Abstract
CYP26A1, which catalyzes the oxidation of all-trans (at)-retinoic acid (RA), is induced moderately by RA in numerous tissues, but is highly responsive in liver. To understand this difference, we have examined the CYP26A1 gene sequence, identified multiple RA response elements (RAREs) and tested them functionally in HepG2 cells as model hepatocytes, and in the liver of vitamin A (VA)-adequate and -deficient rats. Analysis of a 2.2 kbp 5′-flanking region upstream of the CYP26A1 transcription start site (TSS) identified 3 conserved hexameric direct repeat-5 elements, RARE1, -2 and -3, and a half site, RARE4. The full-length promoter containing all 4 elements was sufficient and necessary to increase promoter activity similar to levels of endogenous CYP26A1 mRNA produced in HepG2 cells treated with at-RA. In DNA binding and chromatin immunoprecipitation assays, the binding of RARs to the proximal RARE1 and distal RARE2, -3, and -4 regions of the CYP26A1 promoter was increased in RA-treated HepG2 cells, and greater in VA-sufficient than VA-deficient liver. Moreover, RA increased the binding of RNA polymerase-II in the distal as well as the proximal region, indicating that the distal region may be looped to become positioned close to the TSS, a process favored by retinoic acid receptors. The results support a cooperative model in which the functioning of multiple RAREs may account for the strong inducibility of CYP26A1 in liver, which, in turn, may be important physiologically for restoring retinoid homeostasis when the concentration of RA rises.
Keywords: Gene induction, promoter elements, liver, retinoic acid, Chromatin immunoprecipitation assay
1. Introduction
Vitamin A through its active metabolite, all-trans (at)-retinoic acid (RA), regulates many physiological processes, such as embryonic development, cellular proliferation and differentiation, and immune function. RA is also used clinically in the treatment of leukemia and other diseases (Gallagher, 2002; Reichrath et al., 2007). However, excessive RA is detrimental to tissues, leading to inappropriate gene expression in the short term and overt toxicity if high levels are prolonged. Therefore, mechanisms to control the concentration of RA are essential for the proper functioning of cells, tissues and organs in vivo. Whereas the production of RA from vitamin A (retinol) is regulated through dehydrogenases, its oxidation is mediated by cytochrome P450 activities (Roberts et al., 1980). The cytochrome P450 known as CYP26A1 (P450RA1) is a RA-4-hydroxylase that converts at-RA to polar products including 4-OH-RA, 4-oxo-RA, and 18-OH-RA (Fujii et al., 1997; Ray et al., 1997; White et al., 1997). CYP26A1 is considered to be the major enzyme catalyzing RA oxidation in most tissues. The CYP26A1 gene is well conserved throughout vertebrate species, and the protein is highly specific for the oxidation of at-RA (Lutz et al., 2009). At-RA is not only the specific substrate for CYP26A1 but also a potent inducer of CYP26A1 gene expression, although the regulation is tissue specific (Ray et al., 1997). In the liver, CYP26A1 is very highly regulated (Wang et al., 2002; Cifelli and Ross, 2006). We previously reported that treatment of vitamin A-deficient rats with at-RA resulted dose-dependently in a rapid and dynamic regulation of CYP26A1 mRNA levels, which reached >2000-fold 6 hours after administration of RA, then returned towards basal by 48 hours (Wang et al., 2002). In addition, CYP26A1 mRNA increased progressively with dietary vitamin A intake (Yamamoto et al., 2000) and was highly correlated with liver retinol levels, an indicator of whole-body vitamin A status (Ross, 2003). Almost the entire mRNA induction was due to the activation of the transcriptional process in the liver of adult rats (Zolfaghari et al., 2007; Zolfaghari and Ross, 2009). The liver plays a major role in clearing RA from plasma and in its oxidation, conjugation and elimination (Kurlandsky et al., 1995; Cifelli and Ross, 2006). Thus, understanding the regulation of CYP26A1 in the liver is important for understanding the overall regulation of RA as a potent regulator of gene expression in multiple organ systems.
At-RA functions as an endogenous ligand for nuclear retinoic acid receptors (RARα, β, and γ) which dimerize with retinoid X receptors (RXRα, β, γ) and bind to specific DNA sites, known as retinoic acid response elements (RARE), typically located in the promoters of genes that are transcriptionally regulated by RA (Wei, 2003; Bastien and Rochette-Egly, 2004). The canonical RARE is comprised of a core of two hexameric motifs of PuG(G/T)TCA(X)nPuG(G/T)TCA, generally oriented as a direct repeat (DR) spaced by 2 or 5 nucleotides (n) (Dilworth and Chambon, 2001; Bastien and Rochette-Egly, 2004). The molecular basis of CYP26A1 mRNA induction by RA was first studied by Loudig et al. (Loudig et al., 2000), who characterized the proximal promoter region of the mouse CYP26A1 gene, which is well conserved among human, mouse, and zebrafish. This proximal region contains a classical RARE (referred as RARE1) which cooperated with nearby Sp1/Sp3 binding site, guanine-guanine-rich element, in response of CYP26A1 promoter activation. Later, a second putative RARE (RARE2) located about 2kb upstream of the RARE1 has been identified. Both RARE1 and RARE2 contributed to optimal induction by RA, as tested in mouse teratocarcinoma F19 cells and human breast adenocarcinoma MCF-7 cells, which displayed about a 4 fold increase of reporter activity after treatment with at-RA (Loudig et al., 2005).
The high level of response of CYP26A1 in the liver of RA-treated animals led us to hypothesize that additional elements might exist within the gene’s promoter region that are utilized to drive a high level of expression. Upon further inspection of the upstream of the putative promoter of CYP26A1 we found additional putative elements that might be capable of binding RAR-RXR heterodimers. Thus, in the present studies we have tested their functionality in cell lines representative of human adult liver (HepG2 cells), compared to human embryonic kidney cells (HEK293T cells), and tested the necessity for each element in the liver’s response to RA. Collectively, the results of our studies indicate that liver cells utilize a set of up to 4 RAREs which are all necessary and sufficient for the high-level response to RA in HepG2 liver cells. Moreover, the results suggest a model in which both the proximal and distal regions of the CYP26A1 promoter interact with RNA polymerase II (RNA Pol-II), indicating that the distal region may be looped to become positioned close to the TSS, a process favored by retinoic acid receptors (Yasmin et al., 2004). Additionally, in vivo, CYP26A1 chromatin is more highly associated with RARs and RNA Pol-II in vitamin A-adequate compared to vitamin A-deficient rats. These results advance our understanding of the regulation of RA catabolism in liver in response to conditions, such as drug therapy, when RA concentrations are elevated.
2. Materials and Methods
2.1. Animals
Male Sprague-Dawley rats were fed a nutritional adequate diet deficient in vitamin A from the time of weaning. When they were 45 days of age, they were divided into 2 groups of n=3 rats each, with equal weights, and one group continued to receive the same vitamin A-deficient diet while the other group was fed for one additional week the same diet containing 4 mg retinol/kg of diet as described (Zolfaghari and Ross, 2000). At the end of the experiment, the rats were euthanized by carbon dioxide inhalation and blood and liver tissues were collected for analysis. The experimental protocols for our animal study were approved by the Animal Use and Care Committee of the Pennsylvania State University.
2.2. Cell culture and reagents
HepG2 human hepatocarcinoma cells and HEK293T human embryonic kidney cells were obtained from the American Type Culture Collection (Manassas, VA) and maintained in Dulbecco’s modified eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 0.5% penicillin-streptomycin at 37°C in a 5% CO2-air incubator. For most experiments, the cells were plated at approximately 70% confluency. At-RA (Sigma-Aldrich Chemical Company) was prepared as a concentrated stock in ethanol. Am580 (an RARα agonist (Kagechika, 1994) was provided by Dr. H. Kagechika (Tokyo, Japan). A human CYP26A1 genomic clone containing the extended 5′- and 3′-flanking regions in FOSMID vector was obtained from Children’s Hospital Oakland-BACPAC Resources (Oakland, CA). Purified antibodies specific to RARs, acetylated Histone H4, and RNA Pol-II were purchased from Santa Cruz Biotechnology. For preparation of all other reagents in our laboratory, we followed the protocols reported in the Molecular Cloning Manual (Sambrook and Russell, 2001).
2.3. DNA plasmids
Promoter constructs were based on the human CYP26A1 sequence (from NCBI) and cloned by PCR from CYP26A1 genomic clone in the FOSMID vector into pGL3 Basic Luciferase reporter vector (Promega Corp., Madison, WI). The full length (FL) promoter DNA construct was generated by PCR amplification (30 cycles) of the upstream region of the translation start codon of human CYP26A1 gene, using 5′-agcaagcttGTACAGATAGATTAAAACGT-3′ and aataagcttCACGAAGGTGCAGAGCGC-3′ as the forward and reverse primers, respectively, where lower case letters indicate a HindIII specific nucleotide sequence. Following digestion of the PCR product with HindIII, the fragment was isolated in agarose gel by electrophoresis and ligated into the promoterless pGL3 Basic Luciferase reporter vector. The clone was digested with AscI and AleI and then re-ligated to form the pGL3-FL construct with the FL promoter covering from −2139 to +11 (positions are in relation to TSS) (Ray et al., 1997). For sequence confirmation, the pGL3-FL clone was submitted to sequencing at the Nucleic Acid Facility of the Pennsylvania State University. The human promoter sequence was aligned and compared with those of mouse and rat retrieved from NCBI (www.ncbi.nlm.nih.gov) using ClustalW2 program (www.ebi.ac.uk/Tools/clustalw2/index.html). After amplification, the pGL3-FL was then subjected to digestion by different restriction enzymes, including SmaI, BglII, EcoRI, ApaI and PstI, in order to eliminate specific parts of the full length promoter, and then self religated to generate the elimination constructs (designated E1 to E6). The mutant promoter constructs for RARE nucleotide sequences (M1 to M4) were generated by PCR following the protocol described previously (Zolfaghari and Ross, 2009). Primers used for mutant constructs were: pair 1: 5′-TAGATATCTTTAAAATTGTCTGACCAAGGTAACG-3′ (forward), 5′-TAGATATCTAAAATTCTTTAATTGCGGATTGGGCC-3′ (reverse); pair 2: 5′-TAGATATCTT TATGGCCCGAGGATTGGGAATGG-3′ (forward), 5′-TAGATATCAAAATCCTGCA GGCCGGACCGTG-3′ (reverse); pair 3: 5′-TAGATATCTTAAACACGGTCCGGCCT GCAGG-3′ (forward), 5′-TAGATATCTTAACTGCGGGGCCACCTCGCC-3′ (reverse); pair 4: 5′-TTAGATATCCGTCCCCATTCGTCGGCT-3′ (forward), 5′-TTAGATAT CGGGGCCCATTCCCAATCC-3′ (reverse). Mutated RARE1 was 5′-ATTTTAGAT ATCTTTAA-3′, mutated RARE2 was 5′-GATTTTGATATCTTTAT-3′, mutated RARE3 was 5′-TTAAGATATCCTTAAACA-3′, and mutated RARE4 was 5′-GATATC-3′.
2.4. Transient transfection and luciferase assay
Twenty-four hours before transfection, HEK293T or HepG2 cells were subcultured and 5×105 cells were seeded in each well of 12-well plates in 1 ml of complete culture medium. Two hours before transfection, the complete culture medium was replaced with 5% FBS medium without antibiotics. Transfection was mediated by Lipofectamine™ 2000 (Invitrogen Corp., Carlsbad, CA). After 24 h of transfection, the cells were incubated with full growth medium in the absence or presence of at-RA or Am580 added in a final concentration of 0.1% ethanol. After treatment for 6 h or 24 h, the cells were collected and assayed for luciferase activities using the DRL luciferase assay system (Promega Corp., Madison, WI). pRLTK Renilla luciferase plasmid (Promega) was used as an internal control for transfection. The ratio of Firefly to Renilla luciferase activity was defined as the promoter activity.
2.5. RNA analysis
HepG2 cells and HEK293T cells were cultured in 6-well plates (0.5~1 × 106 cells/well) for various times and treatments, then total cellular RNA was isolated using Rneasy Mini Kit from Qiagen (Valencia, CA). Total RNA was extracted from rat liver or kidney as described (Zolfaghari and Ross, 2000). For Reverse Transcription, 1 μg of total RNA was subjected to reverse transcription using Moloney murine leukemia virus reverse transcriptase (Promega). The diluted reaction product (1/20th of the reaction products) was used for regular PCR or real-time PCR analysis using 2× Real Time SYBR Green/Fluorescein PCR Master Mix (SuperArray Bioscience, Frederick, MD) in a final volume of 20 μl. Glyeraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was amplified at the same time as an internal control. PCR conditions were optimized for denaturation at 94°C, 30 sec, annealing at 60°C, 30 sec, and extension at 72°C, 30 sec for 30 cycles. PCR products were run in 1% agarose by electrophoresis. For regular PCR, the products were analyzed in ethidium bromide containing agarose gel by electrophoresis. The CYP26A1 primers used for PCR were 5′-GTGCCAGTGATTGCTGAAGA-3′ (forward) and 5′-GGAGGTGTCCTCT GGATGAA-3′ (reverse).
2.6. Nuclear extract preparation
Nuclear extracts from HepG2 cells were prepared as previously described (Chen et al., 2002). Briefly, cell pellets was homogenized in a hypotonic buffer [10 mM HEPES, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.2 mM PMSF, 0.5 mM DTT, 1 mM sodium orthovanadate, 0.5% Nonidet P-40] using a Dounce tissue homogenizer. After centrifugation at 2500 × g at 4°C for 10 min, the supernatant (cyptoplasmic fraction) was removed, and then pellets were washed once with hypotonic buffer containing no detergent. Then a hypertonic buffer (20 mM HEPES, pH 7.9, 10% glycerol, 1.5 mM MgCl2, 400 mM KCl, 0.2 mM EDTA, 0.2 mM PMSF, 0.5 mM DTT, 1 mM sodium orthovanadate) was added to extract nuclear protein. After 30 min of incubation on ice, the mixture was centrifuged at 15,000 × g for 30 min. The supernatant was collected as the nuclear extract, divided into aliquots and stored at −80°C.
2.7. Electrophoretic mobility shift assays (EMSA)
Double stranded nucleotide fragments containing putative CYP26A1-RAREs (wild type and mutated fragments) were synthesized as probes. The wild type fragments were labeled with [γ-32P]dATP and incubated with the nuclear protein extract at room temperature for 30 min. For the supershift assay, nuclear proteins were incubated with specific antibodies for 15 min. The reactions were separated on a 5% nondenatured polyacrylamide gel by electrophoresis. After electrophoresis, the gel was dried and subjected to autoradiography.
2.8. Chromatin immunoprecipitation (ChIP) assay
Frozen liver tissue or cultured cells were homogenized in a glass Dounce homogenizer to release nuclei (Zolfaghari et al., 2007), which were then subjected to ChIP assays as described (Hollingshead et al., 2008). Purified antibodies specific to RARs, acetylated Histone H4, and RNA Pol-II were used for immunoprecipitation. The pulled-down DNA was subjected to real-time PCR with oligonucleotide primer pairs designed for different regions of human and rat CYP26A1 promoter and using SYBR Green/Fluorescein PCR Master Mix as described above.
2.9. Statistical analysis
In each experiment, three replicate samples were analyzed for each treatment or time point. In addition, most experiments were further repeated. Statistical analysis was performed by using SuperANOVA software (Abacus Concepts, Berkeley, CA) or GraphPad Prism 5 (GraphPad, LaJolla, CA) for Student’s t-test or one-way analysis of variance. P<0.05 was considered significant.
3. Results
3.1. Three RAREs and a half site are highly conserved in the upstream region of CYP26A1
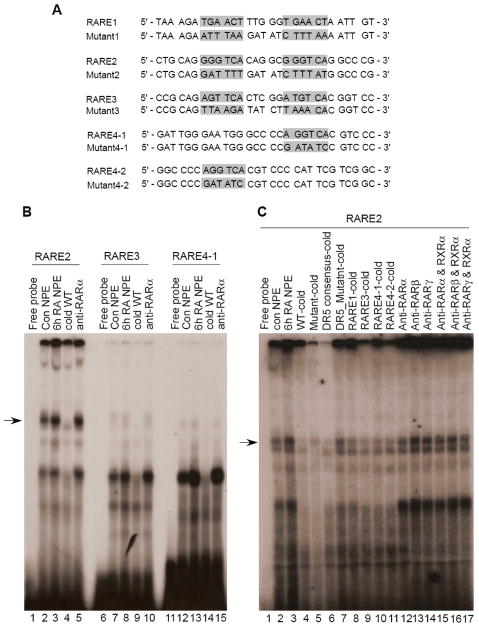
We first analyze the DNA sequence upstream of the ATG translation start codon of the human CYP26A1 gene retrieved from NCBI (www.ncbi.nlm.nih.gov) and found that there are at least three RAREs, each present as DR5 elements, and a half direct repeat site present in the putative promoter region (Fig. 1). Whereas RARE1, reported previously (Loudig et al., 2000), is positioned in reverse direction proximal to the transcription start site just upstream of the putative TATA box sequence the other two elements together with the half site, in a cluster, are located about 2 kbp upstream of the RARE1. All of these elements including the half site are highly conserved among human, mouse and rat (Fig. 1).
Fig. 1. The RARE regions present in the promoter of CYP26A1 gene are highly conserved among human, mouse and rat.
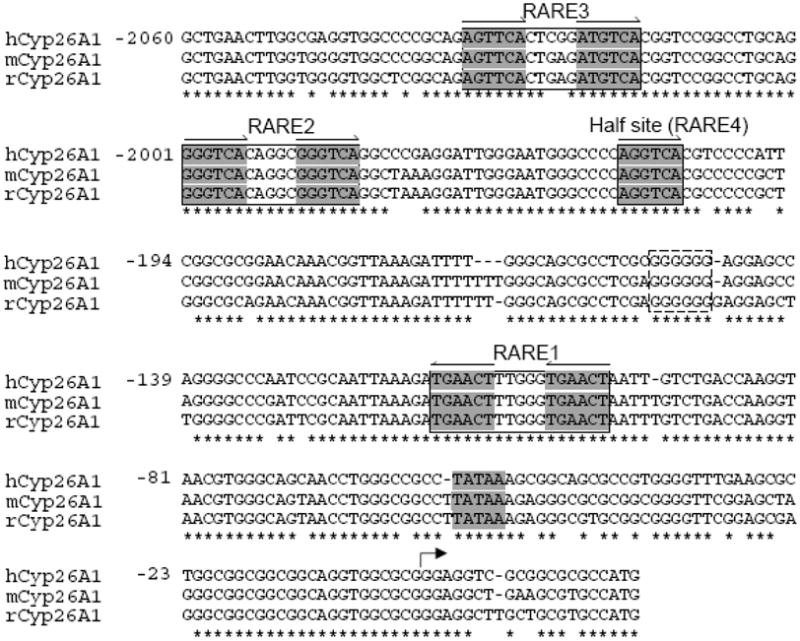
Sequence data from upstream of the coding region of CYP26A1 genes from human, mouse and rat were retrieved from NCBI (www.ncbi.nlm.nih.gov) and aligned using ClustalW2 program (www.ebi.ac.uk/Tools/clustalw2/index.html).
3.2. Relative luciferase activity of the FL construct is correlated with the level of endogenously expressed CYP26A1 mRNA
Since the CYP26A1 gene is active in the liver and highly induced by high vitamin A and at-RA we examined HepG2 human hepatoma cells as a model hepatocyte to analyze the function of these RARE elements including the half site. HepG2 cells expressed very little CYP26A1 mRNA endogenously prior to retinoid treatment; however, upon treatment of the cells with 1 μM of at-RA, CYP26A1 mRNA level was increased by more than 30 times after 6 h and then reduced toward the control level after 24 h treatment (Fig. 2A). This induction was dose-dependent between 1–1000 nM at-RA (Fig. 2B), which includes physiological and pharmacological concentrations, similar to what has been observed in the liver of intact animals (Wang et al., 2002). A similar response was observed when HepG2 cells were treated with Am580, a specific ligand for RARα, although not to the same magnitude as produced by at-RA (Fig. 2A and 2B), which may indicate the possible involvement of more than one RAR subtype in regulation of the CYP26A1 gene.
Fig. 2. Endogenous mRNA levels as well as the promoter activity of the CYP26A1 gene are highly regulated by at-RA and Am580 in HepG2 cells.
Total RNA samples from HepG2 cells treated with either at-RA or Am580 at different time points (A) and concentrations (B) were first reverse-transcribed and then analyzed by real time PCR using specific primers for human CYP26A1 mRNA, and GADPH as a internal control. HepG2 cells transfected with full-length human promoter of CYP26A1 gene (C) pGL3-Basic vector were treated with either at-RA or Am580 at different time points (D) and concentrations (E) and then lysed to measure the luciferase activity as described in the Methods section.
Having shown that HepG2 cells could be an appropriate model for liver to analyze the function of RARE elements present in the putative promoter of CYP26A1 gene, a FL DNA fragment of 2,200 bp spanning the region upstream of the CYP26A1 translation start codon was cloned by PCR from CYP26A1 gene into the promoterless pGL3-basic vector with firefly luciferase reporter gene (Fig. 2C), and cotransfected into HepG2 cells with a pRLTK vector containing Renilla luciferase as the control for transfection efficiency. Following transfection, the cells were treated with either at-RA or Am580 for 6 or 24 h and then lysed for measurement of luciferase activity (Fig. 2D). As expected, the CYP26A1 promoter was very active in the cells and highly induced by at-RA dose-dependently (Fig. 2E) with maximum induction after 24 h as compared to 6 h for the endogenous CYP26A1 mRNA, likely due to the time it takes for the cells to express the active luciferase protein. Again, Am580 was effective in induction of the promoter, but less so than at-RA (Fig. 2D and 2E).
3.3. CYP26A1 promoter activities show cell line specificity
To evaluate the functional contribution of the RARE regions in the activity of CYP26A1 promoter we used both HepG2 cells and HEK293T cells, as non-hepatic cells, for comparison. CYP26A1 mRNA is expressed endogenously in HEK293T cells, however, it is induced just a few fold by at-RA, much less as compared to HepG2 cells (Fig. 3). To assess the active contributions of the RARE regions, first, we deleted different regions of the FL construct by restriction enzymes and re-ligated in the promoterless pGL3-basic vector as shown in Fig. 4A. The FL construct, and those with a series of deletions were transiently transfected individually into HepG2 cells and HEK293T cells and tested for luciferase activity after the cells were incubated with either vehicle or 1 μM at-RA for 24 h. Similar to the expression of CYP26A1 mRNA, the activity of CYP26A1 FL promoter was relatively higher in HEK293T cells than in HepG2 cells without at-RA treatment (data not shown) although the activities were low in both cell types. Upon treatment of the cells with at-RA, the activity of the FL construct increased by more than 20 times in HepG2 cells (Fig. 4B) as compared to 4 fold in HEK293T cells (Fig. 4C). The orientation of the FL promoter construct in the promoterless pGL3-basic vector seems to be essential for full luciferase activity since the reverse position of the promoter resulted in partial activity in HepG2 cells and completely eliminated activity in HEK293T cells both in the absence and presence of at-RA (Fig. 4B and C). The promoter constructs containing the proximal RARE1 alone without the distal ones including the half site were not responsive to at-RA in HepG2 cells but partially responded to at-RA in HEK293T cells. In fact, elimination of the cluster of distal RARE with the half site resulted in an increase in the basal activity in HepG2 cells but no apparent change in HEK293T cells. Elimination of the DNA region between the proximal and distal RARE cluster including the half site (E4), bringing both distal and proximal RARE together in the promoter, completely eliminated the basal activity in HEK293T cells and reduced partially the response of the promoter to at-RA in both cell types. Promoter construct with deleted DNA region containing RARE2 together with half site in the distal region (E5) showed no response to at-RA in HepG2 cells but partial response in HEK293T cells. By deletion analysis, whereas the distal RARE cluster including the half site contributed the major functional activity in the CYP26A1 promoter in response to at-RA in HepG2 cells, the DNA region between the proximal and distal RARE regions was important for basal promoter activity in both cell types (Fig. 4B and 4C).
Fig. 3. CYP26A1 gene is active but less regulated by at-RA in HEK293T cells.
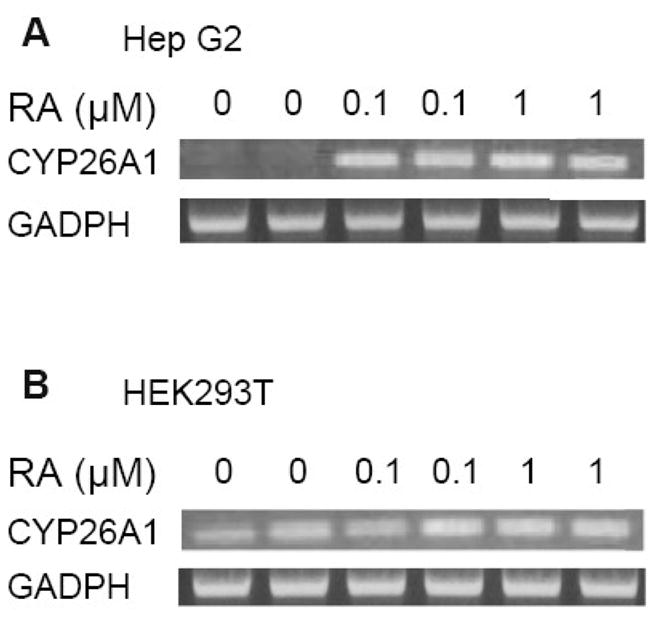
PCR products of the total RNA samples from HepG2 cells (A) and HEK293 cells (B) treated with at-RA at different concentrations for 6 h were analyzed in ethidium bromide containing agarose gel by electrophoresis. GADPH was analyzed as an internal control.
Fig. 4. The full-length promoter of the CYP26A1 gene is sufficient and essential to drive the gene in both HepG2 cells and HEK293T cells in response to at-RA.
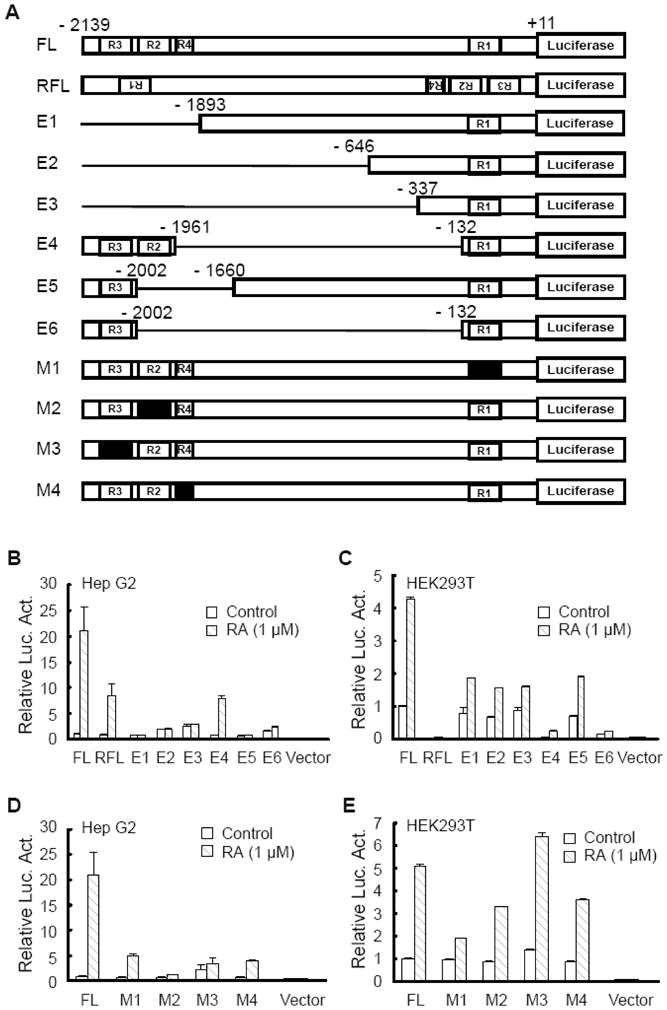
Cells transfected with either the full-length promoter construct of CYP26A1 gene (FL) or with the constructs mutated by polynucleotide elimination or substitutions (A) were treated with 1 μM at-RA for 24 h and then lysed to measure luciferase activities for HepG2 cells (B and D) and HEK293T cells (C and E).
To further evaluate the role played by each specific RARE as well as the half site in the activity of the CYP26A1 promoter, we mutated the individual sites (Fig. 4A) by nucleotide substitution (Zolfaghari and Ross, 2009) in the FL construct and conducted transfection assays in both HepG2 and HEK293T cells. Mutation in the individual RARE sites resulted in a significant decrease in the promoter response to at-RA in HepG2 cells with almost no activity in the FL construct containing the mutated RARE2 (Fig. 4D). The proximal RARE1 was more effective than the distal cluster in the promoter response to at-RA in HEK293T cells (Fig. 4E). Indeed, no significant change was observed in the FL construct with a mutated RARE3 in HEK293T cells. Interestingly, the half site contributed significantly to the promoter response to at-RA in both cell lines since mutation in that site resulted in significant reduction in activity of the promoter response to at-RA (Fig. 4D and 4E).
3.4. Cotransfection of each of the individual retinoic acid receptors (RARα, β, γ) significantly increases promoter activities
Results from earlier experiments on the effect of Am580 on the expression of endogenous CYP26A1 mRNA and promoter activity as compared to at-RA in HepG2 cells (see Fig. 2) may suggest that not only RARα but also RARβ or RARγ, could be involved in the induction of CYP26A1 promoter activity by at-RA. To test such an involvement, we performed cotransfection of the FL constructs including both WT and mutated ones with each of the RAR subtypes, together with RXRα into HepG2 cells and measured the luciferase activity. Cotransfection of wild type FL promoter with any of the three RAR subtypes in the presence of at-RA greatly increased the activity of the wild type FL promoter, about 50 times (for RARγ) to 60 times (for RARα and RARβ) (Fig. 5). Cotransfection of the RAR subtypes with either of the mutated FL constructs had much less effect on the activity of the mutated than the WT promoter. Whereas the lowest activity was observed in the FL construct with mutated RARE2, the highest activity was found in the FL construct with mutated half site (Fig. 5).
Fig. 5. Retinoic acid receptor subtypes all induced the full length promoter of the CYP26A1 gene in HepG2 cells in response to at-RA.
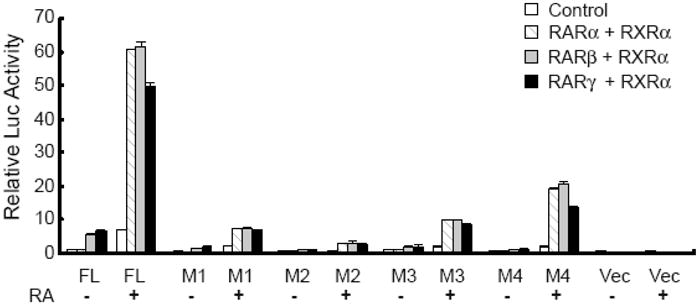
HepG2 cells cotransfected with either wild type or mutated full-length (FL) promoter with RAR subtypes together with RXRα were treated with at-RA for 24 h and then lysed to measure luciferase activities.
3.5. RAREs from CYP26A1 promoter exhibit similar nuclear protein binding patterns and at-RA increases DNA-binding activities
In order to find any complex formed between nuclear proteins and those DNA RARE binding sites identified above we prepared DNA polynucleotide fragments including one wild type and a corresponding one with a mutation in the core RARE region, and use them as a probe or competitor in EMSA assays (Fig. 6A). In this analysis, we focused our attention on the RARE cluster present in the distal region of the CYP26A1 promoter since this region is apparently more involved in the activity of the promoter response to at-RA in HepG2 cells. Specific complexes were formed between nuclear proteins from HepG2 cells and each of the individual distal RAREs (RARE 2, 3 and 4) used as probes, and competed out when the respective unlabeled polynucleotides in excess were used as competitors. Nuclear proteins, from either control or at-RA-treated cells formed more intense binding complexes with RARE2 (the upper bands) than with either RARE3 or RARE4 (Fig. 6B). Nuclear proteins from the cells treated with at-RA formed more intense complexes than those from control cells with all three RARE probes.
Fig. 6. Individual RA response elements form complexes with the proteins in nuclear extract from HepG2 cells.
Nuclear protein extracts from either vehicle or at-RA-treated HepG2 cells were incubated with [32P]-labeled distal RARE (A) as probe and then run on polyacrylamide gel by electrophoresis. The gels were dried and then exposed to X-ray film as described in the Methods section.
To examine further whether the individual RAR subtypes may interact in the formation of complexes in the RARE region present in the CYP26A1 promoter, separate EMSA supershift assays were performed using nuclear extract from either vehicle or at-RA-treated HepG2 cells and antibodies for either individual RAR subtypes alone or in combination with RXRα antibody. Since RARE2 formed stronger complexes with nuclear protein from HepG2 cells (Fig. 6B), this polynucleotide fragment was used as the [32P]-labeled probe for further experiments. Again, nuclear proteins from the cells treated with at-RA formed more intense complexes than those from control-treated cells (lane 2 vs lane 3 in Fig. 6C). The complexes formed seem to be specific since they were competed out by excess cold wild type RARE2 and DR5 polynucleotides, but not by the mutated polynucleotides (lane 3 to 7 in Fig. 6C). Similarly, other cis-acting RAREs, including the proximal one, were competitive as well although not as effectively as wild type RARE2 (lane 8 to 11 in Fig. 6C). Although DR5 might be involved in complex formation as shown by competition results (lane 6 vs lane 3 in Fig. 6C), none of the individual RAR antibodies alone or in combination with the anti-RXRα caused any super-shift retardation. This may be due to the increase in the size of the complexes, which could not passed through the gel. However, the antibodies did show competition with receptor-bound [32P]-labeled RARE2, since the intensities of the complexes were reduced in the presence of the antibodies (lane 12 compared to lane 17 in Fig. 6C). Apparently, anti-RARα antibody produced a stronger competition than the other antibodies both in the absence and presence of anti-RXRα (lane 12 in Fig. 6C). In addition, treatment of the HepG2 cells with at-RA prior to preparation of the nuclear extract markedly increased the DNA-protein binding (lane 2 compared to lane 3 in Fig. 6B and 6C), indicating more of the bound factors were present in at-RA-treated cells compared to control cells.
3.6. Vitamin A status affects the binding of nuclear proteins to the intact CYP26A1 chromatin of the liver of adult rats
Previously, we showed that increase in steady state CYP26A1 mRNA levels by at-RA is mostly, if not entirely, due to increase in the transcription process in the liver of adult rats (Zolfaghari et al., 2007). In this study, we found that at-RA induced the CYP26A1 promoter activity similar to the level of endogenous mRNA, again, indicating that induction of CYP26A1 gene expression by at-RA may, solely, be due to the transcription process. In order to confirm that the RAR receptors are directly involved in the transcriptional activation of CYP26A1 gene, a ChIP assay was employed to evaluate the extent of In vivo binding of the RAR nuclear receptors to the DNA promoter region of CYP26A1 gene. For this, liver tissue samples of vitamin A-deficient and vitamin A-adequate rats were used for analysis of RNA by real-time RT-PCR, and also homogenized to release the nuclei, which were subjected to ChIP assays using different RAR antibodies for immunoprecipitation. The relative expression of CYP26A1 mRNA level was about 5 times lower (P<0.01) in the liver samples of vitamin A-deficient rats as compared to vitamin A-adequate animals (Fig. 7A). For ChIP assays, primer pairs from different regions of the rat promoter of CYP26A1 gene (Fig. 7B) were designed to measure by real-time PCR the DNA samples immunoprecipitated by either anti-RARs antibodies relative to those in the original input DNA prior to pull-down by antibodies. The relative DNA levels were significantly lower (P<0.05) in liver of vitamin A-deficient than vitamin A-adequate control rats when the primer pairs covering the proximal and distal regions of the CYP26A1 promoter were used (Fig. 7B). This may indicate that fewer RAR receptors were bound to those regions in vitamin A-deficient than in vitamin A adequate rats. Since the CYP26A1 promoter was analyzed in HEK293T human embryonic kidney cells, in a separate experiment, to further test the tissue specificity of CYP26A1 inducibility, we studied the effect of vitamin A and at-RA on the mRNA expression levels in the kidney of adult rats, compared to those in the liver. The gene expression level of CYP26A1 was low in both kidney and liver, however, upon treatment of rats with at-RA the expression level was increased by less than 10 times in kidney as compared to about 200 times in the liver (Fig. 7D).
Fig. 7. The bound RARs to the promoter of CYP26A1 gene in intact chromatin were significantly less in the liver of vitamin A deficient- than that in vitamin A adequate adult rats.
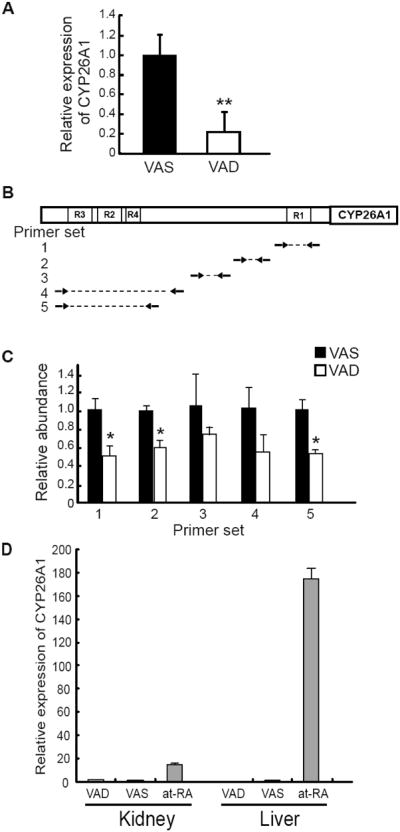
Total RNA was isolated from the liver and used to measure the abundance of CYP26A1 mRNA and ribosomal RNA by real-time RT-PCR (A). ChIP assays were performed for RAR subtypes and real-time PCR was employed to assess the binding in different regions of the promoter of the rat CYP26A1 gene (B and C). Data are the mean ± SD of n=3 rats/group, analyzed by Student’s t-test. *P<0.05, **P<0.01. Total RNA was isolated from the liver and kidney and used to measure the abundance of CYP26A1 mRNA and ribosomal RNA by real-time RT-PCR (D).
3.7. At-RA increases the binding of basic transcription factors in the promoter region of CYP26A1 gene in intact chromatin
To further investigate whether the binding status of the basic transcriptional components in the promoter region of the CYP26A1 gene is significantly affected by at-RA, both HepG2 cells and HEK293T cells were incubated with either ethanol as the vehicle control or at-RA for 1 h and then lysed for ChIP assay using purified anti-acetylated histone H4 and anti-RNA RNA Pol-II antibodies for immunoprecipitation. Primer pairs from different regions of the promoter of CYP26A1 gene (Fig. 8A) were designed to measure by real-time PCR the DNA samples immunoprecipitated by either anti-acetylated histone H4 or anti-RNA Pol-II antibodies relative to those in the original input DNA prior to pull-down by antibodies. Whereas at-RA treatment increased significantly (P<0.05) the binding of both acetylated histone H4 and RNA Pol-II at least in both the proximal and distal regions of the promoter of CYP26A1 gene in HepG2 cells (Fig. 8B and 8C), it induced significantly (P<0.05) only the binding of the RNA Pol-II in the DNA proximal region in HEK293T cells (Fig. 8D and 8E). In fact, the binding of the acetylated histone H4 decreased in the distal region in HEK293T cells (P<0.05).
Fig. 8. At-RA affects the accessibility of transcription factors in the promoter region of CYP26A1 gene in the intact chromatin.

ChIP assay was performed for RNA pol-II and acetylated histone H4. Primer sets were designed (A) to assess by real time PCR the binding in different regions of the promoter of CYP26A1 gene in HepG2 cells (B and C) and HEK293T cells (D and E). Data are the mean ± SD of n=4 replicates, analyzed by Student’s t-test. *P<0.05.
4. Discussion
CYP26A1, believed to be the major cytochrome P450 that catalyzes the oxidation of at-RA, belongs to a group of a few genes of which their expression is directly controlled by at-RA, the pan-agonist ligand for RARα, RARβ, and RARγ. Upon sequence analysis of the 5′ upstream region of CYP26A1 gene we found that there are at least 3 RAREs, present as DR5 elements, together with a half direct repeat present in the promoter of CYP26A1 gene. The proximal RARE1 together with RARE2 at the more distal location in the promoter of mouse gene have been analyzed and shown to be active in response to at-RA in HeLa, COS-1, F19, and MCF-7 cell lines (Loudig et al., 2000; Loudig et al., 2005), but the RARE3 and the half site were not identified and investigated previously. RARE1, oriented in reverse direction, is located proximally to the transcription start site whereas RARE2 and RARE3 together with the half site are located in a cluster present in the distal region about 2 kbp away from the proximal one (Fig. 1). Since the CYP26A1 gene is highly responsive to vitamin A and RA status in the intact liver but not in other tissues including intestine, lung, and kidney (Wang et al, 2002; Ray et al., 1997; Xi and Yang, 2008) in vivo, and moreover, those two RARE (RARE1 and RARE2), alone, are not apparently sufficient to drive CYP26A1 gene at least in the liver, we initiated this study to analyze all those 3 RAREs together with the half site in response to at-RA in a cell model of liver and compared the results to the non-hepatic embryonic cell model. To support our results we also investigated the status of the RAR binding at the 5′ upstream region of the CYP26A1 gene in intact chromatin in the liver of adult rats.
HepG2, a well differentiated human hepatoma cell line (Knowles et al., 1980), appears to be an appropriate model for liver to study the promoter of CYP26A1 gene for 2 reasons. First, the CYP26A1 gene is active in these cells and highly responsive to at-RA, similar to CYP26A1 in the intact liver, and second, the FL promoter of CYP26A1 gene containing both the proximal and distal RAREs including the half site is as active as the endogenously-expressed gene when the cells are treated with retinoids (Fig. 2 and Fig. 3). The strong responses to retinoids appear to be specific to certain cell types since HEK293T cells, a human embryonic kidney cell line, expresses CYP26A1 but the responses to retinoids was an order of magnitude lower than for HepG2 cells. Similarly, relatively low responses to at-RA were observed after the transfection of CYP26A1 promoter into HEK293T cells. Comparatively, the FL promoter responded to at-RA by about 20 to 30 times in HepG2 cells but not more than 4 to 5 times in HEK293T cells.
With mutation either by nucleotide elimination or substitution we demonstrated that the 2.2 kbp DNA promoter is essential and sufficient to drive CYP26A1 gene expression in response to at-RA (Fig. 4). The combination of both proximal and distal RARE regions including the half site contributed significantly to the promoter activity in response to at-RA in HepG2 cells, as mutation of any individual RARE including the half site resulted in a drastic reduction in response of the promoter to at-RA. As a comparison, the proximal RARE region, but not the distal region, seems to be the most significant factor in the relatively lower response to at-RA in HEK293T cells. It is important to note that RARE2 present in the distal region appears to be the most significant contributing factor in the promoter activity of the CYP26A1 gene in response to at-RA in HepG2 cells (Fig. 4C and 4E). Non-RARE regions located between proximal and distal RAREs, which contain several SP1 and SP3 sites appear to play an important role in the basal promoter activity in HEK293T cells only, but in response to at-RA in both cell types. These transcriptional factors have been shown to interact with RARs in driving the expression of several genes in response to RA (Wolf et al., 2005; Khan et al., 2007; Laliotis et al., 2007; Islam et al., 2008; Zolfaghari and Ross, 2009).
Among the retinoic acid receptors, RARα and RXRα are expressed at relatively high levels in the liver of adult rats (Haq et al., 1991; Zolfaghari and Ross, 1995). Whereas RARβ is expressed at low level and its expression is regulated by vitamin A and RA, RARγ is barely detectable in the liver (Zolfaghari and Ross, 1995). We showed here that all of the individual RARs, each in combination with RXRα, were highly active in driving the promoter of CYP26A1 gene in response to at-RA in HepG2 cells, although RARγ was less inducing as compared to either RARα or RARβ (Fig. 5). In contrast, RARγ in combination with RXRα has been reported to be highly active, as compared to the other receptors, in driving the CYP26A1 promoter in P19 cells, a mouse teratocarcinoma cell line (Loudig et al., 2005). Very recently, Tay et al (2010) reported that mRNA expression of the CYP26A1 gene as well as the RARβ gene is highly inducible by specific agonist of RARα but not at all by an RARβ agonist in HepG2 cells. They concluded that endogenous CYP26A1 expression is regulated by RARα but not by RARβ. In our study both RARα and RARβ receptors were equally active on the CYP26A1 promoter in the absence or presence of at-RA, indicating that when RARβ is present in sufficient quantity it is capable of regulating CYP26A1 expression.
The individual receptors were comparatively less inducing in driving the FL promoter containing mutations in any individual RAREs including the half site (Fig. 5). Again, RARE2 was shown to be the most significant element in driving the CYP26A1 promoter in response to at-RA since mutation in this element resulted in the complete lack of response of the FL promoter to all the retinoid receptors in response to at-RA. Since all RAR subtypes individually, when in combination with RXRα, were able to induce the promoter activity of the CYP26A1 gene in response to at-RA, the limiting factor in tissue expression of CYP26A1 gene may not be the RAR types but rather other nuclear transcription factors, since elimination of the non-RARE region of the CYP26A1 promoter resulted in signification reduction in promoter activity. We have shown that Am580, a specific ligand for RARα, was less effective than at-RA in both the induction of the endogenous CYP26A1 gene as well as the FL promoter of CYP26A1 gene in HepG2 cells. This suggests that more than one RAR receptor is involved in driving the expression of the CYP26A1 gene in the liver. Since in the presence of at-RA both RARα and RARβ together with RXRα are expressed at significant levels in liver (Kato et al., 1992; Zolfaghari and Ross, 1995), it is conceivable that they may all cooperate together in the induction of CYP26A1 gene in this organ.
Results from our EMSA study indicated that retinoid receptors in the nuclear protein extract prepared from HepG2 cells did form specific complexes with all of the individual RAREs present in the CYP26A1 promoter, with RARE2 forming a higher intensity complex than with other elements (Fig. 6). Formation of tighter complexes may indicate that RARE2 is contributing more into the overall activity of CYP26A1 promoter in response to at-RA in liver cells. In fact, we showed that mutation of RARE2 alone in the FL promoter of the CYP26A1 gene resulted into the complete lack of the promoter response to at-RA. In ChIP assays, however, more RAR proteins were bound to both the proximal and distal regions of the CYP26A1 promoter present in the intact chromatin of the liver of control vitamin A-adequate as compared to vitamin A-deficient animals (Fig. 7C). Although the distal region is far away from the transcription start site, binding of RNA Pol-II was increased significantly in both the proximal and distal regions of the promoter of the CYP26A1 gene in intact chromatin of HepG2 cells treated with at-RA as compared to the control cells (Fig. 8A). This may indicate that in response to at-RA the distal region is bent over to position the distal RARE cluster closer to the transcription start site in the vicinity of the proximal RARE1. Retinoid receptors have been shown to favor the bending in the promoter of several genes (Yasmin et al., 2004). The results from ChIP experiments are in agreement with those of the transfection study, which showed that both proximal and distal regions contribute significantly in the activity of the promoter of the CYP26A1 gene in HepG2 cells in response to at-RA. On the other hand, the binding of RNA Pol-II was increased in only the proximal, but not the distal region of the promoter, in HEK293T cells in response to at-RA (Fig. 8C). Again, these results agree with those from transfection studies, which showed that the only the proximal region but not the distal may play a significant role in the activity of the RA inducibility of the promoter in HEK293T cells. This may indicate that the bending may not occur significantly in HEK293T cells, probably due to the inactivation of chromatin in the area as indicated by the data on histone4 acetylation by ChIP assay (Fig. 8).
In summary, the results of this study have confirmed the importance of CYP26A1 RARE1 and RARE2 as essential regulatory elements and extended previous work by demonstrating that, for liver cells, each one of three distinct DR5 elements -- RARE1, RARE2, RARE3 -- and in addition the half site located near RARE2 and RARE3, is required for the high-level response of CYP26A1 to at-RA that is observed in liver cells. The interaction of both the proximal and distal regions of the CYP26A1 promoter with RNA Pol-II, detected by the results of our ChIP assays, suggests that looping of DNA between these promoter regions might occur in liver cells as part of the rapid and high-level induction of CYP26A1 expression by at-RA. The high-level response of CYP26A1 in liver may be an adaptive physiological response that serves to lessen the risk of RA toxicity when the concentration of RA rises. However, the highly inducible nature of CYP26A1 in the liver may also limit the pharmacological activity of RA when it is used clinically, due to the rapid oxidation of RA in the liver and the subsequent elimination of polar retinoid metabolites.
Acknowledgments
This research was supported by NIH R01 CA-90214. We thank Michael G. Borland for his helpful advice on ChIP methodology.
Abbreviations
- ChIP
chromatin immunoprecipitation
- EMSA
electrophoretic mobility shift assay
- RNA Pol-II
RNA polymerase II
- RA
retinoic acid
- RAR
retinoic acid receptor
- RARE
retinoic acid response element
- RXR
retinoid X receptor
- TSS
transcription start site
- VA
vitamin A
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Yao Zhang, Email: Yxz152@psu.edu.
Reza Zolfaghari, Email: rxz7@psu.edu.
A. Catharine Ross, Email: acr6@psu.edu.
References
- Bastien J, Rochette-Egly C. Nuclear retinoid receptors and the transcription of retinoid-target genes. Gene. 2004;328:1–16. doi: 10.1016/j.gene.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Chen Q, Ma Y, Ross AC. Opposing cytokine-specific effects of all trans-retinoic acid on the activation and expression of signal transducer and activator of transcription (STAT)-1 in THP-1 cells. Immunology. 2002;107:199–208. doi: 10.1046/j.1365-2567.2002.01485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifelli CJ, Ross AC. All-trans-retinoic acid distribution and metabolism in vitamin A-marginal rats. Am J Physiol Gastrointest Liver Physiol. 2006;291:G195–202. doi: 10.1152/ajpgi.00011.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilworth FJ, Chambon P. Nuclear receptors coordinate the activities of chromatin remodeling complexes and coactivators to facilitate initiation of transcription. Oncogene. 2001;20:3047–54. doi: 10.1038/sj.onc.1204329. [DOI] [PubMed] [Google Scholar]
- Fujii H, Sato T, Kaneko S, Gotoh O, Fujii-Kuriyama Y, Osawa K, Kato S, Hamada H. Metabolic inactivation of retinoic acid by a novel P450 differentially expressed in developing mouse embryos. Embo J. 1997;16:4163–73. doi: 10.1093/emboj/16.14.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher RE. Retinoic acid resistance in acute promyelocytic leukemia. Leukemia. 2002;16:1940–58. doi: 10.1038/sj.leu.2402719. [DOI] [PubMed] [Google Scholar]
- Haq R, Pfahl M, Chytil F. Differential effects of all-trans and 13-cis-retinoic acid on mRNA levels of nuclear retinoic acid receptors in rat lung and liver. Biochem Biophys Res Commun. 1991;180:1137–44. doi: 10.1016/s0006-291x(05)81185-4. [DOI] [PubMed] [Google Scholar]
- Hollingshead HE, Borland MG, Billin AN, Willson TM, Gonzalez FJ, Peters JM. Ligand activation of peroxisome proliferator-activated receptor-beta/delta (PPARbeta/delta) and inhibition of cyclooxygenase 2 (COX2) attenuate colon carcinogenesis through independent signaling mechanisms. Carcinogenesis. 2008;29:169–76. doi: 10.1093/carcin/bgm209. [DOI] [PubMed] [Google Scholar]
- Islam MR, Puri S, Rodova M, Magenheimer BS, Maser RL, Calvet JP. Retinoic acid-dependent activation of the polycystic kidney disease-1 (PKD1) promoter. Am J Physiol Renal Physiol. 2008;295:F1845–54. doi: 10.1152/ajprenal.90355.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagechika H. [Novel synthetic retinoid agonists and antagonists] Yakugaku Zasshi. 1994;114:847–62. doi: 10.1248/yakushi1947.114.11_847. [DOI] [PubMed] [Google Scholar]
- Kato S, Mano H, Kumazawa T, Yoshizawa Y, Kojima R, Masushige S. Effect of retinoid status on alpha, beta and gamma retinoic acid receptor mRNA levels in various rat tissues. Biochem J. 1992;286(Pt 3):755–60. doi: 10.1042/bj2860755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S, Wu F, Liu S, Wu Q, Safe S. Role of specificity protein transcription factors in estrogen-induced gene expression in MCF-7 breast cancer cells. J Mol Endocrinol. 2007;39:289–304. doi: 10.1677/JME-07-0043. [DOI] [PubMed] [Google Scholar]
- Knowles BB, Howe CC, Aden DP. Human hepatocellular carcinoma cell lines secrete the major plasma proteins and hepatitis B surface antigen. Science. 1980;209:497–9. doi: 10.1126/science.6248960. [DOI] [PubMed] [Google Scholar]
- Kurlandsky SB, Gamble MV, Ramakrishnan R, Blaner WS. Plasma delivery of retinoic acid to tissues in the rat. J Biol Chem. 1995;270:17850–7. doi: 10.1074/jbc.270.30.17850. [DOI] [PubMed] [Google Scholar]
- Laliotis GP, Bizelis I, Argyrokastritis A, Rogdakis E. Cloning, characterization and computational analysis of the 5′ regulatory region of ovine glucose 6-phosphate dehydrogenase gene. Comp Biochem Physiol B Biochem Mol Biol. 2007;147:627–34. doi: 10.1016/j.cbpb.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Loudig O, Babichuk C, White J, Abu-Abed S, Mueller C, Petkovich M. Cytochrome P450RAI(CYP26) promoter: a distinct composite retinoic acid response element underlies the complex regulation of retinoic acid metabolism. Mol Endocrinol. 2000;14:1483–97. doi: 10.1210/mend.14.9.0518. [DOI] [PubMed] [Google Scholar]
- Loudig O, Maclean GA, Dore NL, Luu L, Petkovich M. Transcriptional co-operativity between distant retinoic acid response elements in regulation of Cyp26A1 inducibility. Biochem J. 2005;392:241–8. doi: 10.1042/BJ20050874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz JD, Dixit V, Yeung CK, Dickmann LJ, Zelter A, Thatcher JE, Nelson WL, Isoherranen N. Expression and functional characterization of cytochrome P450 26A1, a retinoic acid hydroxylase. Biochem Pharmacol. 2009;77:258–68. doi: 10.1016/j.bcp.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray WJ, Bain G, Yao M, Gottlieb DI. CYP26, a novel mammalian cytochrome P450, is induced by retinoic acid and defines a new family. J Biol Chem. 1997;272:18702–8. doi: 10.1074/jbc.272.30.18702. [DOI] [PubMed] [Google Scholar]
- Reichrath J, Lehmann B, Carlberg C, Varani J, Zouboulis CC. Vitamins as hormones. Horm Metab Res. 2007;39:71–84. doi: 10.1055/s-2007-958715. [DOI] [PubMed] [Google Scholar]
- Roberts AB, Lamb LC, Sporn MB. Metabolism of all-trans-retinoic acid in hamster liver microsomes: oxidation of 4-hydroxy- to 4-keto-retinoic acid. Arch Biochem Biophys. 1980;199:374–83. doi: 10.1016/0003-9861(80)90293-3. [DOI] [PubMed] [Google Scholar]
- Ross AC. Retinoid production and catabolism: role of diet in regulating retinol esterification and retinoic Acid oxidation. J Nutr. 2003;133:291S–296S. doi: 10.1093/jn/133.1.291S. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2001. [Google Scholar]
- Tay S, Dickmann L, Dixit V, Isoherranen N. A comparison of the roles of peroxisome proliferator-activated receptor and retinoic acid receptor on CYP26 regulation. Mol Pharmacol. 2010;77:218–27. doi: 10.1124/mol.109.059071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zolfaghari R, Ross AC. Cloning of rat cytochrome P450RAI (CYP26) cDNA and regulation of its gene expression by all-trans-retinoic acid in vivo. Arch Biochem Biophys. 2002;401:235–43. doi: 10.1016/S0003-9861(02)00043-7. [DOI] [PubMed] [Google Scholar]
- Wei LN. Retinoid receptors and their coregulators. Annu Rev Pharmacol Toxicol. 2003;43:47–72. doi: 10.1146/annurev.pharmtox.43.100901.140301. [DOI] [PubMed] [Google Scholar]
- White JA, Beckett-Jones B, Guo YD, Dilworth FJ, Bonasoro J, Jones G, Petkovich M. cDNA cloning of human retinoic acid-metabolizing enzyme (hP450RAI) identifies a novel family of cytochromes P450. J Biol Chem. 1997;272:18538–41. doi: 10.1074/jbc.272.30.18538. [DOI] [PubMed] [Google Scholar]
- Wolf AT, Medcalf RL, Jern C. The t-PA -7351C>T enhancer polymorphism decreases Sp1 and Sp3 protein binding affinity and transcriptional responsiveness to retinoic acid. Blood. 2005;105:1060–7. doi: 10.1182/blood-2003-12-4383. [DOI] [PubMed] [Google Scholar]
- Xi J, Yang Z. Expression of RALDHs (ALDH1A1s) and CYP26s in human tissues and durig the neural differentiation of P19 embryonal carcinoma stem cell. Gene Expression Patterns. 2008;8:438–42. doi: 10.1016/j.gep.2008.04.003. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Zolfaghari R, Ross AC. Regulation of CYP26 (cytochrome P450RAI) mRNA expression and retinoic acid metabolism by retinoids and dietary vitamin A in liver of mice and rats. Faseb J. 2000;14:2119–27. doi: 10.1096/fj.00-0061com. [DOI] [PubMed] [Google Scholar]
- Yasmin R, Yeung KT, Chung RH, Gaczynska ME, Osmulski PA, Noy N. DNA-looping by RXR tetramers permits transcriptional regulation “at a distance”. J Mol Biol. 2004;343:327–38. doi: 10.1016/j.jmb.2004.08.070. [DOI] [PubMed] [Google Scholar]
- Zolfaghari R, Cifelli CJ, Lieu SO, Chen Q, Li NQ, Ross AC. Lipopolysaccharide opposes the induction of CYP26A1 and CYP26B1 gene expression by retinoic acid in the rat liver in vivo. Am J Physiol Gastrointest Liver Physiol. 2007;292:G1029–36. doi: 10.1152/ajpgi.00494.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolfaghari R, Ross AC. Chronic vitamin A intake affects the expression of mRNA for apolipoprotein A-I, but not for nuclear retinoid receptors, in liver of young and aging Lewis rats. Arch Biochem Biophys. 1995;323:258–64. doi: 10.1006/abbi.1995.9966. [DOI] [PubMed] [Google Scholar]
- Zolfaghari R, Ross AC. Lecithin:retinol acyltransferase from mouse and rat liver. CDNA cloning and liver-specific regulation by dietary vitamin A and retinoic acid. J Lipid Res. 2000;41:2024–34. [PubMed] [Google Scholar]
- Zolfaghari R, Ross AC. An essential set of basic DNA response elements is required for receptor-dependent transcription of the lecithin:retinol acyltransferase (Lrat) gene. Arch Biochem Biophys. 2009;489:1–9. doi: 10.1016/j.abb.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]