Abstract
Genome-wide association studies of lung cancer reported in populations of European background have identified three regions on chromosomes 5p15.33, 6p21.33, and 15q25 that have achieved genome-wide significance with p-values of 10−7 or lower. These studies have been performed primarily in cigarette smokers, raising the possibility that the observed associations could be related to tobacco use, lung carcinogenesis, or both. Since most women in Asia do not smoke, we conducted a genome-wide association study of lung adenocarcinoma in never-smoking females (584 cases, 585 controls) among Han Chinese in Taiwan and found that the most significant association was for rs2736100 on chromosome 5p15.33 (p = 1.30×10−11). This finding was independently replicated in seven studies from East Asia totaling 1,164 lung adenocarcinomas and 1,736 controls (p = 5.38×10−11). A pooled analysis achieved genome-wide significance for rs2736100. This SNP marker localizes to the CLPTM1L-TERT locus on chromosome 5p15.33 (p = 2.60×10−20, allelic risk = 1.54, 95% Confidence Interval (CI) 1.41–1.68). Risks for heterozygote and homozygote carriers of the minor allele were 1.62 (95% CI; 1.40–1.87), and 2.35 (95% CI: 1.95–2.83), respectively. In summary, our results show that genetic variation in the CLPTM1L-TERT locus of chromosome 5p15.33 is directly associated with the risk of lung cancer, most notably adenocarcinoma.
Author Summary
Worldwide, approximately 15% of lung cancer cases occur among nonsmokers. Genome-wide association studies (GWAS) of lung cancer conducted in populations of European background have identified three regions on chromosomes 5, 6, and 15 that harbor genetic variants that confer risk for lung cancer. Prior studies were conducted primarily in cigarette smokers, raising the possibility that the associations could be related to tobacco use, lung carcinogenesis, or both. A GWAS of lung cancer among never-smokers is an optimal setting to discover effects that are independent of smoking. Since most women in Asia do not smoke, we conducted a GWAS of lung adenocarcinoma among never-smoking females (584 cases, 585 controls) in Taiwan, and observed a region on chromosome 5 significantly associated with risk for lung cancer in never-smoking women. The finding was independently replicated in seven studies from East Asia totaling 1,164 lung adenocarcinomas and 1,736 controls. To our knowledge, this study is the first reported GWAS of lung cancer in East Asian women, and together with the replication studies represents the largest genetic association study in this population. The findings provide insight into the genetic contribution of common variants to lung carcinogenesis.
Introduction
To date, several large genome-wide association studies (GWAS) of lung cancer conducted in subjects of European background have identified susceptibility alleles on chromosomes 5p15.33, 6p21.33 and 15q25 [1]–[8]. These studies have shown that statistical evidence that exceeds the threshold of genome wide significance, defined as a p value less than 5×10−7 [9] or 1×10−8 [10]. In each study, the majority of cases and controls were cigarette smokers, making it difficult to determine whether these loci are associated with lung carcinogenesis or tobacco use, or perhaps both [11]. It has been difficult to accrue a sufficiently large set of lung cancer cases with no history of smoking because a high proportion of lung cancer in women as well as men in North America and Europe is directly related to tobacco use. In contrast, a substantial proportion of lung cancer in East Asian women occurs among non-smokers, who interestingly have a relatively high rate of lung cancer [12]. This suggests that genetic and/or environmental factors could account for the observed differences. To investigate this further, we conducted a genome-wide association study with follow-up of notable SNPs in never-smoking women in East Asia. In addition, we genotyped tag SNPs optimized for East Asians for the three regions previously identified by GWAS in European populations.
Results
Genome-wide association scan
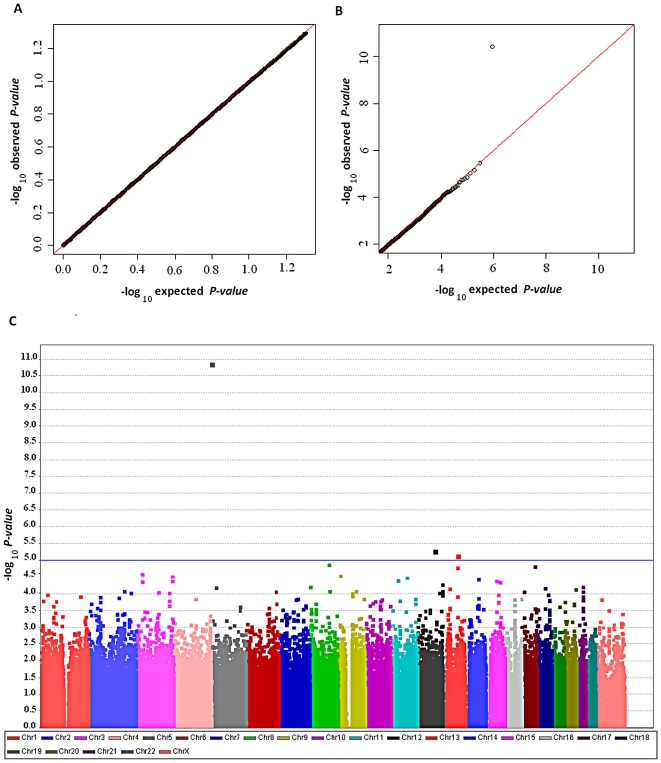
We conducted an initial GWAS of 584 lung cancer cases and 585 controls drawn from a case-control study in Taiwan, the Genetic Epidemiological Study of Lung Adenocarcinoma (GELAC) [13] (Table 1). Cases were restricted to those never-smoking females with a confirmed diagnosis of adenocarcinoma of the lung in GELAC. Controls were drawn from never-smoking female controls in GELAC and frequency-matched by age with cases (see Text S1 for more details). We began with a pilot study in which 54 cases and 54 controls were genotyped with the Illumina HumanCNV370-Duo BeadChip and, based on its success, 550 cases and 549 controls were genotyped on the Illumina HumanHap 610 Quad BeadChip. After quality control metrics were applied to both data sets (see Materials and Methods), the variance inflation factor λ in the genomic control model was found to be 1.013 and the inflation factor λ1000 for an equivalent study of 1000 cases and controls [14] was 1.022; together with the comparison of the observed and expected p-values in the quantile-quantile plot, shown in Figure 1, there is no evidence of a substantial issue related to population substructure but instead, several promising regions in the tail of the distribution are apparent, suitable for follow-up analysis. In fact, the distribution of the bottom 90% of p-values is similar to the expected distribution (Figure 1A) whereas the top 10% p-values displayed a deviation consistent with possible new signals (Figure 1B).
Table 1. Number of cases and controls, and study characteristics, for each participating study center.
| Cases | Controls | Age [mean (std)] | ||||
| Study Center† | Region | (n = 2768) | (n = 3100) | Controls | cases | Study Design |
| GELAC | Taiwan (GWAS) | 584 | 585 | 58.9 (11.2) | 59.0 (11.4) | Case-control |
| GELAC | Taiwan (replication) | 610 | 560 | 57.0 (13.8) | 60.1 (11.6) | Case-control |
| CAMSCH | Mainland China | 287 | 287 | 56.3 (11.6) | 56.3 (10.0) | Case-control |
| SNU | South Korea | 259 | 293 | 61.2 (10.2) | 61.2 (10.5) | Case-control |
| SWHS | Mainland China | 209 | 213 | 58.9 (8.4) | 58.9 (8.3) | Cohort |
| WHLCS | Mainland China | 207 | 207 | 56.9 (9.6) | 56.3 (11.2) | Case-control |
| KNUH | South Korea | 121 | 119 | 60.7 (6.5) | 61.1 (9.4) | Case-control |
| KUMC | South Korea | 95 | 87 | 60.9 (6.4) | 62.7 (10.5) | Case-control |
| GEL-S* | Singapore | 193 | 546 | 64.7 (11.4) | 63.4 (12.1) | Case-control |
| NJLCS* | Mainland China | 203 | 203 | 56.9 (9.8) | 56.8 (9.7) | Case-control |
†: GELAC: Genetic Epidemiological Study of Lung Adenocarcinoma (in Taiwan); CAMSCH: Chinese Academy of Medical Sciences Cancer Hospital Study; SNU: Seoul National University study; SWHS: Shanghai Women's Health Cohort Study; WHLCS: Wuhan Lung Cancer Study; KNUH: Kyungpook National University Study; KUMC: Korea University Study; GEL-S: Genes and Environment in Lung Cancer, Singapore study; NJLCS: Nanjing Lung Cancer Study.
*A subset of data presented in this paper for CAMSCH (Wu et al., (2009) Cancer Res;12:5065–5072), GEL-S (Truong et al., (2010) J Natl Cancer Inst, In press ), and NJLCS (Jin et al., (2009) Carcinogenesis;30:987–990;Wu et al., (2009) Cancer Res;12:5065–5072) was previously published and is noted in Tables S1, S2, S4, S6 and Figure S1.
Figure 1. Genome-wide association results in the GELAC study.
(A) Quantile-quantile plot for lower 90% of −log P-values, (B) upper 10% of −log P-values, and (C) scatter plot of P-values in−log scale from the trend test for 457,504 genotyped variants comparing 584 cases and 585 controls.
As shown in Figure 1C, the scatter plot of p-values on a −log scale for the trend test conducted for 457,504 SNPs used after quality control metrics were applied, only one SNP, rs2736100, was highly associated with lung cancer (p = 1.30 * 10−11) below the threshold of genome-wide significance, namely, p less than 1×10−7 (Figure 2). In an analysis for trend adjusted for age, the allelic odds ratio was 1.83 (1.54–2.18), which is notably higher than the estimates reported in the European studies (Figure 2) [4]. It is remarkable that our finding suggests a higher estimated effect size compared to that which was reported in Europeans, who were primarily smokers. To confirm the signal at rs2736100, the samples in the GWAS were genotyped using an optimized TaqMan assay (ABI, Foster City, CA), which had a concordance of 99.7% between the two platforms [15].
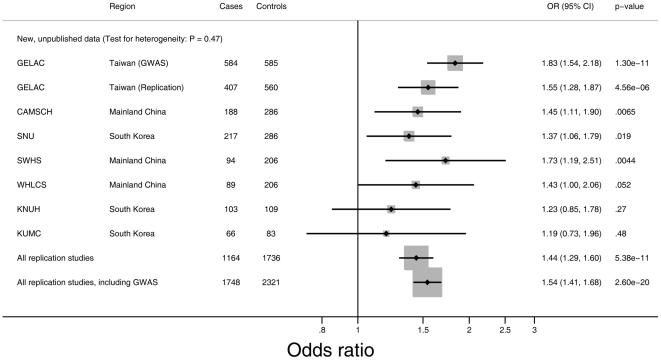
Figure 2. Risk of lung adenocarcinoma associated with rs2736100 for never-smoking female cases and never-smoking female controls from East Asia.
Forest plot representing lung adenocarcinoma risk and the rs2736100 genotype. Odds ratios (OR) and 95% confidence intervals (CI) for lung adenocarcinoma are derived from the per-allele model. All models are adjusted for age and study center. GELAC: Genetic Epidemiological Study of Lung Adenocarcinoma (in Taiwan); CAMSCH: Chinese Academy of Medical Sciences Cancer Hospital Study; SNU: Seoul National University study; SWHS: Shanghai Women's Health Cohort Study; WHLCS: Wuhan Lung Cancer Study; KNUH: Kyungpook National University Study; KUMC: Korea University Study.
Replication of the association of rs2736100 with lung cancer risk
Replication of the strongest signal, rs2736100 was performed in the remaining subjects of the GELAC study [13] as well as six studies of never-smoking Asian women with lung adenocarcinoma in East Asia. A total of 1164 cases with lung adenocarcinoma and 1736 controls were genotyped using an optimized TaqMan assay (shown to have high concordance with the Illumina results as described above). The additional replication studies included the Chinese Academy of Medical Sciences Cancer Hospital study (CAMSCH) [16], Wuhan lung cancer study (WHLCS) [17], Seoul National University study (SNU) [18], Korea University Medical Center study (KUMC) [19], Kyungpook National University Hospital study (KNUH) [20] and Shanghai Women's Health Cohort Study (SWHS) [21], [22] (Table 1). Characteristics of the study subjects from the GWAS and the replication studies are presented in Table 1.
The combined replication study confirmed that rs2736100 is associated with risk for lung adenocarcinoma in never-smoking women in East Asia (p = 5.38 * 10−11; allelic OR = 1.44; 95% CI 1.29–1.60) (Figure 2). In a pooled analysis of the GWAS and replication studies, rs2736100 was conclusively associated with the risk for lung adenocarcinoma in never-smoking females in East Asian populations; the allelic OR is 1.54 (95% CI 1.41–1.68; p = 2.60 * 10−20) (Figure 2 and Table 2). The estimated odds ratios for the heterozygous and homozygous carriers are 1.62 (95% CI 1.40–1.87) and 2.35 (95% CI 1.95–2.83). There was no evidence of heterogeneity between the results of the one cohort study (SWHS) and the pooled analysis of the 6 case-control studies (p = 0.36). Further pooling with two previously published studies, the Nanjing lung cancer study (NJLCS) [23] and the Genes and Environment in Lung Cancer, Singapore study (GEL-S) [24], [25] (Table 1, Table S1), yielded comparable results (p = 1.16 * 10−21) (Table S2, Figure S1). Across all studies, we observed consistently increased risk associated with rs2736100 with no evidence for heterogeneity between studies, measured by the I2 test for heterogeneity (Figure S1). In a subsequent analysis combining all lung cancer cases, rs2736100 was also significantly associated with lung cancer susceptibility (p = 5.50 * 10−20; allelic OR = 1.48; 95% CI 1.36–1.62) (Table 2). This observation is comparable to what is being reported for adenocarcinoma alone, which is not surprising because adenocarcinomas constitute 76% of cases (Table 2).
Table 2. Lung cancer risk associated with rs2736100, among never-smoking females from East Asia, by histology.
| Genotype | Controls | % | Cases | % | OR‡ | 95%CI‡ | p-value |
| all lung cancers | |||||||
| TT | 852 | 36.7 | 599 | 26.0 | |||
| GT | 1132 | 48.8 | 1187 | 51.4 | 1.49 | 1.30–1.70 | 3.76E-09 |
| GG | 337 | 14.5 | 522 | 22.6 | 2.20 | 1.85–2.62 | 2.94E-19 |
| trend | 2321 | 2308 | 1.48 | 1.36–1.62 | 5.50E-20 | ||
| adenocarcinomas | |||||||
| TT | 852 | 36.7 | 428 | 24.5 | |||
| GT | 1132 | 48.8 | 922 | 52.7 | 1.62 | 1.40–1.87 | 8.51E-11 |
| GG | 337 | 14.5 | 398 | 22.8 | 2.35 | 1.95–2.83 | 3.05E-19 |
| trend | 2321 | 1748 | 1.54 | 1.41–1.68 | 2.60E-20 | ||
| squamous cell carcinomas | |||||||
| TT | 852 | 36.7 | 60 | 33.9 | |||
| GT | 1132 | 48.8 | 82 | 46.3 | 1.02 | 0.72–1.44 | 0.91 |
| GG | 337 | 14.5 | 35 | 19.8 | 1.47 | 0.95–2.27 | 0.084 |
| trend | 2321 | 177 | 1.18 | 0.95–1.48 | 0.14 | ||
‡: Odds ratios (OR) and 95% confidence intervals (CI) adjusted for age and study center.
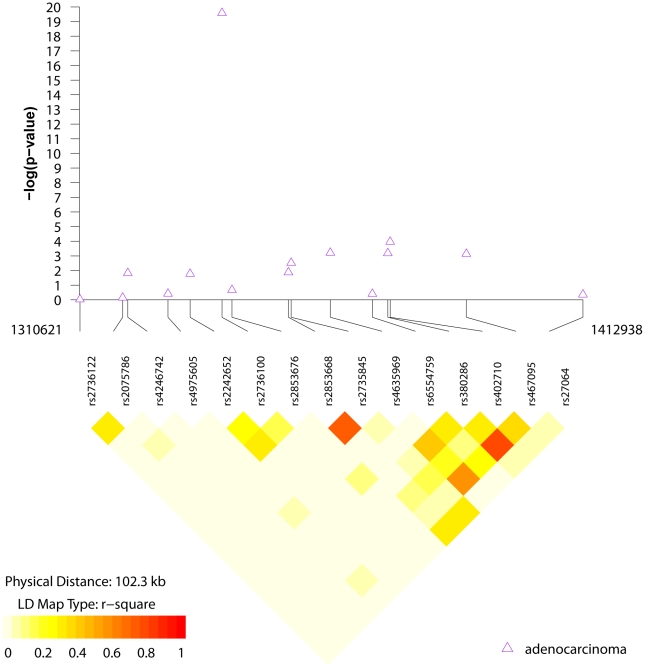
We conducted a first generation fine mapping of this region of chromosome 5p15.33 using 15 tag SNPs optimized in the East Asian studies in HapMap phase 2; the 15 SNPs were chosen using an r2≥0.8 as a threshold and estimated to cover approximately 85% of the known SNPs in HapMap phase 2 (Table S3). We did not identify stronger signals for association with lung adenocarcinoma in the 15 SNPs, as shown in Figure 3 and Table S4. Notably, rs402710, previously reported in GWAS of European ancestry [4], was the second most significant SNP tested in this region but did not achieve genome-wide significance (p = 0.0046) (Table S4). When rs402710 and rs2736100 were analyzed in a multivariable model, the former became non-significant (p = 0.33).
Figure 3. LD structure and association results for the chr5p15 region and lung adenocarcinoma.
LD structure and regional association results for 15 SNPs genotyped in the 5p15 region. SNPs included were tagged with r 2≥0.8 in HapMap CHB [37].
Discussion
In this study of lung adenocarcinoma in East Asian never-smoking women, we report a highly significant association with the common SNP, rs2736100, which localizes to the TERT-CLPTM1L locus on chromosome 5p15.33. Our study is notable because the sample size for never-smoking female cases is substantially larger than previous reports. Moreover, the estimated effect size observed for rs2736100 and adenocarcinoma of the lung (OR = 1.54) is greater than the associations previously reported in European populations (e.g., OR = 1.24 from the largest meta-analysis reported to date [4], [8], p = 0.000046 for difference). Our study provides strong evidence that this locus on chromosome 5p15.33 is directly related to lung carcinogenesis because it has been conclusively shown in non-smoking women.
The SNP marker, rs2736100, is mapped to a region of chromosome 5p15.33 in which common and rare genetic variants have been linked to a spectrum of cancers and related conditions. rs2736100 is localized to intron 2 of the telomerase gene TERT, a reverse transcriptase that is critical for telomere replication and stabilization by controlling telomere length. Variants in the TERT-CLPTM1L locus have been identified by GWAS to harbor susceptibility alleles for cancer of the brain, pancreas and lung [8], [26], [27]. For the latter, a large meta-analysis combined with a new scan indicates that the signal in this locus is most strongly associated with one histology, adenocarcinoma in studies of European subjects [8].
There is further evidence for association of this locus with additional cancers, though the reported results have not yet achieved the genome-wide association threshold; these include cancer of the bladder, prostate, uterine cervix, and skin including basal cell carcinoma and melanoma [4], [5], [7], [27]. Rare variations/mutations in the TERT gene have been described as a risk factor for acute myelogenous leukemia and also explain a proportion of the inherited bone marrow failure family pedigrees with dyskeratosis congenita, a cancer predisposition syndrome [28], [29]. Mutations in the TERT gene have also been described in patients with idiopathic pulmonary fibrosis [30], [31]. Together these findings suggest that the TERT-CLPTM1L 5p15.33 region could be important in the development of a spectrum of cancers. Still, at this time, further studies are needed to fine map the region, based on comprehensive re-sequence analysis in East Asian populations, to narrow the set of genetic variants worthy of functional studies to establish the mechanism underpinning the association marked by the SNP rs2736100 and subsequently compare these findings with comparable analyses in the other diseases.
The plausible mechanisms underlying the association signals across this region of chromosome 5p15.33 are currently under active investigation by many groups. Our findings are particularly interesting because we have identified variants that appear to be directly related to primary carcinogenesis. In this regard, it is critical that future studies evaluate environmental risk factors that may contribute, particularly since there is preliminary data suggesting that smoking as well as other exposures could directly influence telomere lengths [32]. It is noteworthy that lung cancer risk among non-smoking women in East Asia has been linked to indoor air pollution from environmental tobacco smoke [12], fumes produced by high temperature cooking [33], and coal combustion products [34].
Based on the discovery of susceptibility loci on chromosomal regions 6p21.33 and 15q25 first observed in European populations [1]–[3], [35], we attempted to replicate the findings in never-smoking women in East Asia. The strongest SNPs reported in each region plus additional tag SNPs, chosen on the basis of HapMap Phase 2, were genotyped in seven studies. 15 SNPs were selected for 6p21.33, covering an estimated 93% of known SNPs in HapMap phase 2 in the East Asian populations, whereas 24 SNPs were genotyped across 15q25, covering an estimated 83% of known common SNPs in the region (Table S3). In these East Asian never-smoking women, there was no convincing evidence for association at chromosome 6p21.33 or for 15q25 for lung cancer overall or for the adenocarcinoma subtype (Tables S5 and S6).
We report conclusive evidence that common genetic variants in the TERT-CLPTM1L locus on chromosome 5p15.33 are associated with risk for lung adenocarcinoma in non-smoking Asian women. We observed estimated effect sizes that are substantially higher than those previously reported in European smokers, which bears follow-up investigation into the biology of the underlying mechanism of the contribution of this region to primary lung carcinogenesis. Since this region on chromosome 5p15.33 has been implicated in many cancers, our observations should stimulate further investigation of the region that could lead to new insights into carcinogenesis.
Materials and Methods
Studies
A description of each study is provided in Table 1 and Text S1. Lung cancer cases and controls for the GWAS were drawn from the Genetic Epidemiological Study of Lung Adenocarcinoma (GELAC) in Taiwan. A total of 584 never-smoking incident cases and 585 never-smoking controls were included in the GWAS. The replication studies were drawn from seven studies, including additional subjects from the GELAC study [13], the Chinese Academy of Medical Sciences Cancer Hospital study (CAMSCH) [16], the Wuhan lung cancer study (WHLCS) [17], the Seoul National University study (SNU) [18], the Korea University Medical Center study (KUMC) [19], the Kyungpook National University Hospital study (KNUH) [20], and the Shanghai Women's Health Cohort Study (SWHS) [21], [22]. In addition, data were pooled with previously published findings from the Nanjing lung cancer study (NJLCS) [23] and the Genes and Environment in Lung Cancer, Singapore study (GEL-S) [24] (Table 1). All studies are case-control studies with the exception of the SWHS, which is a prospective cohort study. The range of ages is similar in cases and controls across all studies (Table 1).
Ethics statement
All study subjects provided informed consent and each study was approved by its respective institution's IRB.
Genotyping and quality control
Genome-wide association study genotyping and quality control
GWAS genotyping of the GELAC samples was performed in two separate phases. In the pilot phase, 54 cases and 54 controls were genotyped by GeneTech Biotech Co., (Taiwan), using the Illumina HumanCNV370-Duo BeadChip. The cases were never-smoking females diagnosed with lung adenocarcinoma at age ≤51 who had questionnaire data and DNA that passed quality control criteria for scanning. The controls were never-smoking females matched by age (±2 years) to cases.
Cluster definitions were determined using Illumina BeadStudio Genotyping Module v.3.3.4. Genotype calls were based on a quality score (Gene call value) of 0.25 or higher. Four blind duplicate pairs were included, and the concordance of SNP genotype calls between each pair is greater than 99.997%. Quality control metrics for data from the first phase are similar to those for data from the second phase, detailed below.
In the second phase of the GWAS, 550 cases and 549 controls were genotyped with the Illumina HumanHap610 Quad BeadChip on contract at deCODE Genetics, Iceland. The cases were the first never-smoking female lung adenocarcinoma subjects to be enrolled in the study with questionnaire data and DNA that passed quality control for scanning. Cluster definitions were determined using the Illumina BeadStudio Genotyping Module. The median genotype call rate for samples was 99.78%. 95% overall displayed call rates larger than 99.49%; the median call rate for variants is 99.91%, with 95% of variants with call rates above 99.55%. 21 blind duplicate pairs displayed an average concordance greater than 99.99%.
After quality control metrics were applied, 457,504 SNPs were used for the association analysis. SNPs (n = 1,705) were excluded if the call rate was below 90%, (i.e., a missing rate larger than 0.1); SNPs with a minor allele frequency below 0.05 (n = 131,558); SNPs with missing rate between 0.02 and 0.1 and non-random genotype failure with p<0.02 (n = 1,046); and, significant deviation from fitness for Hardy-Weinberg equilibrium (p<0.0001 in controls) (n = 718). 1064 unique samples from phase 2 were used in the association analysis, after two exclusion steps. The first set of exclusions was based on the quality control metrics described above and relatedness among individuals: call rates less than 90% (n = 3); sex discrepancies based on the X chromosome heterozygosity (n = 7); contaminated samples with high heterozygosity scores (n = 4), first or second degree relatives identified using genome-wide pairwise identical by descent (IBD) estimates (n = 9).
We further excluded 12 individuals from phase 2, based on population substructure analysis. In fact, to detect differences in population substructure, pairwise population concordance (PPC) test in PLINK (http://pngu.mgh.harvard.edu/purcell/plink/) [36] were performed with a threshold of 10−20 on two data sets using all autosomal SNPs that had passed the quality control metrics described above. The first data set consists of the 1184 unrelated individuals with high quality genotype data (108 from phase 1 and 1076 from phase 2). The PPC test identified 15 outliers who were distinct from the remaining 1169 (105 from phase 1 and 1064 from phase 2). The eight self-described aborigines (2 in Phase 1 and 6 in Phase 2) were among the outliers. Based on the PPC analysis, the final genome-wide association analysis was conducted using 1169 samples.
To further assess the population homogeneity in our study sample, we conducted additional analyses in our 1184 individuals with HapMap3 release 2 data [37]. The results indicate that for the 1184 unrelated individuals with high quality genotype data, 15 outliers were detected, thus yielding 1169 individuals with homogeneous genetic structure available for follow-up analyses. We seeded the study population with genotype data from hapmap 3 as well as hapmap 2; this included 85 CHD (Chinese in Metropolitan Denver, Colorado), in addition to our 1184 individuals and the hapmap 2 (84 CHB (Han Chinese in Beijing, China) and 86 JPT (Japanese in Tokyo, Japan)). A second analysis included our 1184 study individuals and a larger sample of HapMap3 release 2, namely the CHB, CHD, GIH (Gujarati Indians in Houston, Texas), JPT, LWK (Luhya in Webuye, Kenya), MKK (Maasai in Kinyawa, Kenya), and TSI (Toscani in Italia): the results confirmed that 15 outliers were detected whereas the 1164 represented a homogeneous population.
Although the above PPC tests seem to suggest little population substructure in our 1169 samples, we still used EIGENSTRAT [38] to conduct GWAS analysis to correct possible population stratification. We found that for the SNP rs2736100, the P-value is 1.239×10−11 based on the Armitage trend Chi-square statistic with no stratification correction and the P-value is 2.764×10−11 based on EIGENSTRAT using 10 principal components (the default value) for stratification correction. There was a negligible difference in p-values with and without this correction.
The genotyping cluster plot generated by the Illumina platform for rs2736100 is presented in Figure S2. The adjusted intensities for each allele are plotted, where each color represents a different genotype in the cluster plots. As shown in the figure, clusters of different genotypes are well separated from each other, indicating a high confidence in genotype calling in our study. The genotype call at this locus was confirmed with TaqMan genotyping (concordance of 99.7%).
Replication SNP selection and genotyping
DNA was extracted from blood samples and genotyped at the National Cancer Institute Core Genotyping Facility (CGF) (Http://cgf.nci.nih.gov) for four studies, SNU, KUMC, KNUH, and SWHS. TaqMan genotyping for the GELAC study (including all previously scanned cases and controls plus remaining never-smoking female cases and their matched controls) and the GEL-S studies was conducted in Taiwan and Singapore, respectively. Genotyping for the CAMSCH, WHLCS, and NJLCS studies were conducted at the Cancer Institute and Hospital, Chinese Academy of Medical Science, using TaqMan assays designed and optimized by the CGF (http://snp500cancer.nci.nih.gov).
We selected 54 SNPs optimized for Eastern Asian populations to cover the three chromosomal regions previously reported to show association for lung cancer (i.e., 15 SNPs in 5p15, 15 SNPs in 6p, and 24 SNPs in 15q25) (Table S3). The coordinates for selecting the tag SNPs were based on an r2<0.8 using the CHB samples of HapMap phase 2. The boundaries for the tag SNP selection were as follows: 5p15.33 from 1310620 to 1412939, 6p21.33 from 28782776 to 29018856 and 15q25 from 76593077 to 76702301 (Build 37). We computed genomic coverage using the GLU software package (http://code.google.com/p/glu-genetics/) for common SNPs (MAF≥0.05) based on the most recent build (Build 37) of the HapMap CHB [37] genotype data.
All TaqMan assays (Applied Biosystems Inc., Foster City, CA) for this study were optimized on the ABI 7900HT detection system with high concordance with sequence analysis of 102 individuals as listed on the SNP500Cancer website (http://snp500cancer.nci.nih.gov). All of the genotype frequencies were consistent with Hardy-Weinberg equilibrium except three SNPs (rs402710, rs9368570, and rs9257280) using a chi-square test (P<0.0001, Table S3). All reported genotyped results are based on completion rates of greater than 94% across all studies.
Statistical analysis
Genome-wide association tests
The program PLINK [36] was used to conduct primary statistical tests for association in the discovery phase. Association analyses between individual SNP and the lung cancer risk were carried out using computer packages in PLINK. Q-Q plots analyzed by the trend test are shown in Figure 1. We note that for Phase 1, we imputed the genotypes at the SNPs contained in HumanHap 610 Quad BeadChip but not in HumanCNV370-Duo BeadChip by using IMPUTE developed by Marchini et al. [39] and haplotypes of CHB in HapMap as the reference.
Replication and pooled analyses
Unconditional logistic regression was used to estimate the ORs and 95% CIs, adjusting for age and study center. All p values are two-sided. The most prevalent homozygous genotype was used as the reference group. Tests for trend were conducted by assigning the ordinal values 1, 2, and 3 to the most prevalent genotypes in rank order of wild type, heterozygous, and variant homozygous genotypes, respectively.
Supporting Information
Risk of lung cancer associated with rs2736100 for never-smoking female adenocarcinoma cases and never-smoking female controls from East Asia.
(0.66 MB TIF)
SNP graph of rs2736100 from (A) Illumina 610K (B) Illumina 370K based on Beadstudio Genotyping Module v3.
(1.47 MB TIF)
Lung cancer risk associated with rs2736100 among never-smoking females from East Asia, by study center, including two previously published studies.
(0.04 MB XLS)
Lung cancer risk associated with rs2736100 among never-smoking females from East Asia, by histology, including two previously published studies.
(0.03 MB XLS)
Chromosome 5, 6, and 15 SNPs genotyped.
(0.04 MB XLS)
Lung cancer risk associated with chromosome 5 SNPs, among never-smoking females from East Asia.
(0.05 MB XLS)
Lung cancer risk associated with chromosome 6 SNPs, among never-smoking females from East Asia.
(0.07 MB XLS)
Lung cancer risk associated with chromosome 15 SNPs, among never-smoking females from East Asia.
(0.07 MB XLS)
Supplementary information.
(0.09 MB DOC)
Acknowledgments
We are grateful to the many patients who participated in these studies and to support staff who helped make these studies possible. We also acknowledge the staff of the US NCI Core Genotyping Facility, including Charles Chung, who provided support for graphics; Lan-Chao Wang and Sharon Liu, National Health Research Institutes, Taiwan, for assistance in the analysis of the GWAS data; and Philip Eng, Singapore General Hospital; Swan Swan Leong, National Cancer Center; Alan WK Ng, Tan Tock Seng Hospital; Tow Keang Lim, National University Hospital; and Augustine Tee, Changi General Hospital, for support of the Singapore lung cancer study.
Footnotes
The authors have declared that no competing interests exist.
This study was supported in part by the Intramural Research Program of the NIH, National Cancer Institute. The GWAS based on the GELAC study was supported by the National Research Program on Genomic Medicine (NRPGM) of Taiwan under grants DOH97-TD-G-111-026 and DOH96-TD-G-111-017. The computational support for the GWAS was from grant NSC98-3112-B-400-009. The individual studies were funded by the following sources: The GELAC study was supported by DOH95-TD-G-111-015 from the National Research Program on Genomic Medicine in Taiwan, and 95A1-BSAP01-002 from the National Health Research Institutes, Taiwan. The Chinese Academy of Medical Sciences Cancer Hospital study (CAMSCH) was supported by the China National High Technology Project (2006AA02A401) (DL). The Wuhan lung cancer study (WHLCS) was supported by the National Natural Science Foundation (30872092 and 30525031). The Korea University Medical Center study (KUMC) was supported by a grant of the Korea Health 21 R&D Project, Ministry of Health & Welfare, Republic of Korea (A010250). The Kyungpook National University Hospital study (KNUH) was supported by the National R&D Program for Cancer Control Ministry of Health and Welfare, Republic of Korea (0720550-2). The Shanghai Women's Health Study (SWHS) was supported by the National Cancer Institute extramural research grant [R37 CA70837] and, partially for biological sample collection, National Cancer Institute Intramural Research Program contract NO2-CP-11010 with Vanderbilt University. The Nanjing lung cancer study (NJLCS) was supported by the China National Natural Science Foundation, grant # 30730080 (HS). The GEL-S study was supported by the National Medical Research Council, Singapore (NMRC/0897/2004 and NMRC/1075/2006). Genotyping in GEL-S study was partially funded by US National Institutes of Health (NIH); National Cancer Institute (NCI) (R03 CA133939-01. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40:616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 3.Liu P, Vikis HG, Wang D, Lu Y, Wang Y, et al. Familial aggregation of common sequence variants on 15q24–25.1 in lung cancer. J Natl Cancer Inst. 2008;100:1326–1330. doi: 10.1093/jnci/djn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McKay JD, Hung RJ, Gaborieau V, Boffetta P, Chabrier A, et al. Lung cancer susceptibility locus at 5p15.33. Nat Genet. 2008;40:1404–1406. doi: 10.1038/ng.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rafnar T, Sulem P, Stacey SN, Geller F, Gudmundsson J, et al. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat Genet. 2009;41:221–227. doi: 10.1038/ng.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spitz MR, Amos CI, Dong Q, Lin J, Wu X. The CHRNA5-A3 region on chromosome 15q24–25.1 is a risk factor both for nicotine dependence and for lung cancer. J Natl Cancer Inst. 2008;100:1552–1556. doi: 10.1093/jnci/djn363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Broderick P, Webb E, Wu X, Vijayakrishnan J, et al. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat Genet. 2008;40:1407–1409. doi: 10.1038/ng.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Landi MT, Chatterjee N, Yu K, Goldin LR, Goldstein AM, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet. 2009;85:679–691. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.The Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donnelly P. Progress and challenges in genome-wide association studies in humans. Nature. 2008;456:728–731. doi: 10.1038/nature07631. [DOI] [PubMed] [Google Scholar]
- 11.Chanock SJ, Hunter DJ. Genomics: when the smoke clears. Nature. 2008;452:537–538. doi: 10.1038/452537a. [DOI] [PubMed] [Google Scholar]
- 12.Lam WK. Lung cancer in Asian women-the environment and genes. Respirology. 2005;10:408–417. doi: 10.1111/j.1440-1843.2005.00723.x. [DOI] [PubMed] [Google Scholar]
- 13.Jou YS, Lo YL, Hsiao CF, Chang GC, Tsai YH, et al. Association of an EGFR intron 1 SNP with never-smoking female lung adenocarcinoma patients. Lung Cancer. 2009;64:251–256. doi: 10.1016/j.lungcan.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 14.de Bakker PI, Ferreira MA, Jia X, Neale BM, Raychaudhuri S, et al. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum Mol Genet. 2008;17:R122–R128. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chanock SJ, Manolio T, Boehnke M, Boerwinkle E, Hunter DJ, et al. Replicating genotype-phenotype associations. Nature. 2007;447:655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- 16.Wu C, Hu Z, Yu D, Huang L, Jin G, et al. Genetic variants on chromosome 15q25 associated with lung cancer risk in Chinese populations. Cancer Res. 2009;69:5065–5072. doi: 10.1158/0008-5472.CAN-09-0081. [DOI] [PubMed] [Google Scholar]
- 17.Bai Y, Xu L, Yang X, Hu Z, Yuan J, et al. Sequence variations in DNA repair gene XPC is associated with lung cancer risk in a Chinese population: a case-control study. BMC Cancer. 2007;7:81. doi: 10.1186/1471-2407-7-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim JH, Kim H, Lee KY, Choe KH, Ryu JS, et al. Genetic polymorphisms of ataxia telangiectasia mutated affect lung cancer risk. Hum Mol Genet. 2006;15:1181–1186. doi: 10.1093/hmg/ddl033. [DOI] [PubMed] [Google Scholar]
- 19.Jung HY, Whang YM, Sung JS, Shin HD, Park BL, et al. Association study of TP53 polymorphisms with lung cancer in a Korean population. J Hum Genet. 2008;53:508–514. doi: 10.1007/s10038-008-0278-y. [DOI] [PubMed] [Google Scholar]
- 20.Park JY, Park SH, Choi JE, Lee SY, Jeon HS, et al. Polymorphisms of the DNA repair gene xeroderma pigmentosum group A and risk of primary lung cancer. Cancer Epidemiol Biomarkers Prev. 2002;11:993–997. [PubMed] [Google Scholar]
- 21.Zhang Y, Shu XO, Gao YT, Ji BT, Yang G, et al. Family history of cancer and risk of lung cancer among nonsmoking Chinese women. Cancer Epidemiol Biomarkers Prev. 2007;16:2432–2435. doi: 10.1158/1055-9965.EPI-07-0398. [DOI] [PubMed] [Google Scholar]
- 22.Zheng W, Chow WH, Yang G, Jin F, Rothman N, et al. The Shanghai Women's Health Study: rationale, study design, and baseline characteristics. Am J Epidemiol. 2005;162:1123–1131. doi: 10.1093/aje/kwi322. [DOI] [PubMed] [Google Scholar]
- 23.Jin G, Xu L, Shu Y, Tian T, Liang J, et al. Common genetic variants on 5p15.33 contribute to risk of lung adenocarcinoma in a Chinese population. Carcinogenesis. 2009;30:987–990. doi: 10.1093/carcin/bgp090. [DOI] [PubMed] [Google Scholar]
- 24.Truong T, Hung RJ, Amos CI, Wu X, Bickeböller H, et al. Replication of lung cancer susceptibility loci at chromosomes 15q25, 5p15, and 6p21: a pooled analysis from the International Lung Cancer Consortium. J Natl Cancer Inst. 2010 doi: 10.1093/jnci/djq178. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang L, Lim W, Eng P, Leong SS, Lim TK, et al. Lung cancer in Chinese women: evidence for an interaction between tobacco smoking and exposure to inhalants in the indoor environment. Environ Health Perspect Environ Health Perspect. 2010 doi: 10.1289/ehp.0901587. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petersen GM, Amundadottir L, Fuchs CS, Kraft P, Stolzenberg-Solomon RZ, et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat Genet. 2010;42:224–228. doi: 10.1038/ng.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shete S, Hosking FJ, Robertson LB, Dobbins SE, Sanson M, et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet. 2009;41:899–904. doi: 10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamaguchi H, Calado RT, Ly H, Kajigaya S, Baerlocher GM, et al. Mutations in TERT, the gene for telomerase reverse transcriptase, in aplastic anemia. N Engl J Med. 2005;352:1413–1424. doi: 10.1056/NEJMoa042980. [DOI] [PubMed] [Google Scholar]
- 29.Calado RT, Regal JA, Hills M, Yewdell WT, Dalmazzo LF, et al. Constitutional hypomorphic telomerase mutations in patients with acute myeloid leukemia. Proc Natl Acad Sci U S A. 2009;106:1187–1192. doi: 10.1073/pnas.0807057106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mushiroda T, Wattanapokayakit S, Takahashi A, Nukiwa T, Kudoh S, et al. A genome-wide association study identifies an association of a common variant in TERT with susceptibility to idiopathic pulmonary fibrosis. J Med Genet. 2008;45:654–656. doi: 10.1136/jmg.2008.057356. [DOI] [PubMed] [Google Scholar]
- 31.Tsakiri KD, Cronkhite JT, Kuan PJ, Xing C, Raghu G, et al. Adult-onset pulmonary fibrosis caused by mutations in telomerase. Proc Natl Acad Sci U S A. 2007;104:7552–7557. doi: 10.1073/pnas.0701009104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valdes AM, Andrew T, Gardner JP, Kimura M, Oelsner E, et al. Obesity, cigarette smoking, and telomere length in women. Lancet. 2005;366:662–664. doi: 10.1016/S0140-6736(05)66630-5. [DOI] [PubMed] [Google Scholar]
- 33.Wakelee HA, Chang ET, Gomez SL, Keegan TH, Feskanich D, et al. Lung cancer incidence in never smokers. J Clin Oncol. 2007;25:472–478. doi: 10.1200/JCO.2006.07.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lan Q, Chapman RS, Schreinemachers DM, Tian L, He X. Household stove improvement and risk of lung cancer in Xuanwei, China. J Natl Cancer Inst. 2002;94:826–835. doi: 10.1093/jnci/94.11.826. [DOI] [PubMed] [Google Scholar]
- 35.Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 39.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Risk of lung cancer associated with rs2736100 for never-smoking female adenocarcinoma cases and never-smoking female controls from East Asia.
(0.66 MB TIF)
SNP graph of rs2736100 from (A) Illumina 610K (B) Illumina 370K based on Beadstudio Genotyping Module v3.
(1.47 MB TIF)
Lung cancer risk associated with rs2736100 among never-smoking females from East Asia, by study center, including two previously published studies.
(0.04 MB XLS)
Lung cancer risk associated with rs2736100 among never-smoking females from East Asia, by histology, including two previously published studies.
(0.03 MB XLS)
Chromosome 5, 6, and 15 SNPs genotyped.
(0.04 MB XLS)
Lung cancer risk associated with chromosome 5 SNPs, among never-smoking females from East Asia.
(0.05 MB XLS)
Lung cancer risk associated with chromosome 6 SNPs, among never-smoking females from East Asia.
(0.07 MB XLS)
Lung cancer risk associated with chromosome 15 SNPs, among never-smoking females from East Asia.
(0.07 MB XLS)
Supplementary information.
(0.09 MB DOC)