. Body plan evolution of ascomycetes, as inferred from an RNA polymerase II phylogeny. Proc Natl Acad Sci U S A. 2004;101:4507–12. doi: 10.1073/pnas.0400938101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magan N, Aldred D. Why do fungi produce mycotoxins? In: Dijksterhuis J, Samson RA, editors. New Challenges in Food Mycology. Taylor & Francis; Boca Raton, FL: 2006. pp. 121–133. [Google Scholar]
- Maiya S, et al. The fumitremorgin gene cluster of Aspergillus fumigatus: identification of a gene encoding brevianamide F synthetase. Chembiochem. 2006;7:1062–9. doi: 10.1002/cbic.200600003. [DOI] [PubMed] [Google Scholar]
- Malz S, et al. Identification of a gene cluster responsible for the biosynthesis of aurofusarin in the Fusarium graminearum species complex. Fungal Genet Biol. 2005;42:420–33. doi: 10.1016/j.fgb.2005.01.010. [DOI] [PubMed] [Google Scholar]
- McDonagh A, et al. Sub-Telomere Directed Gene Expression during Initiation of Invasive Aspergillosis. PLoS Pathog. 2008;4:e1000154. doi: 10.1371/journal.ppat.1000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry J, Finn R. Pfam: a domain-centric method for analyzing proteins and proteomes. Methods Mol Biol. 2007;396:43–58. doi: 10.1007/978-1-59745-515-2_4. [DOI] [PubMed] [Google Scholar]
- Nierman WC, et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature. 2005;438:1151–6. doi: 10.1038/nature04332. [DOI] [PubMed] [Google Scholar]
- Palaez F. Biological activities of fungal metabolites. In: An Z, editor. Handbook of industrial microbiology. Marcel Dekker; New York: 2005. pp. 49–92. [Google Scholar]
- Perrin RM, et al. Transcriptional regulation of chemical diversity in Aspergillus fumigatus by LaeA. PLoS Pathog. 2007;3:e50. doi: 10.1371/journal.ppat.0030050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor RH, et al. Co-expression of 15 contiguous genes delineates a fumonisin biosynthetic gene cluster in Gibberella moniliformis. Fungal Genet Biol. 2003;38:237–49. doi: 10.1016/s1087-1845(02)00525-x. [DOI] [PubMed] [Google Scholar]
- Reeves EP, et al. A nonribosomal peptide synthetase (Pes1) confers protection against oxidative stress in Aspergillus fumigatus. Febs J. 2006;273:3038–53. doi: 10.1111/j.1742-4658.2006.05315.x. [DOI] [PubMed] [Google Scholar]
- Reiber K, et al. The expression of selected non-ribosomal peptide synthetases in Aspergillus fumigatus is controlled by the availability of free iron. FEMS Microbiol Lett. 2005;248:83–91. doi: 10.1016/j.femsle.2005.05.028. [DOI] [PubMed] [Google Scholar]
- Schrettl M, et al. Siderophore biosynthesis but not reductive iron assimilation is essential for Aspergillus fumigatus virulence. J Exp Med. 2004;200:1213–9. doi: 10.1084/jem.20041242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwecke T, et al. Nonribosomal peptide synthesis in Schizosaccharomyces pombe and the architectures of ferrichrome-type siderophore synthetases in fungi. Chembiochem. 2006;7:612–22. doi: 10.1002/cbic.200500301. [DOI] [PubMed] [Google Scholar]
- Selengut JD, et al. TIGRFAMs and Genome Properties: tools for the assignment of molecular function and biological process in prokaryotic genomes. Nucleic Acids Res. 2007;35:D260–4. doi: 10.1093/nar/gkl1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shwab EK, et al. Histone deacetylase activity regulates chemical diversity in Aspergillus. Eukaryot Cell. 2007;6:1656–64. doi: 10.1128/EC.00186-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DJ, et al. Cloning and heterologous expression of the penicillin biosynthetic gene cluster from penicillum chrysogenum. Biotechnology (N Y) 1990;8:39–41. doi: 10.1038/nbt0190-39. [DOI] [PubMed] [Google Scholar]
- Song Z, et al. Fusarin C biosynthesis in Fusarium moniliforme and Fusarium venenatum. Chembiochem. 2004;5:1196–1203. doi: 10.1002/cbic.200400138. [DOI] [PubMed] [Google Scholar]
- Sonnhammer EL, et al. Pfam: multiple sequence alignments and HMM-profiles of protein domains. Nucleic Acids Res. 1998;26:320–2. doi: 10.1093/nar/26.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starcevic A, et al. ClustScan: an integrated program package for the semi-automatic annotation of modular biosynthetic gene clusters and in silico prediction of novel chemical structures. Nucleic Acids Res. 2008;36:6882–92. doi: 10.1093/nar/gkn685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczyk E, et al. Identification and characterization of the asperthecin gene cluster of Aspergillus nidulans. Appl Environ Microbiol. 2008;74:7607–12. doi: 10.1128/AEM.01743-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend CA. Structural studies of natural product biosynthetic proteins. Chem Biol. 1997;4:721–30. doi: 10.1016/s1074-5521(97)90310-0. [DOI] [PubMed] [Google Scholar]
- Tsai HF, et al. A developmentally regulated gene cluster involved in conidial pigment biosynthesis in Aspergillus fumigatus. J Bacteriol. 1999;181:6469–77. doi: 10.1128/jb.181.20.6469-6477.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unsold IA, Li SM. Overproduction, purification and characterization of FgaPT2, a dimethylallyltryptophan synthase from Aspergillus fumigatus. Microbiology. 2005;151:1499–505. doi: 10.1099/mic.0.27759-0. [DOI] [PubMed] [Google Scholar]
- Unsold IA, Li SM. Reverse prenyltransferase in the biosynthesis of fumigaclavine C in Aspergillus fumigatus: gene expression, purification, and characterization of fumigaclavine C synthase FGAPT1. Chembiochem. 2006;7:158–64. doi: 10.1002/cbic.200500318. [DOI] [PubMed] [Google Scholar]
- Weber T, et al. CLUSEAN: a computer-based framework for the automated analysis of bacterial secondary metabolite biosynthetic gene clusters. J Biotechnol. 2009;140:13–7. doi: 10.1016/j.jbiotec.2009.01.007. [DOI] [PubMed] [Google Scholar]
- Williams RB, et al. Epigenetic remodeling of the fungal secondary metabolome. Org Biomol Chem. 2008;6:1895–7. doi: 10.1039/b804701d. [DOI] [PubMed] [Google Scholar]
- Yu J, et al. Gene profiling for studying the mechanism of aflatoxin biosynthesis in Aspergillus flavus and A. parasiticus. Food Addit Contam. 2007;24:1035–42. doi: 10.1080/02652030701513800. [DOI] [PubMed] [Google Scholar]
- Zaleta-Rivera K, et al. A bidomain nonribosomal peptide synthetase encoded by FUM14 catalyzes the formation of tricarballylic esters in the biosynthesis of fumonisins. Biochemistry. 2006;45:2561–9. doi: 10.1021/bi052085s. [DOI] [PubMed] [Google Scholar]
- Zhang S, et al. Indole-diterpene gene cluster from Aspergillus flavus. Appl Environ Microbiol. 2004;70:6875–83. doi: 10.1128/AEM.70.11.6875-6883.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. The key steps in the SMURF algorithm. The vertical arrows represent each subsequent step in the algorithm. The horizontal strings of squares and circles show intermediate outputs of these steps. Backbone genes (NRPSs, PKSs, Hybrids, and DMATSs) are shown as black squares; SM positive decorating genes as shown as black circles; and SM negative genes are shown as gray circles.