Abstract
Caenorhabditis elegans SKN-1 (ortholog of mammalian Nrf1/2/3) is critical for oxidative stress resistance and promotes longevity under reduced insulin/IGF-1–like signaling (IIS), dietary restriction (DR), and normal conditions. SKN-1 inducibly activates genes involved in detoxification, protein homeostasis, and other functions in response to stress. Here we used genome-scale RNA interference (RNAi) screening to identify mechanisms that prevent inappropriate SKN-1 target gene expression under non-stressed conditions. We identified 41 genes for which knockdown leads to activation of a SKN-1 target gene (gcs-1) through skn-1-dependent or other mechanisms. These genes correspond to multiple cellular processes, including mRNA translation. Inhibition of translation is known to increase longevity and stress resistance and may be important for DR–induced lifespan extension. One model postulates that these effects derive from reduced energy needs, but various observations suggest that specific longevity pathways are involved. Here we show that translation initiation factor RNAi robustly induces SKN-1 target gene transcription and confers skn-1-dependent oxidative stress resistance. The accompanying increases in longevity are mediated largely through the activities of SKN-1 and the transcription factor DAF-16 (FOXO), which is required for longevity that derives from reduced IIS. Our results indicate that the SKN-1 detoxification gene network monitors various metabolic and regulatory processes. Interference with one of these processes, translation initiation, leads to a transcriptional response whereby SKN-1 promotes stress resistance and functions together with DAF-16 to extend lifespan. This stress response may be beneficial for coping with situations that are associated with reduced protein synthesis.
Author Summary
The nematode C. elegans has proven to be an invaluable organism for elucidating mechanisms that influence aging. Here we used genome-scale RNA interference screening in C. elegans to identify mechanisms that regulate a set of genes that defend against oxygen radicals and other stresses. These genes are activated by the SKN-1 protein, which promotes longevity. We found that many biological processes influence the regulation of SKN-1–dependent stress defenses. These processes include mRNA translation, the mechanism by which proteins are synthesized. Previous work showed that reductions in translation slow aging, an effect that may be important in conserved longevity pathways. One model postulates that this increased longevity derives from reduced energy requirements, but here we determined that SKN-1–dependent stress gene expression and oxidative stress resistance are increased dramatically when translation initiation is inhibited. This effect is accompanied by increased longevity that depends largely upon SKN-1 acting in concert with DAF-16, a gene regulator that is widely implicated in longevity. We conclude that reductions in translation result in a stress gene activation response that increases both stress resistance and lifespan and may help the organism cope with situations that are associated with decreased protein synthesis.
Introduction
Small molecules that react with proteins, lipids, and nucleic acids can damage cells catastrophically. Oxidative stress refers to damage caused by reactive oxygen species (ROS), but other reactive molecules are produced during metabolism of endogenous (endobiotic) or exogenous (xenobiotic) compounds. Oxidative or xenobiotic stress is central to the pathogenesis of diabetes, atherosclerosis, cirrhosis, and many other syndromes, and has been implicated in aging [1]–[6]. Eukaryotic cells handle reactive compounds through a detoxification system in which lipophilic molecules are solubilized (Phase 1), and reactive species that include ROS and products of the Phase 1 system are inactivated (Phase 2) and may be transported out of the cell (Phase 3) [7]–[9].
Many Phase 2 detoxification genes are induced coordinately in response to oxidative or xenobiotic stress. This stress response is important in the liver and several other tissues in mammals, in which it is mediated by the Nrf1/2/3 (NF-E2-related factor) transcription factors [9], [10]. In the nematode C. elegans, this conserved stress response is mediated by the Nrf protein ortholog SKN-1 [11]. In the intestine, which is the major detoxification organ in C. elegans, SKN-1 accumulates in nuclei and activates target genes in response to various stresses [11], [12]. The relationship between SKN-1 and its targets is more complicated than a simple on/off stress response, however. Under non-stressed conditions SKN-1 up- or down-regulates a wide range of genes, including Phase 1, Phase 2, and Phase 3 detoxification, membrane, lysosomal, proteasomal, metabolic, and regulatory protein genes, many of which seem to be direct targets [12]. SKN-1 responds to stress by upregulating narrower sets of detoxification genes, and under certain conditions some SKN-1 target genes are activated by SKN-1-independent mechanisms [12]–[14]. It remains to be determined how cellular processes and regulatory inputs modulate expression of these overlapping groups of SKN-1-regulated genes.
C. elegans has been particularly advantageous for identifying mechanisms that influence aging. It was discovered in C. elegans that lifespan is increased by reductions in insulin/IGF-1-like signaling (IIS), a pathway that has since been implicated in longevity in Drosophila, mammals, and possibly humans [15], [16]. In C. elegans, this increased longevity requires the FOXO ortholog DAF-16, which is inhibited by IIS. SKN-1 is inhibited by IIS in parallel to DAF-16, contributes to the increases in lifespan and stress resistance that derive from reduced IIS, and promotes longevity under normal conditions [17]. While these activities involve SKN-1 expression in the intestine, SKN-1 is also found in the ASI chemosensory neurons, which sense food availability and influence metabolism [11]. SKN-1 expression in these neurons is required for lifespan to be increased by dietary restriction (DR), a condition that extends lifespan in essentially every species examined [18]. SKN-1 is not required for interference with mitochondrial function to extend lifespan, however, indicating that it is not essential in all longevity pathways [17].
In species as diverse as yeast and rodents, longevity is also increased when mRNA translation is inhibited [19]. It is particularly important to understand how this occurs, because reductions in translation are involved in conserved mechanisms that promote longevity. From yeast to mice, lifespan is increased by inhibition of the TOR (target of rapamycin) signaling pathway, which integrates growth and nutrient availability cues and promotes translation [19], [20]. TOR signaling activates the ribosomal S6 protein kinase (S6K), which upregulates translation, and inhibits eIF4E-binding protein (4E-BP), an inhibitor of cap-dependent translation. In Drosophila, reversing these effects is required for rapamycin treatment to extend lifespan, and increased 4E-BP activity is important for lifespan to be extended by DR, a pathway that may involve TOR signaling [20], [21]. Moreover, reduction of S6K activity increases lifespan in yeast, C. elegans, Drosophila, and mice [19], [22]–[25]. While lower levels of translation might promote longevity simply by decreasing the energy requirements of protein synthesis [26], recent evidence indicates that specific regulatory mechanisms are involved. In yeast and Drosophila, reductions in overall translation levels lead to preferential translation of beneficial genes [21], [27]. Furthermore, some C. elegans studies have reported that DAF-16 is needed for lifespan to be extended when translation initiation is inhibited by RNAi or mutation of general translation factors [23], [28], [29], although other analyses of initiation factors suggest that DAF-16 is not required [22], [26], [30], [31]. Given that DAF-16 and SKN-1 are inhibited in parallel by IIS, and cooperate to regulate some target genes [17], it is an intriguing question whether SKN-1 might act in parallel to DAF-16 to promote longevity in response to reduced translation initiation.
Here we have employed genome-scale RNAi screening in C. elegans to identify mechanisms that prevent inappropriate expression of SKN-1-dependent stress defense genes. We identified 41 genes for which knockdown robustly activated a SKN-1-responsive promoter in the intestine, in most cases dependent upon skn-1. These genes represented multiple cellular processes that are monitored by SKN-1-dependent stress defenses. As several of these genes are involved in mRNA translation and protein synthesis, we investigated the involvement of skn-1 in the effects of inhibiting translation initiation. We found that inhibition of genes involved in two different steps in translation initiation induced a robust transcriptional stress response, resulting in increased oxidative stress resistance that required SKN-1 but not DAF-16. In contrast, the accompanying longevity increases were mediated largely by the combined action of DAF-16 and SKN-1, indicating that these transcription factors are each crucial for the beneficial effects of translation suppression.
Results
Identification of genes that prevent constitutive SKN-1 target activation
SKN-1 is inhibited from functioning constitutively in the intestine through phosphorylation by the IIS pathway kinases and glycogen synthase kinase-3 (GSK-3) [17], [32], but it is otherwise largely unknown how SKN-1 target genes are regulated. To identify mechanisms and cellular functions that limit expression of SKN-1 targets in the absence of stress, we used RNAi to screen for genes that prevent the Phase 2 gene gcs-1 from being active constitutively in the intestine (Figure 1 and Figure S1A) [11]. gcs-1 (γ-Glutamyl-Cysteine Synthetase heavy chain) is rate-limiting for glutathione (GSH) synthesis, and is induced by SKN-1/Nrf proteins in diverse eukaryotes. In the intestine gcs-1 is expressed at low levels under normal conditions, and is upregulated dramatically by oxidative stress [11], [12]. This regulation can be visualized using a reporter in which the gcs-1 promoter drives expression of the green fluorescent protein (GFP) gene (gcs-1p::GFP; Figure S1A).
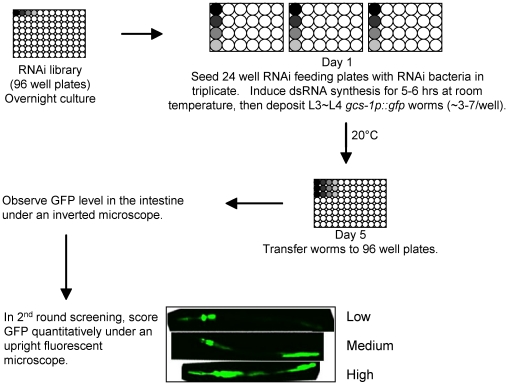
Figure 1. RNAi screening overview.
After overnight culture, RNAi bacteria were seeded onto 24-well RNAi feeding plates in triplicate. dsRNA synthesis was induced for 5–6 hours, then 3–7 L3 or L4 gcs-1p::GFP worms were deposited into each well. After four days growth at 20°C, the triplicate worm samples were transferred to 96 well plates for assessment of the GFP signal in the intestine. Approximately 300 candidates were identified in the first screening round, in which a worm population was assessed rapidly as to whether the intestinal GFP signal was elevated. Those cDNA clones were analyzed in a quantitative second round of screening, in which intestinal GFP signal was scored as High, Medium, or Low as described [11], [17], [32] (Materials and Methods). Genes were scored as positive if gcs-1 upregulation was robust in all three trials in the second round (Figure 2A; Table 1). Four distinct RNAi clones are represented by different shading in individual wells, with the remainder of the plates being arbitrarily left blank.
C. elegans is an advantageous organism for genome-scale RNAi screening, because RNAi can be performed in living animals by feeding [33]. We screened a C. elegans ORFeome library that consists of 11,511 full-length curated cDNA clones, or approximately 57% of the expressed genome (Figure 1) [34]. Two rounds of screening confirmed 37 “positive” genes for which RNAi resulted in robust and consistent expression of gcs-1::GFP in the intestine (Figure 1 and Figure 2A; Table 1). Our screen inevitably missed genes that are associated with developmental defects or modest RNAi-mediated gcs-1 induction, such as akt-1, or -2 [17]. However, it was reassuring that we identified two genes that are involved in GSH production (glutathione reductase: C46F11.2 and the GCS regulatory subunit: E01A2.1, Table 1), because conditions that decrease GSH levels would be expected to upregulate gcs-1 [11], [12]. We also identified wdr-23, which encodes an apparent ubiquitin ligase subunit that binds SKN-1 and may trigger its degradation [35]. Together, these last findings strongly support the validity of our screen.
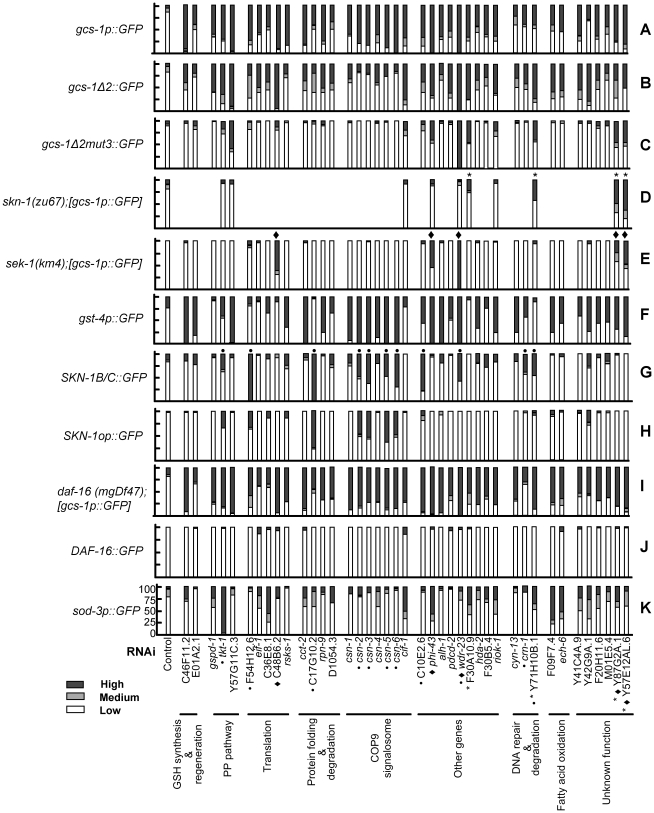
Figure 2. Analysis of genes that prevent constitutive gcs-1 expression.
Confirmed RNAi screening positives and additional COP9 signalosome subunits were examined by RNAi knockdown for effects on the indicated GFP reporters in L4 stage C. elegans. Reporters were scored for levels of nuclear GFP localization (SKN-1B/C::GFP, SKN-1op::GFP, DAF-16::GFP) or GFP expression in the intestine as High, Medium, or Low (Figure 1; Materials and Methods). Percentages of worms in each group were plotted on the Y axis in each panel. In each case a representative example of at least three RNAi experiments is shown (n>30 for each experiment). (A) gcs-1p::GFP expression. (B) Expression of the gcs-1Δ2::GFP reporter, which lacks a SKN-1-independent pharyngeal regulatory sequence and serves as a control for (C) (Figure S1A) [11]. (C) Expression of gcs-1Δ2mut3::GFP, in which an important SKN-1 binding site is mutated (Figure S1A) [11]. (D) gcs-1p::GFP expression in skn-1(zu67) mutants. Two independent transgenic lines each gave similar results. (E) gcs-1p::GFP expression in the sek-1(km4) mutant, in which stress-induced p38 signaling is blocked [41]. (F) Expression of the gst-4p::GFP promoter, a SKN-1 target [12], [40]. (G) Levels of nuclear SKN-1 expressed from SKN-1B/C::GFP, which encodes SKN-1 isoforms b and c (Figure S1B) [11]. (H) Nuclear accumulation of SKN-1 expressed from SKN-1op::GFP, which encodes all three SKN-1 isoforms (Figure S1B) [17]. (I) Expression of gcs-1p::GFP in daf-16(mgDf47) animals. (J) Presence of DAF-16::GFP (Table S6) in intestinal nuclei. (K) Activity of the DAF-16 target sod-3 in the intestine. Black diamonds and asterisks indicate genes for which gcs-1 was induced independently of sek-1 or skn-1, respectively (summarized in Figure 3). Dots indicate genes that were associated with unambiguous accumulation of SKN-1::GFP in intestinal nuclei.
Table 1. List of genes for which RNAi induced gcs-1 expression in the intestine.
| Functional group | Gene ID | Function (NCBI-KOGs description) |
| Glutathione regeneration | C46F11.2 | mitochondrial glutathione reductase |
| Glutathione synthesis | E01A2.1 | Glutamate-cysteine ligase regulatory subunit |
| Pentose phosphate pathway | B0035.5 | gspd-1, Glucose-6-phosphate 1-dehydrogenase |
| Pentose phosphate pathway | F01G10.1 | tkt-1,Transketolase |
| Pentose phosphate pathway | Y57G11C.3 | 6-phosphogluconolactonase - like protein |
| Fatty acid oxidation | F09F7.4 | Enoyl-CoA hydratase |
| Fatty acid oxidation | T05G5.6 | ech-6, Enoyl-CoA hydratase |
| Translation | F54H12.6 | Elongation factor 1 beta/delta chain |
| Translation | T27F7.3b | eif-1, Translation initiation factor 1 (eIF-1/SUI1) |
| Translation | C36E8.1 | RNA polymerase I transcription factor |
| Translation | C48B6.2 | U3 small nucleolar ribonucleoprotein (snoRNP) component |
| Translation | Y47 D3A.16 | rsks-1, Ribosomal protein S6 kinase |
| Protein folding & degradation | T21B10.7 | cct-2, Chaperonin complex component |
| Protein folding & degradation | C17G10.2 | Hsp90 co-chaperone CNS1 (contains TPR repeats) |
| Protein folding & degradation | T06D8.8 | rpn-9, 26S proteasome regulatory complex |
| Protein folding & degradation | D1054.3 | Suppressor of G2 allele of skp1 |
| COP9 signalosome | Y59A8A.1 | csn-1 |
| COP9 signalosome | B0025.2 | csn-2 |
| COP9 signalosome | Y38C1AA.2 | csn-3 |
| COP9 signalosome | Y55F3AM.15 | csn-4 |
| COP9 signalosome | B0547.1 | csn-5 |
| COP9 signalosome | Y67H2A.6 | csn-6 |
| COP9 signalosome | K08F11.3 | cif-1, COP9 Signalosome and eIF3 complex shared subunit |
| DNA repair & degradation | Y116A8C.34 | cyn-13, Cyclophilin-type peptidyl-prolyl cis-trans isomerase |
| DNA repair & degradation | Y47G6A.8 | crn-1, 5′-3′ exonuclease |
| DNA repair & degradation | Y71H10B.1 | IMP-GMP specific 5′-nucleotidase |
| Other genes | C10E2.6 | Monocarboxylate transporter |
| Other genes | K10C2.4 | phi-43, Fumarylacetoacetase |
| Other genes | F54D8.3 | alh-1, Aldehyde dehydrogenase |
| Other genes | R07E5.10 | pdcd-2, mammalian Programmed Cell Death Protein homolog |
| Other genes | D2030.9 | wdr-23, WD40 repeat-containing protein |
| Other genes | F30A10.9 | Predicted nucleic-acid-binding protein, contains PIN domain |
| Other genes | C08B11.2 | hda-2, Histone Deacetylase |
| Other genes | F30B5.4 | Similar to oxidative stress-induced growth inhibitor 2 in H. sapiens |
| Other genes | M01B12.5 | riok-1, Similar to serine/threonine kinase RIO1. |
| Unknown function | Y41C4A.9 | Uncharacterized conserved protein |
| Unknown function | Y42G9A.1 | Unknown function |
| Unknown function | F20H11.6 | Unknown function |
| Unknown function | M01E5.4 | Unknown function |
| Unknown function | Y87G2A.1 | Unknown function |
| Unknown function | Y57E12AL.6 | Unknown function |
The RNAi screen identified 37 of these genes, and four were identified subsequently by virtue of their being COP9 signalosome subunits (csn-2, csn-3, csn-6, cif-1; see text).
Most of the genes we identified are conserved across metazoa (Table 1, not shown), suggesting that the screen is likely to have identified conserved mechanisms that affect Phase 2 gene expression. These genes correspond to a variety of biological processes, including metabolism, mRNA translation, lipid oxidation, DNA degradation and repair, transcription, and protein folding and degradation (Table 1). Three genes (csn-1, csn-4, and csn-5) encode subunits of the COP9 signalosome, a complex that regulates cullin-based ubiquitin ligases by removing the NEDD8 modification from cullins [36]–[38]. Knockdown of the four other C. elegans COP9 signalosome subunits [39], which were not present in our library, also resulted in robust gcs-1 activation (Figure 2A; Table 1). This brought the total number of genes that we analyzed further to 41. Most of our positive genes also influenced expression of the SKN-1 target gene gst-4 (Figure 2F) [12], [40], suggesting that they may broadly affect SKN-1-dependent stress defenses.
Multiple pathways regulate gcs-1 expression
We next investigated how the gcs-1 promoter was activated by RNAi knockdown of the 41 genes identified in the screen. To determine whether SKN-1 was required for gcs-1 induction, we first tested whether RNAi affected expression of a gcs-1 promoter mutant that lacks an important SKN-1 binding site (gcs-1Δ2mut3::GFP)(Figure 2C and Figure S1A) [11]. If this mutated reporter was induced, we examined whether RNAi upregulated gcs-1p::GFP in a skn-1 genetic mutant. For four genes we observed clear skn-1 independent induction of the gcs-1 promoter (F30A10.9: predicted nucleic acid binding protein, Y71H10B.1: IMP-GMP specific 5′-nucleotidase, Y87G2A.1 and Y57E12AL.6: unknown function) (Figure 2D, marked with an asterisk, and Figure 3). Phosphorylation of SKN-1 in response to p38 stress-activated mitogen-activated kinase (MAPK) signaling is generally required for SKN-1 to accumulate in intestinal nuclei and activate target genes [17], [32], [41]. For most of our screening positives, gcs-1 was not induced in animals that lack the MAPK kinase SEK-1, which is essential for p38 signaling [41] (Figure 2E). In contrast, and consistent with a recent study [35], wdr-23 knockdown robustly activated gcs-1p::GFP in the sek-1 null background (Figure 2E, marked with a diamond). This was also true for four other genes (C48B6.2: snoRNP component, phi-43: Fumarylacetoacetase, Y87G2A.1 and Y57E12AL.6: unknown function). Knockdown of the last two genes also activated gcs-1 independently of skn-1. Thus, although intestinal gcs-1 expression is generally SKN-1-dependent, gcs-1 can be induced through pathways that are independent of SKN-1, p38 signaling, or both mechanisms (Figure 3).
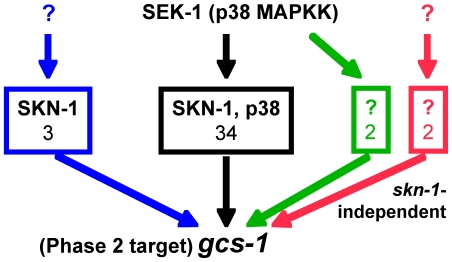
Figure 3. Pathways of gcs-1 activation in the intestine.
RNAi against 34 of the 41 genes we identified in this study resulted in gcs-1 promoter activation through a canonical mechanism that required both skn-1 and p38 MAPK signaling, as illustrated by requirement for the p38 MAPKK SEK-1 (black box)(Figure 2) [41]. In many of these cases the levels of SKN-1::GFP in intestinal nuclei were not dramatically increased (Figure 2), implying that gcs-1 may be activated by SKN-1 through mechanisms besides increasing the overall levels of nuclear SKN-1 (see text). For three genes (C48B6.2: snoRNP component, phi-43 and wdr-23; blue box) RNAi-induced gcs-1 activation required SKN-1, but not p38 MAPK signaling (as revealed by sek-1-independence). For two genes (F30A10.9: nucleic acid binding protein, and Y71H10B.1: IMP-GMP specific 5′-nucleotidase), induction required SEK-1 but not SKN-1 (green box), implying that a different transcription factor was involved. In two cases (Y87G2A.1 and Y57E12AL.6), gcs-1 was activated independently of both SKN-1 and SEK-1 (red box).
Our screen was designed to identify mechanisms that regulate SKN-1 itself, or might influence parallel processes that limit gcs-1 expression. To test whether the genes we identified inhibit nuclear accumulation of SKN-1, we performed RNAi in two strains that carry transgenes in which SKN-1 isoforms are fused to GFP (Figure S1B). Interestingly, only a minority of the genes that regulated gcs-1 through a skn-1-dependent mechanism clearly affected the levels of SKN-1 in intestinal nuclei, including multiple COP9 signalosome subunits, tkt-1, F54H12.6: eEF1, C17G10.2: HSP-90 co-chaperone, C10E2.6: Monocarboxylate Transporter, and wdr-23 (Figure 2G and 2H, marked with a dot). Presumably other genes that regulate gcs-1 in a skn-1-dependent manner act through a mechanism other than simply increasing nuclear SKN-1 levels. We also investigated whether our positives influence daf-16-dependent functions, because DAF-16 regulates many stress defense genes [15], [16]. For each gene, RNAi robustly activated the gcs-1 reporter in a daf-16 null mutant (Figure 2I), confirming our previous finding that gcs-1 is expressed independently of daf-16 [17]. We did not detect robust accumulation of DAF-16::GFP in intestinal nuclei, but for several genes we observed induction of the skn-1-independent DAF-16 target reporter sod-3 (superoxide dismutase), suggesting that some DAF-16-dependent genes were affected (Figure 2J and 2K) [17], [42]. Some genes we identified thus appear to influence stress defense pathways that include targets of both SKN-1 and DAF-16.
Many genes that inhibit gcs-1 expression limit stress resistance
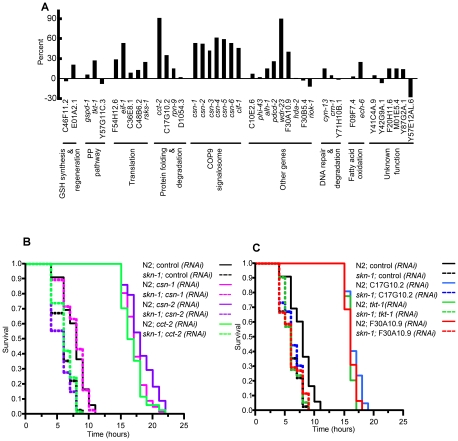
Our screen should identify genes for which RNAi activated the gcs-1 promoter as a consequence of increased oxidative stress, but we also expected to identify regulatory genes and mechanisms that prevent gcs-1 from being expressed constitutively. In the latter case, RNAi knockdown of these genes might increase oxidative stress resistance. Accordingly, for many of the genes we identified, RNAi dramatically enhanced resistance to treatment with the organoperoxide tert-butyl hydrogen peroxide (TBHP) (Figure 4A and Figure S2; Table S1). In addition to wdr-23, which has been implicated in stress resistance [35], robust effects were observed for many genes involved in translation, protein folding or degradation, and the COP9 signalosome. We observed comparable increases in TBHP resistance when a group of these genes was analyzed in a daf-16 null mutant, indicating that daf-16 is not required (Figure S2 and Figure S3; Table S2). We next asked whether a set of genes that had the greatest effects on stress resistance in N2 and daf-16 animals could promote stress resistance in a skn-1 mutant. In each case, RNAi largely failed to increase oxidative stress resistance when skn-1 was lacking (Figure 4B and 4C; Table S3; see below). We conclude that many of the genes we identified are involved in mechanisms that limit oxidative stress resistance by modulating activity of SKN-1-dependent stress responses.
Figure 4. Effects of gcs-1-regulatory genes on stress resistance.
(A) Effects on tert-butyl hydrogen peroxide (TBHP) resistance in wild-type (N2) animals. L4 worms were placed on RNAi or control bacteria for three days at 20°C, then transferred to assay plates containing a lawn of OP50 and 9.125 mM TBHP. Survival was then scored over a time-course. A bar graph shows the percent change in mean survival time for each gene compared to control RNAi. Representative experiments are shown here and plotted in Figure S2. Where only 2 experiments were performed, the experiment in which RNAi gave the less robust effect is graphed. Results of individual experiments, numbers of worms analyzed, and statistical analyses are presented in Table S1. (B, C) Effects on TBHP resistance in skn-1(zu67) animals. RNAi assays of selected genes were performed and analyzed as in (A), but using 15.4 mM TBHP. Representative experiments are presented as plots of proportional survival over time, with results and statistical analysis of individual experiments provided in Table S3. Note that in each case, the increase in stress resistance deriving from RNAi of these genes was almost completely dependent upon skn-1.
Translation inhibition induces a transcriptional stress response that involves SKN-1
It was intriguing that five of our initial screening positives are involved in mRNA translation (Table 1), because several studies have reported that C. elegans lifespan and stress resistance are increased when genes that encode general translation factors or ribosomal proteins are inhibited by RNAi during adulthood [22], [23], [26], [28]–[31], [43]. Those longevity genes included two that we identified in our screen: the initiation factor eIF1 (eif-1) and the S6K ortholog rsks-1 (Table 1). Our findings suggested that interference with mRNA translation might result in induction of SKN-1-dependent stress responses, and that SKN-1 might be involved in the stress resistance and lifespan extensions that derive from reduced translation. Accordingly, although our screen identified many interesting genes and candidate mechanisms that influence SKN-1-dependent stress responses, we directed our further efforts towards investigating the relationship between mRNA translation and SKN-1 function.
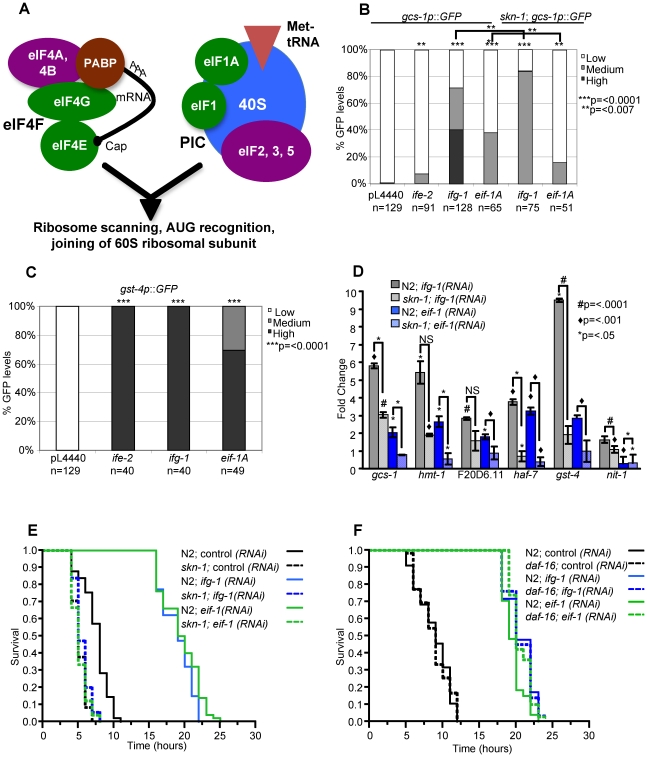
We focused our analyses of translation on initiation factors because their lifespan phenotypes have been examined extensively, and because some studies indicated that their effects on lifespan involve DAF-16, which is inhibited by IIS in parallel to SKN-1 and may have some overlapping functions with SKN-1 (see Introduction). It is well established that mutation or adulthood RNAi knockdown of either eIF4G (IFG-1) or the somatically-expressed eIF4E isoform IFE-2 results in decreased protein synthesis, and increased lifespan and stress resistance [22], [23], [26], [31]. These longevity extensions appear to occur independently of any effects of translation inhibition on fecundity [23], [26]. Each of these factors is a subunit of the eIF4F complex, which circularizes and translationally activates mRNAs by linking their 5′ cap to poly-A-binding protein (Figure 5A) [44]. The eIF4F complex promotes binding of mRNA by the translation pre-initiation complex (PIC), which includes the 40S ribosomal subunit, a different set of initiation factors, and the methionyl tRNA that mediates initiation (Figure 5A) [44]. Here we have further examined stress and lifespan phenotypes associated with the eIF4F components IFE-2 and IFG-1, along with EIF-1 (Table 1) and EIF-1A (H06H21.3). The latter two factors are components of the PIC, and are each involved in ribosome scanning and translation start codon selection [44]. Our experiments therefore investigated the effects of impairing two distinct mechanisms involved in translation initiation (Figure 5A).
Figure 5. Induction of SKN-1–dependent target gene expression and stress resistance in response to translation initiation factor RNAi.
(A) Translation initiation factors that were examined in this study. The eIF4F complex stabilizes capped mRNAs and activates them for translation by interacting with their 5′ cap and poly-A-binding protein (PABP) [44]. This interaction promotes binding of these mRNAs by the translation pre-initiation complex (PIC), which includes the 40 S ribosome subunit and the initiator tRNA. Subsequent steps in initiation follow this binding event. Initiation factors that we examined in this study are shown in green. (B) Activation of the gcs-1p::GFP reporter. N2 or skn-1(zu67) worms that carry the gcs-1p::GFP reporter were exposed to the indicated RNAi or control bacteria beginning at the L2 stage. They were scored for GFP fluorescence at day one of adulthood as in Figure 2, at which time the worms appeared normal and were laying eggs that hatched. p-values indicated above individual bars correspond to comparison with control RNAi. Similar reporter induction was observed after three days of initiation factor RNAi that began at adulthood day one, and no reporter activity was observed when control RNAi was performed in skn-1(zu67) animals (not shown). p values were calculated by the Chi2 method. (C) Activation of the gst-4 reporter, scored as in (B). (D) Induction of endogenous SKN-1 target gene expression in response to translation initiation factor RNAi, analyzed by quantitative RT-PCR (qRT-PCR) performed in triplicate. RNAi was performed as in (B). Each gene assayed is upregulated under stress conditions [12]. A representative experiment is shown, in which Fold Change and p-values above individual bars refer to comparison to control RNAi. Additional qRT-PCR experiments and statistical analyses are described in Table S4. (E) Induction of skn-1-dependent stress resistance. After exposure to the indicated RNAi bacteria as in Figure 4, N2 or skn-1(zu67) worms were placed on plates containing 15.4 mM TBHP, then scored for survival over time. In each case, the worms appeared normal and were laying eggs when they were transferred to TBHP plates. In N2 but not skn-1 mutant worms, stress resistance was dramatically enhanced by prior exposure to translation initiation factor RNAi. All experiments and statistics are provided in Table S3. (F) Comparison of TBHP resistance in N2 and daf-16 mutant worms, performed and analyzed as in (E). daf-16 was not required for the increases in oxidative resistance that derive from translation initiation factor RNAi.
We first investigated whether, in general, RNAi knockdown of translation initiation factors activates SKN-1-dependent stress responses. Initially we examined how initiation factor RNAi affected SKN-1 target gene promoter activity, as in the eif-1(PIC) analyses performed for our screen (Figure 2). Transcription from the transgenic gcs-1 promoter was induced robustly by ifg-1(eIF4F) RNAi, and modestly by RNAi against ife-2(eIF4F) or eif-1A(PIC) (Figure 5B). This gcs-1 induction was partially dependent upon skn-1 (Figure 5B). In each case, translation factor RNAi also strongly activated the well-characterized SKN-1 target promoter gst-4 (Figure 5C). We also analyzed effects on endogenous SKN-1-regulated gene expression, focusing on one factor each from eIF4F and the PIC (IFG-1 and EIF-1, respectively). We assayed mRNA production from two genes that are skn-1-dependent under both normal and oxidative stress conditions (gst-4 and nit-1), along with other genes that are upregulated by SKN-1 in response to stress [12] (Figure 5D). Importantly, RNAi against either of these initiation factors dramatically increased expression of multiple endogenous SKN-1-regulated genes (Figure 5D; Table S4). When RNAi was performed in a skn-1 mutant, this induction was much less robust or did not occur at all (Figure 5D; Table S4). We conclude that impairment of either of these two translation initiation complexes results in transcription-mediated stress responses in which SKN-1 plays a critical role.
We next examined how translation initiation factor RNAi affects oxidative stress resistance. RNAi against either ifg-1(eIF4F) or eif-1(PIC) dramatically increased TBHP resistance in either wild type or daf-16 mutant animals (Figure 5E and 5F; Table S3). In contrast, these increases in stress resistance were essentially abolished in a skn-1 mutant (Figure 5E and Table S3). Similar results were obtained in analyses of ife-2(eIF4F) and eif-1A(PIC) RNAi (Table S3). We conclude that the dramatic increase in SKN-1 target gene transcription that occurs after translation initiation factor RNAi results in oxidative stress resistance that depends upon skn-1, but not daf-16.
Translation inhibition extends lifespan through daf-16- and skn-1–dependent mechanisms
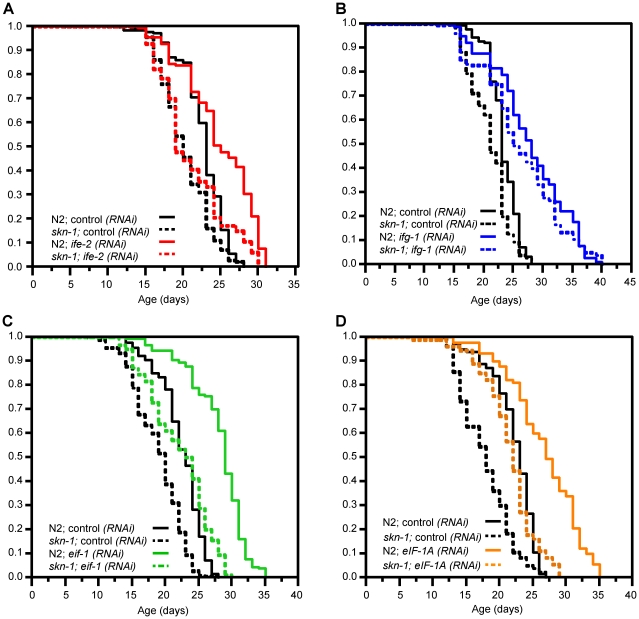
To investigate whether skn-1 contributes to the longevity increases that derive from RNAi knockdown of these translation initiation factors, we compared the effects of performing RNAi in the wild type strain N2, and two skn-1 loss-of-function mutants (skn-1(zu135) and skn-1(zu67)). ife-2(eIF4F) RNAi did not consistently extend lifespan in these skn-1 mutants, in contrast to results obtained in wild type animals (Figure 6A; Table 2 and Table S5). The mean lifespan associated with ifg-1(eIF4F) RNAi was only slightly reduced by lack of SKN-1, however (Figure 6B; Table 2 and Table S5). RNAi against either eif-1(PIC) or eif-1A(PIC) increased the mean lifespan of N2 worms to approximately the extent observed for ifg-1(eIF4F) RNAi (Figure 6C and 6D; Table 2 and Table S5). Lifespan was also increased when these last two genes were knocked down in skn-1 mutants, but in each case the mean lifespan was markedly shorter than when the corresponding RNA was performed in N2 animals (Figure 6C and 6D; Table 2 and Table S5). In addition, the percent increase in mean lifespan associated with eif-1(PIC) RNAi was reduced in skn-1(zu135) mutants compared to N2 (12% vs. 26%, Table 2). The extent to which skn-1 is required for RNAi-associated lifespan extension thus varies among these initiation factor genes.
Figure 6. Importance of SKN-1 for lifespan extension deriving from translation initiation factor RNAi.
(A) Survival plot showing effects of ife-2(eIF4F) RNAi. This lifespan extension was greatly reduced by the skn-1(zu135) mutation, which was used in all experiments in this figure. (B) Longevity extension by ifg-1(eIF4F) RNAi. Survival of ifg-1(RNAi) worms was not substantially decreased by skn-1 mutation. (C,D) Longevity extension by eif-1(PIC) and eif-1A(PIC) RNAi. The longevity associated with inhibiting these genes was decreased but not eliminated by skn-1 mutation. Note that overall survival of these RNAi animals is nevertheless significantly impaired in the skn-1 background compared to N2. All longevity analyses were performed at 20°C, with lifespans measured from hatching and RNAi initiated at day 1 of adulthood. Each panel shows a composite of multiple experiments in which the populations shown were analyzed in parallel, with the proportion surviving indicated on the y-axis. These data are summarized in Table 2, with individual experiments described in Table S5.
Table 2. Lifespan analyses.
| Strain | Mean Lifespan ± SEM 20° (days) | Median Lifespan | 75th Percentile 20°C (days) | p value (log-rank) against Control | % Lifespan Extension | N2 | No. of Exp. | Figure |
| N2; control(RNAi) | 22.64±0.2 | 23 | 25 | - | 264/320 | 3 | 6A | |
| skn-1(zu135); control(RNAi) | 20.29±0.2 | 20 | 23 | - | 348/382 | 3 | 6A | |
| N2; ife-2(RNAi) | 24.86±0.4 | 25 | 29 | <.0001a | 10 | 165/181 | 3 | 6A |
| skn-1(zu135); ife-2(RNAi) | 21.11±0.4 | 19 | 24 | <.0003b | 4 | 153/173 | 3 | 6A |
| N2; control(RNAi) | 22.30±0.2 | 23 | 25 | - | 225/271 | 4 | 6B | |
| skn-1(zu135); control(RNAi) | 21.02±0.2 | 21 | 23 | - | 236/260 | 3 | 6B | |
| N2; ifg-1(RNAi) | 27.91±0.5 | 28 | 33 | <.0001a | 25 | 186/242 | 4 | 6B |
| skn-1(zu135); ifg-1(RNAi) | 26.21±0.5 | 25 | 32 | <.0001b | 25 | 175/204 | 3 | 6B |
| N2; control(RNAi) | 22.43±0.2 | 23 | 25 | - | 280/296 | 5 | 6C | |
| skn-1(zu135); control(RNAi) | 19.87±0.3 | 20 | 22 | - | 211/220 | 4 | 6C | |
| N2; eif-1(RNAi) | 28.16±0.2 | 29 | 31 | <.0001a | 26 | 285/298 | 5 | 6C |
| skn-1(zu135); eif-1(RNAi) | 22.18±0.3 | 23 | 26 | <.0001b | 12 | 207/210 | 4 | 6C |
| N2; control(RNAi) | 22.28±0.3 | 23 | 25 | - | 120/122 | 2 | 6D | |
| skn-1(zu135); control(RNAi) | 17.94±0.4 | 18 | 21 | - | 97/100 | 2 | 6D | |
| N2; eif-1A(RNAi) | 26.84±0.5 | 27 | 31 | <.0001a | 20 | 130/140 | 2 | 6D |
| skn-1(zu135); eif-1A(RNAi) | 21.62±0.4 | 22 | 24 | <.0001b | 21 | 106/107 | 2 | 6D |
| skn-1(zu67); control(RNAi) | 18.33±0.4 | 17 | 22 | - | 111/122 | 2 | N.A. | |
| skn-1(zu67); ife-2(RNAi) | 18.31±0.4 | 17 | 23 | 0.915 c | 0 | 123/126 | 2 | N.A. |
| skn-1(zu67); ifg-1(RNAi) | 25.88±0.6 | 26 | 31 | <.0001 c | 41 | 101/111 | 2 | N.A. |
| N2; control(RNAi) | 21.40±0.3 | 21 | 23 | - | 99/100 | 2 | N.A. | |
| skn-1(zu67); control(RNAi) | 17.68±0.3 | 17 | 21 | - | 209/221 | 4 | N.A. | |
| N2; eif-1(RNAi) | 26.23±0.6 | 27 | 30 | <.0001a | 23 | 105/112 | 2 | N.A. |
| skn-1(zu67); eif-1(RNAi) | 20.42±0.4 | 19 | 24 | <.0001c | 15 | 183/201 | 4 | N.A. |
| N2; control(RNAi) | 22.51±0.3 | 22 | 24 | - | 65/70 | 1 | 7B | |
| daf-16(mgDf47); control(RNAi) | 19.74±0.4 | 20 | 23 | - | 113/116 | 2 | 7B | |
| daf-16(mgDf47); ife-2(RNAi) | 19.77±0.4 | 20 | 24 | 0.4388d | 0 | 125/131 | 2 | 7B |
| N2; control(RNAi) | 23.55±0.2 | 24 | 25 | - | 130/135 | 2 | 7C, 7E | |
| daf-16(mgDf47); control(RNAi) | 21.07±0.3 | 22 | 24 | - | 168/171 | 3 | 7C | |
| daf-16(mgDf47);skn-1(zu67); control(RNAi) | 16.91±0.3 | 17 | 19 | - | 141/160 | 3 | 7E | |
| daf-16(mgDf47); ifg-1(RNAi) | 23.61±0.4 | 25 | 28 | <.0001d | 12 | 165/168 | 3 | 7C |
| daf-16(mgDf47); skn-1(zu67) ifg-1(RNAi) | 18.72±0.4 | 18 | 23 | <.0001e | 11 | 154/167 | 3 | 7E |
| N2; control(RNAi) | 22.62±0.2 | 24 | 25 | - | 229/235 | 4 | 7D, 7F | |
| daf-16(mgDf47); control(RNAi) | 18.88±0.2 | 19 | 21 | - | 275/276 | 5 | 7D | |
| daf-16(mgDf47);skn-1(zu67); control(RNAi) | 16.16±0.2 | 16 | 19 | - | 194/206 | 4 | 7F | |
| daf-16(mgDf47); eif-1(RNAi) | 20.91±0.3 | 21 | 24 | <.0001d | 11 | 279/289 | 5 | 7D |
| daf-16(mgDf47);skn-1(zu67); eif-1(RNAi) | 16.42±0.2 | 16 | 18 | 0.2232e | 2 | 193/204 | 4 | 7F |
| N2; control(RNAi) – glc | 24.40±0.2 | 25 | 26 | - | 122/126 | 2 | 7A | |
| N2; control(RNAi) + glc | 20.19±0.2 | 20 | 22 | <.0001f | −17 | 178/178 | 3 | 7A |
| N2; ife-2(RNAi) + glc | 19.96±0.2 | 20 | 22 | 0.2536g | −1 | 188/191 | 3 | 7A |
| N2; ifg-1(RNAi) + glc | 19.49±0.2 | 19 | 22 | 0.2941g | −3 | 109/110 | 3 | 7A |
| N2; eif-1(RNAi) + glc | 19.85±0.2 | 20 | 22 | 0.3771g | −2 | 176/176 | 3 | 7A |
These combined results were derived from individual experiments that are described in Table S5. RNAi experiments are grouped and graphed in the indicated figures with controls that were performed in parallel. Lifespan extensions correspond to parallel control RNAi experiments. Numbers of animals are indicated as the total assayed (minus exclusions) over the total at the start of the experiment. p values refer to the following pL4440 RNAi controls: N2a, skn-1(zu135) b, skn-1(zu67) c, daf-16(mgDf47) d, daf-16(mgDf47); skn-1(zu67) e, N2-glcf, N2+glcg. glc means glucose.
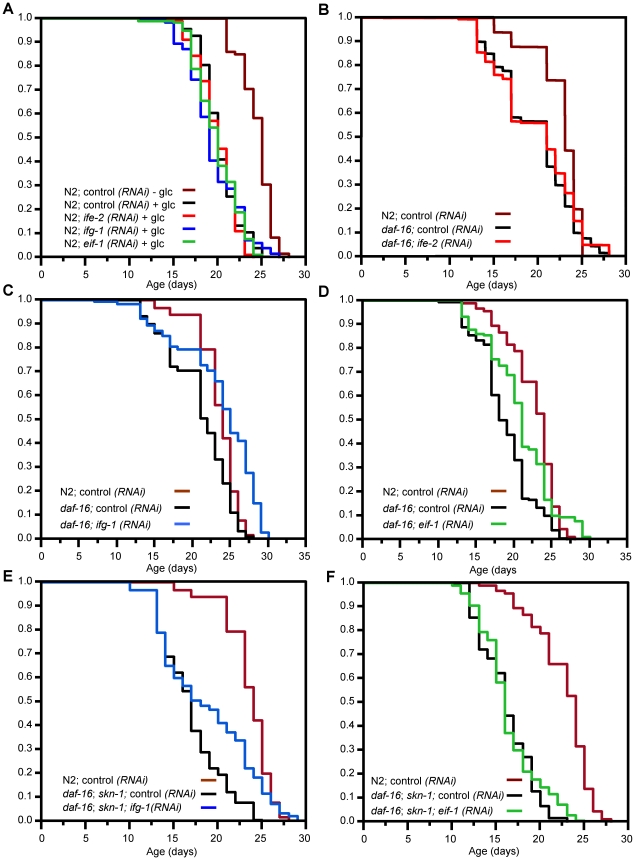
As noted in the Introduction, results among laboratories differ with respect to whether daf-16 is needed for the lifespan extensions associated with translation initiation factor RNAi. While these apparent discrepancies might derive simply from differences in experimental conditions, this is an important question to explore further. If daf-16 is not involved in these effects, for example, it would suggest that translational suppression affects lifespan through novel mechanisms that do not intersect with the IIS pathway [26]. Our finding that SKN-1 contributes to these lifespan extensions in some cases suggests that those increases might involve DAF-16 and SKN-1 acting together. To test this idea, we first investigated whether these lifespan extensions occur under conditions of glucose feeding. In C. elegans glucose feeding inhibits DAF-16 by increasing IIS pathway activity, and thereby largely prevents mutations that reduce IIS from extending C. elegans lifespan [45], [46]. Given that IIS inhibits both DAF-16 and SKN-1 [17], glucose feeding should also reduce SKN-1 function in parallel to DAF-16, and therefore should block the pro-longevity effects of translation initiation factor RNAi if these two transcription factors are required. Consistent with published findings [46], we found that glucose feeding shortened the lifespan of N2 worms that were fed control RNAi (Figure 7A; Table 2). Importantly, the mean lifespans of ife-2(RNAi)(eIF4F), ifg-1(RNAi)(eIF4F), and eif-1(RNAi)(PIC) animals were even more dramatically decreased by glucose feeding, which prevented RNAi from extending lifespan in each case (Figure 7A; Table 2). This last finding suggests that the longevity benefits of inhibiting translation initiation are abrogated by upregulation of IIS signaling, supporting the idea that they may be largely dependent upon SKN-1 and DAF-16.
Figure 7. Lifespan extension in response to translation initiation factor RNAi is mediated primarily by DAF-16 and SKN-1.
(A) Survival after exposure to translation initiation factor RNAi and 2% glucose. Glucose feeding increases IIS, which inhibits both DAF-16 and SKN-1 [17], [45]. Lifespan extension by translation factor inhibition is eliminated under these conditions. (B) ife-2(eIF4F) RNAi fails to extend lifespan in a daf-16(mgDf47) mutant. (C, D) Modest lifespan extension in response to ifg-1(eIF4F) or eif-1(PIC) RNAi in daf-16(mgDf47) animals. Note that survival of these RNAi animals is impaired in the daf-16 background compared to N2 (Figure 6B and 6C). (E, F) Survival of daf-16(mgDf47); skn-1(zu67) double mutants exposed to ifg-1(eIF4F) or eif-1(PIC) RNAi. In this genetic background ifg-1(eIF4F) RNAi extends lifespan exclusively among the longest-lived worms, and eif-1(PIC) RNAi has only a negligible effect. Experiments were performed and plotted as in Figure 6 and are summarized in Table 2, with individual experiments described in Table S5.
To test the above model directly, we investigated how the lifespan extensions associated with RNAi against ife-2(eIF4F), ifg-1(eIF4F), and eif-1(PIC) are affected by mutation of skn-1 and daf-16, either individually or simultaneously. Under our conditions the prolongevity effect of ife-2(eIF4F) RNAi was essentially prevented by mutation of daf-16 (Figure 7B; Table 2), consistent with a previous report [23]. Knockdown of either ifg-1(eIF4F) or eif-1(PIC) extended the mean lifespan of a daf-16 mutant, but to a lesser extent than was characteristic of N2 animals (Figure 7C and 7D; Table 2). This result is largely consistent with previous evidence that the ifg-1(RNAi) lifespan extension requires daf-16 [23], [28]. A homozygous daf-16; skn-1 double mutant develops into adults that appear normal, but are characterized by a slightly reduced lifespan compared to either single mutant allele (Table 2). In this double mutant ifg-1(eIF4F) RNAi resulted in a modest lifespan increase that was confined largely to the longest-lived animals (compare median and 75%-ile lifespans; Figure 7E; Table 2). While mutation of skn-1 on its own affected the mean lifespan of ifg-1(RNAi) animals only minimally (Figure 6B; Table 2), in the daf-16 mutant concurrent loss of skn-1 activity decreased ifg-1(RNAi) mean lifespan and altered the shape of the survival curve (compare results obtained in daf-16 and daf-16; skn-1 mutants; Figure 7C and 7F; Table 2). This suggests that lack of daf-16 not only blunted the beneficial effects of ifg-1(eIF4F) RNAi, but also sensitized these animals to lack of skn-1. Importantly, eif-1(PIC) RNAi essentially failed to extend lifespan in the daf-16; skn-1 double mutant (Figure 7F; Table 2). When our individual trials were combined, the average percent increase in lifespan associated with eif-1(PIC) RNAi in the daf-16; skn-1 double mutant (2%) was significantly different from that seen in N2 (p = .0028, Student's t-test), skn-1(zu67)(p = .0937), or daf-16(mgDf47)(p = .0469)(Table 2 and S5). These analyses indicated that DAF-16 and SKN-1 each contributed significantly to the lifespan extension associated with eif-1(PIC) RNAi. Together, our findings provide strong support for the idea that DAF-16 is critical for the effects of translation initiation factor RNAi on lifespan, and additionally indicate that for some factors SKN-1 plays an important and possibly overlapping role.
Discussion
Various biological processes limit the activity of SKN-1/Nrf–dependent stress defenses
In this study we have used RNAi screening to identify mechanisms that prevent SKN-1-dependent stress response genes from being expressed inappropriately under normal conditions. The list of genes we detected is not complete, because we screened only about 60% of the expressed genome and employed stringent criteria for positive selection, and would have missed genes that prevented development. Nevertheless, our screen revealed that a variety of mechanisms and biological processes influence activity of SKN-1 and its target genes. For example, the proportion of these genes for which RNAi clearly increased the levels of SKN-1::GFP in intestinal nuclei was surprisingly low (about 25%, Figure 2). These particular positives could regulate SKN-1 itself, or processes that influence SKN-1 directly. Importantly, most of the remaining positives affected gcs-1 activity in a skn-1-dependent manner, suggesting that they influence mechanisms that act on target genes in parallel to SKN-1, or might modify SKN-1 to increase its activity but not its concentration in the nucleus. We also identified genes that affect gcs-1 expression independently of skn-1, p38 signaling, or both mechanisms (Figure 2 and Figure 3). In other species, considerable attention has been focused on regulation of the SKN-1 ortholog Nrf2 by the ubiquitin ligase and possible redox sensor Keap1, which targets Nrf2 for degradation in the absence of stress [9], [10], [47]. However, the related mammalian proteins Nrf1 and Nrf3 do not appear to be regulated by Keap1, a sequence ortholog of which seems to be lacking in C. elegans. Our results predict that Nrf proteins and their target genes, like SKN-1 and gcs-1, are likely to be regulated by a complex web of cellular processes and signaling pathways.
Many of the genes we identified are involved in metabolic processes (Table 1), which is not surprising given that SKN-1-regulated genes defend against stress deriving from excess levels of ROS or other reactive compounds [12]. For example, we identified several genes in the pentose phosphate pathway, which produces the critical reductant NADPH. Lack of PHI-43, which catalyzes the last step in tyrosine degradation, results in lethality that derives from accumulation of toxic tyrosine metabolites [48]. Similarly, monocarboxylate transporters (C10E2.6) prevent excessive accumulation of small molecules such as pyruvate, lactate, and ketone bodies. SKN-1 regulates numerous genes under normal conditions, and responds to stresses by inducing overlapping sets of stress defense genes [12]. Several of the genes we identified in this screen are themselves upregulated transcriptionally by SKN-1 (C46L11.2-glutathione reductase, E01A2.1-GCS regulatory subunit, phi-43, alh-1, rpn-9, Table 1) [12], suggesting that SKN-1 is involved in homeostatic feedback regulation of various cellular processes. It may be important to regulate SKN-1 target gene activity tightly for many reasons: metabolite levels profoundly influence metabolism, IIS and other signaling pathways are affected by redox conditions [49], and excessive GSH could upset protein folding by inhibiting disulfide bond formation [50].
Several of our screening positives are involved in protein folding or degradation, many of which affected SKN-1 nuclear accumulation. WDR-23 appears to target SKN-1 directly for degradation (Table 1) [35]. This would seem to provide a model for how SKN-1 could be affected by the COP9 signalosome, which sustains cullin activity [36]–[38]. However, p38 signaling is required for gcs-1 to be upregulated by loss of COP9 signalosome genes, in contrast to wdr-23, suggesting that the COP9 signalosome regulates SKN-1 at a different step (Figure 2E) [35]. We observed particularly strong effects on SKN-1 nuclear accumulation after knockdown of an HSP-90 co-chaperone (C17G10.2)(Figure 2G and 2H), but knockdown of the chaperonin cct-2 and the proteasome lid subunit rpn-9 upregulated SKN-1 target genes in a skn-1-dependent manner without detectably increasing the presence of SKN-1 in nuclei (Figure 2). These various genes associated with protein homeostasis thus may influence SKN-1 target gene expression through multiple pathways. RNAi against a set of these genes increased oxidative stress resistance in a manner that was almost completely dependent upon skn-1 (csn-1, csn-2, cct-2, C17G10.2; Figure 4B and 4C; Table S3). Previous studies have shown that SKN-1 upregulates many proteasomal and other genes involved in protein turnover, including rpn-9 [12], and that knockdown of several other proteasome or chaperonin subunits results in skn-1-dependent gst-4 induction, or SKN-1 nuclear accumulation [40]. Perhaps SKN-1 helps maintain the proteasome and other mechanisms that promote protein homeostasis. The SKN-1 ortholog Nrf1 is required for inducible upregulation of proteasome genes in mouse fibroblasts [51], suggesting that this might be a conserved function of SKN-1/Nrf proteins.
SKN-1 mediates effects of translation inhibition on stress resistance and longevity
Having identified screening positives that are involved in mRNA translation or ribosome function, including two known longevity genes (eif-1(PIC) and rsks-1, Table 1) [22], [23], [31], we investigated whether SKN-1 contributes to the increases in stress resistance and lifespan that derive from inhibiting translation initiation. The dramatic increases in oxidative stress resistance that accompanied translation initiation factor RNAi did not require daf-16 but were eliminated in a skn-1 mutant (Figure 5E and 5F; Table S3), indicating that SKN-1 plays a critical role in the effects of translation inhibition on stress resistance. In contrast, the increases in lifespan that derive from inhibiting translation initiation seemed to depend largely upon the activity of both daf-16 and skn-1 (Table 2). DAF-16 on its own contributed to these increases for each gene that we analyzed (Table 2), consistent with several previous analyses of the effects of translation on aging (see Introduction). SKN-1 was less critical than DAF-16 for these longevity benefits, but nevertheless still played an important role. Most notably, SKN-1 contributed to the percent longevity increases associated with knockdown of ife-2 and eif-1, and SKN-1 and DAF-16 together mediated the longevity increase that derived from eif-1 knockdown, which was essentially eliminated in daf-16; skn-1 double mutants (Table 2). Glucose feeding had a similar effect for each gene we examined, also consistent with DAF-16 and SKN-1 being important. Previous studies have disagreed with respect to the importance of daf-16 for longevity deriving from inhibiting translation initiation (see Introduction), possibly because of differences in experimental conditions. By analyzing requirements for both DAF-16 and SKN-1, we have obtained strong support for the notion that these longevity benefits are mediated through effects on specific regulatory pathways, and not simply through reducing the consumption of resources by protein synthesis.
Notably, these effects of translation inhibition do not simply require these transcription factors to be present and functioning as they would under normal conditions, but also involve induction of stress response gene transcription. RNAi knockdown of each initiation factor we examined led to activation of SKN-1 target gene promoters (ife-2(eIF4F), ifg-1(eIF4F), eif-1(PIC), and eif-1A(PIC); Figure 2A, Figure 5B and 5C), and knockdown of either ifg-1(eIF4F) or eif-1(PIC) dramatically upregulated transcription of endogenous SKN-1 target genes (Figure 5D). These gene induction events were largely but not completely dependent upon skn-1 (Figure 5B and 5D), suggesting that SKN-1 and additional stress defense regulators are involved. Consistent with this idea, knockdown of the initiation factor eIF2Bδ was reported to increase expression of a set of stress response genes, an effect that was partially dependent upon daf-16 [29]. RNAi against translation initiation factors could potentially increase DAF-16 and SKN-1 activity simply by inhibiting IIS. However, SKN-1 accumulates in intestinal nuclei when IIS is decreased [17], and this did not occur after RNAi against the initiation factor genes we studied (Figure 2G and 2H; not shown). This suggests that translation initiation may not induce skn-1-dependent gene expression simply by reducing IIS or promoting nuclear accumulation of SKN-1, and instead may affect signaling or transcription pathways that function synergistically with SKN-1. Elucidation of these pathways may ultimately reveal why the requirements for daf-16 and skn-1 for longevity extension varied among the translation factors we examined (Table 2).
Several lines of evidence indicate that suppression of translation is important for the longevity extensions associated with reductions in TOR signaling, and possibly DR [19], [52]. In Drosophila, S6K downregulation and 4E-BP are required for Drosophila lifespan to be extended by treatment with rapamycin, which inhibits the TORC1 form of TOR kinase [20]. Furthermore, DR extension of Drosophila lifespan involves an increase in 4E-BP activity, which allows mitochondrial genes to be translated preferentially by virtue of their shorter 5′ untranslated regions [21]. If the latter mechanism is conserved in C. elegans, our results predict that reductions in translation would trigger this pathway in addition to the transcriptional effects we have described. We observed some remaining longevity extension associated with ifg-1(eIF4F) RNAi when both skn-1 and daf-16 were lacking, implying that an additional longevity-promoting mechanism was activated (Figure 7E; Table 2). In C. elegans the lifespan extensions associated with TOR inhibition require the PHA-4 transcription factor, but TOR, S6K, and ribosomal proteins appear to modulate lifespan independently of DAF-16, suggesting that multiple overlapping processes might be involved [22], [23], [53], [54]. One intriguing possibility is that the SKN-1-dependent transcriptional response we have observed here is induced as a consequence of particular genes being translated preferentially. It will be interesting to determine whether this transcriptional response is associated with other situations where translation is reduced, including DR.
Why would interference with translation initiation direct SKN-1 and DAF-16 to enhance stress resistance and longevity? Translation is reduced in response to nutrient deprivation, a condition under which it is presumably adaptive to mobilize mechanisms that promote stress resistance and survival. It might be beneficial to activate SKN-1-dependent antioxidant defenses simply because protein synthesis is reduced, since the highly reactive sulfur within methionine residues in cellular proteins may be an important protective antioxidant [55]. Alternatively, a reduction in protein synthesis might perturb metabolic processes so that reactive metabolites accumulate, making it helpful to increase the activity of small molecule detoxification mechanisms [12]. In addition, at least 30% of nascent polypeptides are normally degraded co-translationally by the proteasome because of inefficient folding or translation errors [56], [57]. If interference with translation initiation increased the fraction of polypeptides that were subject to degradation, upregulation of SKN-1 target genes involved in protein homeostasis could be adaptive. DAF-16-dependent processes are also likely to be beneficial for coping with translation perturbation, because when DAF-16 activity is very high C. elegans larvae enter a diapause state in which metabolic needs are sharply reduced, and stress resistance is elevated [58]. On the other hand, it may be advantageous to hold SKN-1- and DAF-16-dependent oxidative stress defenses in check under growth conditions, when IIS and translation rates are higher, because phosphatases that inhibit IIS are themselves inhibited by oxidation [49]. Irrespective of the biological rationale, our results show that interference with translation initiation triggers mechanisms that stimulate SKN-1-dependent transcription of stress defense genes, making it of considerable importance to identify those mechanisms.
Materials and Methods
C. elegans strains
Unless otherwise indicated, worms were cultured at 20°C on NGM plates that were seeded with a lawn of E. coli strain OP50-1 (Caenorhabditis genetics center). The C. elegans strains used are described in Table S6. The Ex003[gcs-1p::GFP] transgenic array expresses GFP driven by the gcs-1 promoter (Figure S1) [11]. Strains in which this array was integrated into the genome were generated by UV treatment using a Stratalinker 2400 (Stratagene) set at 400 (×100 µJoules). Three independent gcs-1p::GFP integrated lines were generated, two of which were crossed into skn-1(zu67) to create strains LD1173 and LD1175. The extrachromosomal Ex003[gcs-1p::gfp] array was introduced into daf-16 and sek-1 mutant backgrounds by crossing. The following mutants were used:
skn-1(zu135)
A nonsense mutation that would prevent DNA binding by all three SKN-1 isoforms [59].
skn-1(zu67)
A nonsense mutation that lies within the coding regions of SKN-1a and SKN-1c but not SKN-1b (Figure S1), but is associated with all known skn-1 phenotypes [11], [17], [18], [59].
daf-16(mgDf47)
A deficiency that removes daf-16 coding regions [60].
sek-1(km4)
A putative null mutation that removes most of the coding region [41].
Genome-scale RNAi screening
The C.elegans orfeome RNAi library that was screened consists of 11,511 distinct genes [34]. Screening RNAi was performed in 24 well plates in which NGM agar was supplemented with 50 µg/mL carbenicillin and 2 mM IPTG (Isopropyl β-D-1-thiogalactopyranoside)(Figure 1). Unseeded plates were stored in the cold room for < one week. RNAi bacteria were expanded in 96-well flat bottom blocks (QIAGEN, Valencia, CA) overnight at 37°C in 600 µL LB with 50 µg/mL carbenicillin. After seeding of individual bacterial clones, the RNAi plates were dried in a laminar flow hood and left at room temperature for 5–6 hours to induce dsRNA synthesis, then L3 or early L4 stage gcs-1p::GFP worms were deposited into each well (Day 1). After incubation for 4 days at 20°C (Day 5), worms were washed off with M9 containing 6 mM sodium azide (for immobilization), then transferred to 96-well black clear bottom plates (Corning) for observation under an inverted fluorescent microscope. This low azide concentration did not affect gcs-1p::GFP expression (not shown). For this 1st round screen, RNAi for each gene was performed in triplicate. Clones were scored as positive if gcs-1p::GFP upregulation was unambiguously observed in at least one of the triplicate wells.
Approximately 300 candidate positives that were identified in the first round were examined for gcs-1 reporter induction in a 2nd screen in which (i) feeding RNAi was performed in 6-well plates, (ii) mothers were removed on day 3 by picking, and (iii) gcs-1p::GFP reporter expression in their progeny was scored on day 5 using an upright fluorescence microscope. Worms were transferred to a 2% agarose pad on a slide in M9, and covered with a glass slip prior to scoring. To discriminate intestinal autofluorescence from GFP, a triple band emission filter set (Chroma 6100) was used in conjunction with a narrow-band excitation filter (484/14 nm) [11]. Worms were scored for High, Medium, and Low gcs-1p::GFP expression as described below (Figure 1). At least 3 analyses of more than 30 worms each were performed for each RNAi clone. Positive genes for which robust gcs-1 reporter activation was observed in all RNAi replicates were confirmed by sequencing. Additional COP9 signalosome subunit genes (csn-2, -3, -6 and cif-1) were not present in the screening library but were subcloned from a later ORFeome version by standard Gateway reactions.
RNAi
Unless otherwise indicated, feeding RNAi was carried out essentially as described, with HT115 carrying the empty pL4440 vector used as the control [61]. RNAi clones were grown with 12.5 µg/ml tetracycline and 100 µg/ml ampicillin. On the following day, cultures were diluted and grown to OD600 of 1 and induced with 0.6 mM IPTG. This culture was used to seed plates containing tetracycline, ampicillin and 0.6 mM IPTG.
GFP reporter scoring
Essentially the same published scoring procedure was used to score intestinal GFP fluorescence for gcs-1p::GFP and other reporters (Figure 1) [11], [17], [32]. For promoter reporters, “High” indicates that GFP signal was detected at high levels throughout most of the intestine, while “Medium” refers to animals in which robust GFP signal was present only anteriorly or posteriorly. For the SKN-1::GFP fusion reporters, High indicated that a strong SKN-1:: GFP signal was present in all intestinal nuclei, and Medium that nuclear SKN-1:: GFP was present at high levels anteriorly, posteriorly or both, but barely visible midway through the intestine, or that a weak signal was observed in all intestinal nuclei. In general, L2 stage animals were placed on RNAi plates and allowed to develop to the L4 or early adult stage prior to scoring. p values were determined from a Chi2 test.
RNA isolation and quantitative PCR
L2 stage larvae were fed RNAi or control bacteria until day 1 of adulthood. Animals were picked onto clean plates to minimize contamination, then total RNA was extracted from approximately 60 animals suspended in 25 µl of M9. RNA was extracted using Trizol (Sigma), and cDNA was synthesized using the Superscript Reverse Transcription Kit (Life Technologies). qRT-PCR was performed on an ABI 7700 instrument using the SYBER Green Real Time PCR kit (Life Technologies), the comparative Ct method, and normalization to act-1.
Stress resistance assays
For TBHP resistance, L4 stage worms were fed with RNAi or control bacteria for three days at 20°C, then transferred to NGM plates that contained either 9.125 mM or 15.4 mM TBHP (Sigma) and were seeded with E.coli OP50. These plates were prepared two hours before transferring worms by adding TBHP (Sigma) to molten agar at 50–55°C. Each plate contained 20 worms, and the assay was performed in triplicate at 20°C. Worms were scored as dead when they did not respond to repeated gentle prodding with a platinum wire pick. All data were analyzed using JMP software.
Lifespan analysis
Animals were maintained for at least two generations to assure health prior to analysis. Hermaphrodites were synchronized by timed egg laying for 8 hours and allowed to develop at 16°C on control RNAi. At day 1 of adulthood they were transferred to NGM plates containing 100 µg/ml FuDR and either RNAi or control pL4440 bacteria, with which they were fed throughout life. Lifespan assays were carried out at 20°C, with animals scored as dead or alive daily by gentle prodding with a pick. For glucose feeding, 2% glucose was included in the agar. Animals that crawled off the plate, ruptured, or died from internal hatching of progeny were excluded from analysis. Lifespans were measured from hatching. Survival plots, p values (Log-Rank), and proportional hazards were determined using JMP software.
Supporting Information
(A) Diagram of the gcs-1 promoter transgenes used in this study, which were described previously in [11]. The gcs-1Δ2 promoter lacks a region that confers skn-1-independent pharyngeal expression. An SKN-1 binding site that is required for most SKN-1-dependent promoter activity is mutated in the gcs-1(Δ2mut3)::GFP transgene. (B) SKN-1 isoforms (Wormbase). The three SKN-1 isoforms (SKN-1a (623aa), b (310aa) and c (533aa)) all share the same C-terminus, to which GFP has been attached. SKN-1b and SKN-1c are expressed from the SKN-1B/C::GFP transgene, which rescues all known skn-1 phenotypes [11], [18], and all three isoforms are expressed from SKN-1op::GFP, which includes upstream operon sequences that drive SKN-1a expression [17].
(0.15 MB TIF)
Survival plots of representative TBHP resistance assays involving wild type N2 and daf-16(mgDf47) worms, performed as described in Figure 4A. Data were analyzed by JMP and plotted with EXCEL. Statistical analyses are shown in Table S2.
(0.60 MB TIF).
TBHP resistance deriving from translation initiation factor RNAi is daf-16-independent. A survival assay that was performed and analyzed as in Figure 4A. Percent increase in mean survival compared to control is graphed. Representative experiments are shown here and plotted in Figure S2. All experiments and statistics are provided in Table S2. When analyzed side-by-side, N2 and daf-16 worms were roughly comparable with respect to TBHP resistance (see Figure 5E and 5F; Table S3).
(0.18 MB TIF)
Effects of RNAi clones on resistance of wild-type (N2) worms to TBHP. Individual experiments are listed that were performed as in Figure 4A. Representative survival plots are shown in Figure S2.
(0.13 MB DOC)
Effects of RNAi clones on resistance of daf-16 mutant worms to TBHP. Individual experiments are listed that were performed as in Figure 4A. Representative survival plots are shown in Figure S2.
(0.08 MB DOC)
skn-1-dependence of TBHP resistance. Individual stress exposure experiments were performed as in Figure 4B and 4C. In each experiment, survival times were compared to pL4440 RNAi control. Note that the increases in stress resistance associated with translation initiation factor RNAi were consistently almost completely dependent upon skn-1, but did not require daf-16. Worms were censored if they bagged, escaped, or ruptured. p values were calculated by log-rank.
(0.08 MB DOC)
qRT-PCR analyses of SKN-1 target gene expression. Analyses of endogenous SKN-1 target gene mRNA levels were performed as described in Figure 5D, and Materials and Methods. In each experiment, fold change refers to the relative RNA levels detected in RNAi-treated versus pL4440 control worms. Note that the extent of induction was generally decreased in skn-1 mutants. Each value was obtained through a qRT-PCR analysis that was performed in triplicate. p values were calculated by Student's t test.
(0.08 MB DOC).
Summary and statistical analysis of individual lifespan experiments. Data presented in Table 2, Figure 6, and Figure 7 were compiled from these experiments. In each case, RNAi treatment was performed in parallel with a pL4440 RNAi control sample, with the percent mean lifespan extension indicated. Worms were censored that bagged, escaped or ruptured. p values were calculated by log-rank.
(0.11 MB DOC).
Strains used in this study, with references.
(0.04 MB DOC)
Acknowledgments
We thank Chris Link and Tom Johnson for generously providing strains and Blackwell lab members and Malene Hansen for advice or critically reading the manuscript.
Footnotes
The authors have declared that no competing interests exist.
This work was supported by NIH grants GM62891 (TKB), HG001715 (MV), and CA81658 (MV); by the Joslin Diabetes Center DERC (DK036836); and by a fellowship from the Juvenile Diabetes Research Foundation (www.jdrf.org)(JW). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Brownlee M. Biochemistry and molecular cell biology of diabetic complications. Nature. 2001;414:813–820. doi: 10.1038/414813a. [DOI] [PubMed] [Google Scholar]
- 2.Lithgow GJ, Walker GA. Stress resistance as a determinate of C. elegans lifespan. Mech Ageing Dev. 2002;123:765–771. doi: 10.1016/s0047-6374(01)00422-5. [DOI] [PubMed] [Google Scholar]
- 3.Droge W. Free radicals in the physiological control of cell function. Physiol Rev. 2002;82:47–95. doi: 10.1152/physrev.00018.2001. [DOI] [PubMed] [Google Scholar]
- 4.Gems D, McElwee JJ. Broad spectrum detoxification: the major longevity assurance process regulated by insulin/IGF-1 signaling? Mech Ageing Dev. 2005;126:381–387. doi: 10.1016/j.mad.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 5.Kregel KC, Zhang HJ. An integrated view of oxidative stress in aging: basic mechanisms, functional effects, and pathological considerations. Am J Physiol Regul Integr Comp Physiol. 2007;292:R18–36. doi: 10.1152/ajpregu.00327.2006. [DOI] [PubMed] [Google Scholar]
- 6.Lenzen S. Oxidative stress: the vulnerable beta-cell. Biochem Soc Trans. 2008;36:343–347. doi: 10.1042/BST0360343. [DOI] [PubMed] [Google Scholar]
- 7.Xu C, Li CY, Kong AN. Induction of phase I, II and III drug metabolism/transport by xenobiotics. Arch Pharm Res. 2005;28:249–268. doi: 10.1007/BF02977789. [DOI] [PubMed] [Google Scholar]
- 8.Hayes JD, McMahon M. Molecular basis for the contribution of the antioxidant responsive element to cancer chemoprevention. Cancer Lett. 2001;174:103–113. doi: 10.1016/s0304-3835(01)00695-4. [DOI] [PubMed] [Google Scholar]
- 9.Kobayashi M, Yamamoto M. Nrf2-Keap1 regulation of cellular defense mechanisms against electrophiles and reactive oxygen species. Adv Enzyme Regul. 2006;46:113–140. doi: 10.1016/j.advenzreg.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 10.Sykiotis GP, Bohmann D. Stress-activated cap‘n’collar transcription factors in aging and human disease. Sci Signal. 2010;3:re3. doi: 10.1126/scisignal.3112re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.An JH, Blackwell TK. SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev. 2003;17:1882–1893. doi: 10.1101/gad.1107803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oliveira RP, Porter Abate J, Dilks K, Landis J, Ashraf J, et al. Condition-adapted stress and longevity gene regulation by Caenorhabditis elegans SKN-1/Nrf. Aging Cell. 2009;8:524–541. doi: 10.1111/j.1474-9726.2009.00501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park SK, Tedesco PM, Johnson TE. Oxidative stress and longevity in Caenorhabditis elegans as mediated by SKN-1. Aging Cell. 2009;8:258–269. doi: 10.1111/j.1474-9726.2009.00473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olahova M, Taylor SR, Khazaipoul S, Wang J, Morgan BA, et al. A redox-sensitive peroxiredoxin that is important for longevity has tissue- and stress-specific roles in stress resistance. Proc Natl Acad Sci U S A. 2008;105:19839–19844. doi: 10.1073/pnas.0805507105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- 16.Antebi A. Genetics of aging in Caenorhabditis elegans. e129PLoS Genet. 2007;3 doi: 10.1371/journal.pgen.0030129. doi: 10.1371/journal.pgen.0030129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tullet JM, Hertweck M, An JH, Baker J, Hwang JY, et al. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bishop NA, Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447:545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- 19.Kennedy BK, Kaeberlein M. Hot topics in aging research: protein translation, 2009. Aging Cell. 2009;8:617–623. doi: 10.1111/j.1474-9726.2009.00522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bjedov I, Toivonen JM, Kerr F, Slack C, Jacobson J, et al. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 2010;11:35–46. doi: 10.1016/j.cmet.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zid BM, Rogers AN, Katewa SD, Vargas MA, Kolipinski MC, et al. 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell. 2009;139:149–160. doi: 10.1016/j.cell.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan KZ, Palter JE, Rogers AN, Olsen A, Chen D, et al. Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. . Aging Cell. 2007;6:111–119. doi: 10.1111/j.1474-9726.2006.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansen M, Taubert S, Crawford D, Libina N, Lee SJ, et al. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- 24.Selman C, Tullet JM, Wieser D, Irvine E, Lingard SJ, et al. Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science. 2009;326:140–144. doi: 10.1126/science.1177221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, et al. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr Biol. 2004;14:885–890. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Syntichaki P, Troulinaki K, Tavernarakis N. eIF4E function in somatic cells modulates ageing in Caenorhabditis elegans. Nature. 2007;445:922–926. doi: 10.1038/nature05603. [DOI] [PubMed] [Google Scholar]
- 27.Steffen KK, MacKay VL, Kerr EO, Tsuchiya M, Hu D, et al. Yeast life span extension by depletion of 60 s ribosomal subunits is mediated by Gcn4. Cell. 2008;133:292–302. doi: 10.1016/j.cell.2008.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henderson ST, Bonafe M, Johnson TE. daf-16 protects the nematode Caenorhabditis elegans during food deprivation. J Gerontol A Biol Sci Med Sci. 2006;61:444–460. doi: 10.1093/gerona/61.5.444. [DOI] [PubMed] [Google Scholar]
- 29.Tohyama D, Yamaguchi A, Yamashita T. Inhibition of a eukaryotic initiation factor (eIF2Bdelta/F11A3.2) during adulthood extends lifespan in Caenorhabditis elegans. Faseb J. 2008;22:4327–4337. doi: 10.1096/fj.08-112953. [DOI] [PubMed] [Google Scholar]
- 30.Chen D, Pan KZ, Palter JE, Kapahi P. Longevity determined by developmental arrest genes in Caenorhabditis elegans. Aging Cell. 2007;6:525–533. doi: 10.1111/j.1474-9726.2007.00305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Curran SP, Ruvkun G. Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet. 2007;3:e56. doi: 10.1371/journal.pgen.0030056. doi: 10.1371/journal.pgen.0030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.An JH, Vranas K, Lucke M, Inoue H, Hisamoto N, et al. Regulation of the Caenorhabditis elegans oxidative stress defense protein SKN-1 by glycogen synthase kinase-3. Proc Natl Acad Sci U S A. 2005;102:16275–16280. doi: 10.1073/pnas.0508105102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- 34.Rual JF, Ceron J, Koreth J, Hao T, Nicot AS, et al. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 2004;14:2162–2168. doi: 10.1101/gr.2505604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choe KP, Przybysz AJ, Strange K. The WD40 repeat protein WDR-23 functions with the CUL4/DDB1 ubiquitin ligase to regulate nuclear abundance and activity of SKN-1 in Caenorhabditis elegans. Mol Cell Biol. 2009;29:2704–2715. doi: 10.1128/MCB.01811-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cope GA, Deshaies RJ. COP9 signalosome: a multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell. 2003;114:663–671. doi: 10.1016/s0092-8674(03)00722-0. [DOI] [PubMed] [Google Scholar]
- 37.Pick E, Pintard L. In the land of the rising sun with the COP9 signalosome and related Zomes. Symposium on the COP9 signalosome, Proteasome and eIF3. EMBO reports. 2009;10:343–348. doi: 10.1038/embor.2009.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chamovitz DA. Revisiting the COP9 signalosome as a transcriptional regulator. EMBO reports. 2009;10:352–358. doi: 10.1038/embor.2009.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luke-Glaser S, Roy M, Larsen B, Le Bihan T, Metalnikov P, et al. CIF-1, a shared subunit of the COP9/signalosome and eukaryotic initiation factor 3 complexes, regulates MEL-26 levels in the Caenorhabditis elegans embryo. Mol Cell Biol. 2007;27:4526–4540. doi: 10.1128/MCB.01724-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kahn NW, Rea SL, Moyle S, Kell A, Johnson TE. Proteasomal dysfunction activates the transcription factor SKN-1 and produces a selective oxidative-stress response in Caenorhabditis elegans. Biochem J. 2008;409:205–213. doi: 10.1042/BJ20070521. [DOI] [PubMed] [Google Scholar]
- 41.Inoue H, Hisamoto N, An JH, Oliveira RP, Nishida E, et al. The C. elegans p38 MAPK pathway regulates nuclear localization of the transcription factor SKN-1 in oxidative stress response. Genes Dev. 2005;19:2278–2283. doi: 10.1101/gad.1324805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Honda Y, Honda S. The daf-2 gene network for longevity regulates oxidative stress resistance and Mn-superoxide dismutase gene expression in Caenorhabditis elegans. Faseb J. 1999;13:1385–1393. [PubMed] [Google Scholar]
- 43.Hamilton B, Dong Y, Shindo M, Liu W, Odell I, et al. A systematic RNAi screen for longevity genes in C. elegans. Genes Dev. 2005;19:1544–1555. doi: 10.1101/gad.1308205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee SJ, Murphy CT, Kenyon C. Glucose shortens the life span of C. elegans by downregulating DAF-16/FOXO activity and aquaporin gene expression. Cell Metab. 2009;10:379–391. doi: 10.1016/j.cmet.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Porter Abate J, Blackwell TK. Life is short, if sweet. Cell Metab. 2009;10:338–339. doi: 10.1016/j.cmet.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sykiotis GP, Bohmann D. Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev Cell. 2008;14:76–85. doi: 10.1016/j.devcel.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fisher AL, Page KE, Lithgow GJ, Nash L. The Caenorhabditis elegans K10C2.4 gene encodes a member of the fumarylacetoacetate hydrolase family: a Caenorhabditis elegans model of type I tyrosinemia. J Biol Chem. 2008;283:9127–9135. doi: 10.1074/jbc.M708341200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Loh K, Deng H, Fukushima A, Cai X, Boivin B, et al. Reactive oxygen species enhance insulin sensitivity. Cell Metab. 2009;10:260–272. doi: 10.1016/j.cmet.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sevier CS, Kaiser CA. Ero1 and redox homeostasis in the endoplasmic reticulum. Biochim Biophys Acta. 2008;1783:549–556. doi: 10.1016/j.bbamcr.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 51.Radhakrishnan SK, Lee CS, Young P, Beskow A, Chan JY, et al. Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol Cell. 2010;38:17–28. doi: 10.1016/j.molcel.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mair W, Dillin A. Aging and survival: the genetics of life span extension by dietary restriction. Annu Rev Biochem. 2008;77:727–754. doi: 10.1146/annurev.biochem.77.061206.171059. [DOI] [PubMed] [Google Scholar]
- 53.Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, et al. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- 54.Sheaffer KL, Updike DL, Mango SE. The Target of Rapamycin pathway antagonizes pha-4/FoxA to control development and aging. Curr Biol. 2008;18:1355–1364. doi: 10.1016/j.cub.2008.07.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Netzer N, Goodenbour JM, David A, Dittmar KA, Jones RB, et al. Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature. 2009;462:522–526. doi: 10.1038/nature08576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schubert U, Anton LC, Gibbs J, Norbury CC, Yewdell JW, et al. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- 57.Turner GC, Varshavsky A. Detecting and measuring cotranslational protein degradation in vivo. Science. 2000;289:2117–2120. doi: 10.1126/science.289.5487.2117. [DOI] [PubMed] [Google Scholar]
- 58.Fielenbach N, Antebi A. C. elegans dauer formation and the molecular basis of plasticity. Genes Dev. 2008;22:2149–2165. doi: 10.1101/gad.1701508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bowerman B, Eaton BA, Priess JR. skn-1, a maternally expressed gene required to specify the fate of ventral blastomeres in the early C. elegans embryo. Cell. 1992;68:1061–1075. doi: 10.1016/0092-8674(92)90078-q. [DOI] [PubMed] [Google Scholar]
- 60.Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, et al. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 61.Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2:RESEARCH0002. doi: 10.1186/gb-2000-2-1-research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Link CD, Johnson CJ. Reporter transgenes for study of oxidant stress in Caenorhabditis elegans. Methods Enzymol. 2002;353:497–505. doi: 10.1016/s0076-6879(02)53072-x. [DOI] [PubMed] [Google Scholar]
- 63.Essers MA, de Vries-Smits LM, Barker N, Polderman PE, Burgering BM, et al. Functional interaction between beta-catenin and FOXO in oxidative stress signaling. Science. 2005;308:1181–1184. doi: 10.1126/science.1109083. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Diagram of the gcs-1 promoter transgenes used in this study, which were described previously in [11]. The gcs-1Δ2 promoter lacks a region that confers skn-1-independent pharyngeal expression. An SKN-1 binding site that is required for most SKN-1-dependent promoter activity is mutated in the gcs-1(Δ2mut3)::GFP transgene. (B) SKN-1 isoforms (Wormbase). The three SKN-1 isoforms (SKN-1a (623aa), b (310aa) and c (533aa)) all share the same C-terminus, to which GFP has been attached. SKN-1b and SKN-1c are expressed from the SKN-1B/C::GFP transgene, which rescues all known skn-1 phenotypes [11], [18], and all three isoforms are expressed from SKN-1op::GFP, which includes upstream operon sequences that drive SKN-1a expression [17].
(0.15 MB TIF)
Survival plots of representative TBHP resistance assays involving wild type N2 and daf-16(mgDf47) worms, performed as described in Figure 4A. Data were analyzed by JMP and plotted with EXCEL. Statistical analyses are shown in Table S2.
(0.60 MB TIF).
TBHP resistance deriving from translation initiation factor RNAi is daf-16-independent. A survival assay that was performed and analyzed as in Figure 4A. Percent increase in mean survival compared to control is graphed. Representative experiments are shown here and plotted in Figure S2. All experiments and statistics are provided in Table S2. When analyzed side-by-side, N2 and daf-16 worms were roughly comparable with respect to TBHP resistance (see Figure 5E and 5F; Table S3).
(0.18 MB TIF)
Effects of RNAi clones on resistance of wild-type (N2) worms to TBHP. Individual experiments are listed that were performed as in Figure 4A. Representative survival plots are shown in Figure S2.
(0.13 MB DOC)
Effects of RNAi clones on resistance of daf-16 mutant worms to TBHP. Individual experiments are listed that were performed as in Figure 4A. Representative survival plots are shown in Figure S2.
(0.08 MB DOC)
skn-1-dependence of TBHP resistance. Individual stress exposure experiments were performed as in Figure 4B and 4C. In each experiment, survival times were compared to pL4440 RNAi control. Note that the increases in stress resistance associated with translation initiation factor RNAi were consistently almost completely dependent upon skn-1, but did not require daf-16. Worms were censored if they bagged, escaped, or ruptured. p values were calculated by log-rank.
(0.08 MB DOC)
qRT-PCR analyses of SKN-1 target gene expression. Analyses of endogenous SKN-1 target gene mRNA levels were performed as described in Figure 5D, and Materials and Methods. In each experiment, fold change refers to the relative RNA levels detected in RNAi-treated versus pL4440 control worms. Note that the extent of induction was generally decreased in skn-1 mutants. Each value was obtained through a qRT-PCR analysis that was performed in triplicate. p values were calculated by Student's t test.
(0.08 MB DOC).
Summary and statistical analysis of individual lifespan experiments. Data presented in Table 2, Figure 6, and Figure 7 were compiled from these experiments. In each case, RNAi treatment was performed in parallel with a pL4440 RNAi control sample, with the percent mean lifespan extension indicated. Worms were censored that bagged, escaped or ruptured. p values were calculated by log-rank.
(0.11 MB DOC).
Strains used in this study, with references.
(0.04 MB DOC)