Abstract
Nanotechnology is increasingly using both materials and nano-objects synthesized by living beings, most of them produced by microbial cells. Emerging technologies and highly integrative approaches (such as 'omics and systems biology), that have been largely proven successful for the production of proteins and secondary metabolites are now expected to become fully adapted for the improved biological production of nanostructured materials with tailored properties. The so far underestimated potential of microbial cell factories in nanotechnology and nanomedicine is expected to emerge, in the next years, in the context of novel needs envisaged in the nanoscience universe. This should prompt a careful revisiting of the microbial cell factories as the most versatile biological platforms to supply functional materials for nanotechnological applications.
Generally speaking, Nanotechnology refers to the fabrication, manipulation and utilization of submicron objects, particularly those between 1 and 100 nm. Physical and chemical sciences have developed tools and procedures to fabricate nanoscale entities with intriguing applications in electronics, material sciences and medicine. In the biomedical context, the relevance of Nanotechnology relies on the particular biophysical properties of nanoscale objects and their particular interaction with living beings such as high diffusion in organs and tissues, high surface-volume ratio, efficient uptake by mammalian cells and high biological impact in biological interfaces through mecano-transduction signaling [1] and a spectrum of alternative cell activities and responses [2]. The extraordinary bio-effectiveness of nanoparticles has in turn derived into a strong debate about their potential toxicity, when directly exposed to the human body or released to the environment [3], which is still unsolved. The biological impact of these manmade nanoscale entities and their suitability to be functionalized for specific binding or to act as carriers for therapeutics empowers a spectrum of specific applications in diagnosis and therapy, including imaging, biosensing, regenerative medicine, drug delivery and gene therapy. The clinically-oriented fabrication, tailoring and application of bio-active nanoparticles conceptually sustains the Nanomedicine framework.
Bionanotechnology (as well as Nanobiotechnolgy) are rather fuzzy terms whose overlapping meanings are under continuous refining, as their associated technologies and applications keep evolving. They are often understood as the generation of hybrid materials (deriving from chemical and biological synthesis), or bio-inspired materials [4]. In a different reading frame, Bionanotechnology can be observed as "Nanotechnology through Biotechnology" [5], that is, the bio-fabrication of nano-objects, or bi-functional macromolecules usable as tools to construct or manipulate nano-objects. Because of their wide physiological diversity, small size, genetic manipulability and controlled culturability, microbial cells are ideal producers of a diversity of nanostructures, materials and instruments for Nanosciences, ranging from fully natural products such as viruses, polymers and magnetosomes, to engineered proteins or protein constructs such as virus-like particles (VLPs), and peptide-displaying phages or cells and tailored metal particles (Figure 1).
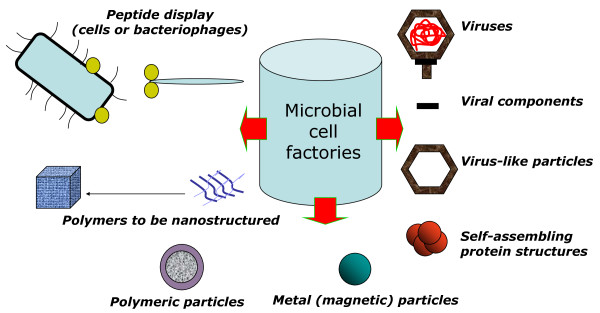
Figure 1.
Biosynthetic potential of Microbial Cell Factories in Nanosciences. Bacteria and other microbes are good producers of particulate entities with values in Nanotechnology in general and in Nanomedicine in particular. From top left, clockwise: First, cells themselves and their infecting viruses are used for peptide display (cell surface display or phage display technologies, respectively). Among other applications including selecting ligands for receptor mediated drug delivery, biosensing or imaging [27-31], peptide display show important promises in molecular biomimetics, to generate molecular links between synthetic and biological components of hybrid materials [32,33]. Also, animal, plant and bacterial viruses, being manageable at the nanoscale, are used as scaffolds for nanofabrication of electronic components [34] and as building blocks for functionalized surfaces [35,36], apart from their more conventional application as vehicles for the delivery of nucleic acids in gene therapy [37,38]. Interestingly, viral components as the DNA-packaging motor of phi29 bacteriophage [39] have been explored as vehicles in drug delivery. VLPs, produced in both eukaryotic microbes and in bacteria, apart from their conventional application in vaccination show promising potential as nano-containers for drug delivery [40]. Other protein self assembling complexes produced in bacteria such as flagella, explored to generate nanomotors or as templates for nanofabrication [41-43], or inclusion bodies, used as soft-matter scaffolds in tissue engineering [44-46] or as functional materials [47] are under deep exploration and further development. Magnetosomes from magnetotactic bacterial have shown exciting potentials in drug delivery, imaging and tissue engineering [48-53], while a diversity of metal nanoparticles produced in bacteria, whose properties can be tuned during production, are in use or under development for nano-electronics, therapy and imaging [54-60]. Main microbial polymers including polysaccharides, polyesters and polyamides can be nanostructured upon isolation from producing cells for uses in material sciences, while others, such as gellan, dextran, PHA and HA, are naturally produced as nanoparticulate materials [61-65], than can be further functionalized in vivo by the genetic engineering of producing cells [66,67].
In summary, microbial cells are natural producers of (or they can be easily adapted to synthesize) nano-sized entities with relevant biomedical applications, being the cell factories themselves promising tools for the emerging technologies and biomedical applications related with the use of nanosized entities. Microbial Call Factories, the journal, has addressed some of these relevant topics through primary research papers and reviews. For instance, Chen and coauthors [6] have recently shown how polyhydroxyalkanoate (PHA) nanosized granules are convenient instruments for protein purification, while other authors [7] have improved the production of clinically relevant microbial materials suitable for nanoparticle in vitro fabrication including alginate, hyaluronic acid (HA) and PHA [8,9].
Moreover, cell surface display technologies [10,11] as well as the engineering of bacterial particulate materials (such as spores) for peptide display [12] have been represented in our article list. Furthermore, the journal has often addressed protein quality issues [13-19], that are highly relevant to the design and production of protein complexes and protein nanoparticles, among which virus like particles (VLPs) [20], self-assembling silk-like proteins [21] and bacterial inclusion bodies [22-26] have been particularly studied.
However, the number of submissions dealing with Nanotechnological applications or instruments deriving from microbial cells and the spectrum of coverage of Nanoscience-related topics are still low. Therefore, the editorial board of Microbial Cell Factories is pleased to encourage all the authors working in microbiology, biomedicine and biomaterial sciences to consider the potential of the Cell Factory platforms and to submit their primary research data and reviews to the journal. As a fully settled but highly dynamic journal, Microbial Cell Factories is committed to fully cover emerging technologies and scientific areas of hot interest from which microbial products are relevant, provided the biological aspects of the production (the Cell Factory concept) of the model particles, structures or materials are conveniently stressed.
References
- Curtis AS, Dalby M, Gadegaard N. Cell signaling arising from nanotopography: implications for nanomedical devices. Nanomed. 2006;1:67–72. doi: 10.2217/17435889.1.1.67. [DOI] [PubMed] [Google Scholar]
- Jiang W, Kim BY, Rutka JT, Chan WC. Nanoparticle-mediated cellular response is size-dependent. Nat Nanotechnol. 2008;3:145–150. doi: 10.1038/nnano.2008.30. [DOI] [PubMed] [Google Scholar]
- Sanvicens N, Marco MP. Multifunctional nanoparticles--properties and prospects for their use in human medicine. Trends Biotechnol. 2008;26:425–433. doi: 10.1016/j.tibtech.2008.04.005. [DOI] [PubMed] [Google Scholar]
- Taylor PM. Biological matrices and bionanotechnology. Philos Trans R Soc Lond B Biol Sci. 2007;362:1313–1320. doi: 10.1098/rstb.2007.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarikaya M, Tamerler C, Jen AK, Schulten K, Baneyx F. Molecular biomimetics: nanotechnology through biology. Nat Mater. 2003;2:577–585. doi: 10.1038/nmat964. [DOI] [PubMed] [Google Scholar]
- Zhang S, Wang ZH, Chen GQ. Microbial polyhydroxyalkanote synthesis repression protein PhaR as an affinity tag for recombinant protein purification. Microb Cell Fact. 2010;9:28. doi: 10.1186/1475-2859-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez JA, Montemayor MI, Fraguas J, Murado MA. Hyaluronic acid production by Streptococcus zooepidemicus in marine by-products media from mussel processing wastewaters and tuna peptone viscera. Microb Cell Fact. 2010;9:46. doi: 10.1186/1475-2859-9-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M, Patel SK, Kalia VC. Bacillus subtilis as potential producer for polyhydroxyalkanoates. Microb Cell Fact. 2009;8:38. doi: 10.1186/1475-2859-8-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo E, Pena C, Nunez C, Segura D, Espin G. Molecular and bioengineering strategies to improve alginate and polydydroxyalkanoate production by Azotobacter vinelandii. Microb Cell Fact. 2007;6:7. doi: 10.1186/1475-2859-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao X, Jiang M, Yu Z, Cai H, Li L. Surface display of heterologous proteins in Bacillus thuringiensis using a peptidoglycan hydrolase anchor. Microb Cell Fact. 2009;8:48. doi: 10.1186/1475-2859-8-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford N, Mourez M. Surface display of proteins by gram-negative bacterial autotransporters. Microb Cell Fact. 2006;5:22. doi: 10.1186/1475-2859-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinc K, Isticato R, Dembek M, Karczewska J, Iwanicki A, Peszynska-Sularz G. Expression and display of UreA of Helicobacter acinonychis on the surface of Bacillus subtilis spores. Microb Cell Fact. 2010;9:2. doi: 10.1186/1475-2859-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Alonso M, Gonzalez-Montalban N, Garcia-Fruitos E, Villaverde A. Learning about protein solubility from bacterial inclusion bodies. Microb Cell Fact. 2009;8:4. doi: 10.1186/1475-2859-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Fruitos E, Gonzalez-Montalban N, Morell M, Vera A, Ferraz RM, Aris A. Aggregation as bacterial inclusion bodies does not imply inactivation of enzymes and fluorescent proteins. Microb Cell Fact. 2005;4:27. doi: 10.1186/1475-2859-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Marco A. Strategies for successful recombinant expression of disulfide bond-dependent proteins in Escherichia coli. Microb Cell Fact. 2009;8:26. doi: 10.1186/1475-2859-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolaj O, Spada S, Robin S, Wall JG. Use of folding modulators to improve heterologous protein production in Escherichia coli. Microb Cell Fact. 2009;8:9. doi: 10.1186/1475-2859-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen HP, Mortensen KK. Soluble expression of recombinant proteins in the cytoplasm of Escherichia coli. Microb Cell Fact. 2005;4:1. doi: 10.1186/1475-2859-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer-Miralles N, Domingo-Espin J, Corchero JL, Vazquez E, Villaverde A. Microbial factories for recombinant pharmaceuticals. Microb Cell Fact. 2009;8:17. doi: 10.1186/1475-2859-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser B, Saloheimo M, Rinas U, Dragosits M, Rodriguez-Carmona E, Baumann K. Protein folding and conformational stress in microbial cells producing recombinant proteins: a host comparative overview. Microb Cell Fact. 2008;7:11. doi: 10.1186/1475-2859-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurramkonda C, Adnan A, Gabel T, Lunsdorf H, Ross A, Nemani SK. Simple high-cell density fed-batch technique for high-level recombinant protein production with Pichia pastoris: Application to intracellular production of Hepatitis B surface antigen. Microb Cell Fact. 2009;8:13. doi: 10.1186/1475-2859-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibel T. Spider silks: recombinant synthesis, assembly, spinning, and engineering of synthetic proteins. Microb Cell Fact. 2004;3:14. doi: 10.1186/1475-2859-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peternel S, Grdadolnik J, Gaberc-Porekar V, Komel R. Engineering inclusion bodies for non denaturing extraction of functional proteins. Microb Cell Fact. 2008;7:34. doi: 10.1186/1475-2859-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabate R, Espargaro A, Saupe SJ, Ventura S. Characterization of the amyloid bacterial inclusion bodies of the HET-s fungal prion. Microb Cell Fact. 2009;8:56. doi: 10.1186/1475-2859-8-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrilli E, Giuliani M, Marino G, Tutino ML. Influence of production process design on inclusion bodies protein: the case of an Antarctic flavohemoglobin. Microb Cell Fact. 2010;9:19. doi: 10.1186/1475-2859-9-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurgen B, Breitenstein A, Urlacher V, Buttner K, Lin H, Hecker M. Quality control of inclusion bodies in Escherichia coli. Microb Cell Fact. 2010;9:41. doi: 10.1186/1475-2859-9-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lethanh H, Neubauer P, Hoffmann F. The small heat-shock proteins IbpA and IbpB reduce the stress load of recombinant Escherichia coli and delay degradation of inclusion bodies. Microb Cell Fact. 2005;4:6. doi: 10.1186/1475-2859-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petty NK, Evans TJ, Fineran PC, Salmond GP. Biotechnological exploitation of bacteriophage research. Trends Biotechnol. 2007;25:7–15. doi: 10.1016/j.tibtech.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Deutscher SL. Phage display in molecular imaging and diagnosis of cancer. Chem Rev. 2010;110:3196–3211. doi: 10.1021/cr900317f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balestrieri ML, Napoli C. Novel challenges in exploring peptide ligands and corresponding tissue-specific endothelial receptors. Eur J Cancer. 2007;43:1242–1250. doi: 10.1016/j.ejca.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Aina OH, Liu R, Sutcliffe JL, Marik J, Pan CX, Lam KS. From combinatorial chemistry to cancer-targeting peptides. Mol Pharm. 2007;4:631–651. doi: 10.1021/mp700073y. [DOI] [PubMed] [Google Scholar]
- Sidhu SS. Phage display in pharmaceutical biotechnology. Curr Opin Biotechnol. 2000;11:610–616. doi: 10.1016/S0958-1669(00)00152-X. [DOI] [PubMed] [Google Scholar]
- Sarikaya M, Tamerler C, Jen AK, Schulten K, Baneyx F. Molecular biomimetics: nanotechnology through biology. Nat Mater. 2003;2:577–585. doi: 10.1038/nmat964. [DOI] [PubMed] [Google Scholar]
- Tamerler C, Sarikaya M. Molecular biomimetics: nanotechnology and bionanotechnology using genetically engineered peptides. Philosophical Transactions of the Royal Society A-Mathematical Physical and Engineering Sciences. 2009;367:1705–1726. doi: 10.1098/rsta.2009.0018. [DOI] [PubMed] [Google Scholar]
- Mao C, Solis DJ, Reiss BD, Kottmann ST, Sweeney RY, Hayhurst A. Virus-based toolkit for the directed synthesis of magnetic and semiconducting nanowires. Science. 2004;303:213–217. doi: 10.1126/science.1092740. [DOI] [PubMed] [Google Scholar]
- Steinmetz NF, Bize A, Findlay KC, Lomonossoff GP, Manchester M, Evans DJ. Site-specific and Spatially Controlled Addressability of a New Viral Nanobuilding Block: Sulfolobus islandicus Rod-shaped Virus 2. Advanced Functional Materials. 2008;18:3478–3486. doi: 10.1002/adfm.200800711. [DOI] [Google Scholar]
- Steinmetz NF, Shah SN, Barclay JE, Rallapalli G, Lomonossoff GP, Evans DJ. Virus-templated silica nanoparticles. Small. 2009;5:813–816. doi: 10.1002/smll.200801348. [DOI] [PubMed] [Google Scholar]
- Edelstein ML, Abedi MR, Wixon J. Gene therapy clinical trials worldwide to 2007--an update. J Gene Med. 2007;9:833–842. doi: 10.1002/jgm.1100. [DOI] [PubMed] [Google Scholar]
- Evans DJ. Exploitation of plant and archaeal viruses in bionanotechnology. Biochem Soc Trans. 2009;37:665–670. doi: 10.1042/BST0370665. [DOI] [PubMed] [Google Scholar]
- Lee TJ, Schwartz C, Guo P. Construction of bacteriophage phi29 DNA packaging motor and its applications in nanotechnology and therapy. Ann Biomed Eng. 2009;37:2064–2081. doi: 10.1007/s10439-009-9723-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundy BC, Franciszkowicz MJ, Swartz JR. Escherichia coli-based cell-free synthesis of virus-like particles. Biotechnol Bioeng. 2008;100:28–37. doi: 10.1002/bit.21716. [DOI] [PubMed] [Google Scholar]
- Martel S, Felfoul O, Mohammadi M, Mathieu JB. Interventional procedure based on nanorobots propelled and steered by flagellated magnetotactic bacteria for direct targeting of tumors in the human body. Conf Proc IEEE Eng Med Biol Soc. 2008;2008:2497–2500. doi: 10.1109/IEMBS.2008.4649707. [DOI] [PubMed] [Google Scholar]
- Deplanche K, Woods RD, Mikheenko IP, Sockett RE, Macaskie LE. Manufacture of stable palladium and gold nanoparticles on native and genetically engineered flagella scaffolds. Biotechnol Bioeng. 2008;101:873–880. doi: 10.1002/bit.21966. [DOI] [PubMed] [Google Scholar]
- van den Heuvel MG, Dekker C. Motor proteins at work for nanotechnology. Science. 2007;317:333–336. doi: 10.1126/science.1139570. [DOI] [PubMed] [Google Scholar]
- Garcia-Fruitos E, Seras-Franzoso J, Vazquez E, Villaverde A. Tunable geometry of bacterial inclusion bodies as substrate materials for tissue engineering. Nanotechnology. 2010;21:205101. doi: 10.1088/0957-4484/21/20/205101. [DOI] [PubMed] [Google Scholar]
- Diez-Gil C, Krabbenborg S, Garcia-Fruitos E, Vazquez E, Rodriguez-Carmona E, Ratera I. The nanoscale properties of bacterial inclusion bodies and their effect on mammalian cell proliferation. Biomaterials. 2010;31:5805–5812. doi: 10.1016/j.biomaterials.2010.04.008. [DOI] [PubMed] [Google Scholar]
- Garcia-Fruitos E, Rodriguez-Carmona E, Diez-Gil C, Ferraz RM, Vazquez E, Corchero JL. Surface Cell Growth Engineering Assisted by a Novel Bacterial Nanomaterial. Advanced Materials. 2009;21:4249–4253. doi: 10.1002/adma.200900283. [DOI] [Google Scholar]
- Garcia-Fruitos E, Villaverde A. Friendly production of bacterial inclusion bodies. Korean Journal of Chemical Engineering. 2010;27:385–389. [Google Scholar]
- Corchero JL, Villaverde A. Biomedical applications of distally controlled magnetic nanoparticles. Trends Biotechnol. 2009;27:468–476. doi: 10.1016/j.tibtech.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Ding Y, Li J, Liu J, Yang J, Jiang W, Tian J. Deletion of the ftsZ-like gene results in the production of superparamagnetic magnetite magnetosomes in Magnetospirillum gryphiswaldense. J Bacteriol. 2010;192:1097–1105. doi: 10.1128/JB.01292-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohuchi S, Schuler D. In vivo display of a multisubunit enzyme complex on biogenic magnetic nanoparticles. Appl Environ Microbiol. 2009;75:7734–7738. doi: 10.1128/AEM.01640-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JB, Duan JH, Dai SL, Ren J, Guo L, Jiang W. Preparation and anti-tumor efficiency evaluation of doxorubicin-loaded bacterial magnetosomes: magnetic nanoparticles as drug carriers isolated from Magnetospirillum gryphiswaldense. Biotechnol Bioeng. 2008;101:1313–1320. doi: 10.1002/bit.22011. [DOI] [PubMed] [Google Scholar]
- Sun JB, Zhao F, Tang T, Jiang W, Tian JS, Li Y. High-yield growth and magnetosome formation by Magnetospirillum gryphiswaldense MSR-1 in an oxygen-controlled fermentor supplied solely with air. Appl Microbiol Biotechnol. 2008;79:389–397. doi: 10.1007/s00253-008-1453-y. [DOI] [PubMed] [Google Scholar]
- Lang C, Schuler D, Faivre D. Synthesis of Magnetite Nanoparticles for Bio- and Nanotechnology: Genetic Engineering and Biomimetics of Bacterial Magnetosomes. Macromol Biosci. 2007;7:144–151. doi: 10.1002/mabi.200600235. [DOI] [PubMed] [Google Scholar]
- Vaidyanathan R, Gopalram S, Kalishwaralal K, Deepak V, Pandian SR, Gurunathan S. Enhanced silver nanoparticle synthesis by optimization of nitrate reductase activity. Colloids Surf B Biointerfaces. 2010;75:335–341. doi: 10.1016/j.colsurfb.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Gurunathan S, Kalishwaralal K, Vaidyanathan R, Venkataraman D, Pandian SR, Muniyandi J. Biosynthesis, purification and characterization of silver nanoparticles using Escherichia coli. Colloids Surf B Biointerfaces. 2009;74:328–335. doi: 10.1016/j.colsurfb.2009.07.048. [DOI] [PubMed] [Google Scholar]
- Chen YL, Tuan HY, Tien CW, Lo WH, Liang HC, Hu YC. Augmented biosynthesis of cadmium sulfide nanoparticles by genetically engineered Escherichia coli. Biotechnol Prog. 2009;25:1260–1266. doi: 10.1002/btpr.199. [DOI] [PubMed] [Google Scholar]
- Kalishwaralal K, Deepak V, Pandian SRK, Gurunathan S. Biological synthesis of gold nanocubes from Bacillus licheniformis. Bioresource Technology. 2009;100:5356–5358. doi: 10.1016/j.biortech.2009.05.051. [DOI] [PubMed] [Google Scholar]
- Kalimuthu K, Suresh BR, Venkataraman D, Bilal M, Gurunathan S. Biosynthesis of silver nanocrystals by Bacillus licheniformis. Colloids Surf B Biointerfaces. 2008;65:150–153. doi: 10.1016/j.colsurfb.2008.02.018. [DOI] [PubMed] [Google Scholar]
- Husseiny MI, El-Aziz MA, Badr Y, Mahmoud MA. Biosynthesis of gold nanoparticles using Pseudomonas aeruginosa. Spectrochim Acta A Mol Biomol Spectrosc. 2007;67:1003–1006. doi: 10.1016/j.saa.2006.09.028. [DOI] [PubMed] [Google Scholar]
- Gericke M, Pinches A. Microbial production of gold nanoparticles. Gold Bulletin. 2006;39:22–28. [Google Scholar]
- Chen CQ. A microbial polyhydroxyalkanoates (PHA) based bio- and materials industry. Chem Soc Rev. 2009;38(8):2434–2446. doi: 10.1039/b812677c. Ref Type: Generic. [DOI] [PubMed] [Google Scholar]
- Atwood JA, Rehm BH. Protein engineering towards biotechnological production of bifunctional polyester beads. Biotechnol Lett. 2009;31:131–137. doi: 10.1007/s10529-008-9836-9. [DOI] [PubMed] [Google Scholar]
- Ossipov DA. Nanostructured hyaluronic acid-based materials for active delivery to cancer. Expert Opin Drug Deliv. 2010;7(6):681–703. doi: 10.1517/17425241003730399. [DOI] [PubMed] [Google Scholar]
- Ibrahim HK, El-Leithy IS, Makky AA. Mucoadhesive Nanoparticles as Carrier Systems for Prolonged Ocular Delivery of Gatifloxacin/Prednisolone Bitherapy. Mol Pharm. 2010;7:576–585. doi: 10.1021/mp900279c. [DOI] [PubMed] [Google Scholar]
- Mellors R, Benzeval I, Eisenthal R, Hubble J. Preparation of self-assembled microspheres and their potential for drug delivery. Pharmaceutical Development and Technology. 2010;15:105–111. doi: 10.3109/10837450903036163. [DOI] [PubMed] [Google Scholar]
- Yao YC, Zhan XY, Zhang J, Zou XH, Wang ZH, Xiong YC. A specific drug targeting system based on polyhydroxyalkanoate granule binding protein PhaP fused with targeted cell ligands. Biomaterials. 2008;29:4823–4830. doi: 10.1016/j.biomaterials.2008.09.008. [DOI] [PubMed] [Google Scholar]
- Grage K, Jahns AC, Parlane N, Palanisamy R, Rasiah IA, Atwood JA. Bacterial Polyhydroxyalkanoate Granules: Biogenesis, Structure, and Potential Use as Nano-/Micro-Beads in Biotechnological and Biomedical Applications. Biomacromolecules. 2009;10:660–669. doi: 10.1021/bm801394s. [DOI] [PubMed] [Google Scholar]