Abstract
The IL23/IL17 pathway is pivotal in the development of chronic mucosal inflammation seen in Crohn’s disease (CD). Genetic variants in the IL23R and IL12B have been associated with CD susceptibility. We investigated ten genes within the IL23/IL17 pathway in a case-control study of 763 CD cases and 254 healthy controls. We identified novel association in haplotypes in IL17A (empirical p value = 0.02), IL17RA (p = 0.001), IL17RD (p = 0.001), IL12RB1 (p = 0.003) and IL12RB2 (p = 0.001) as well as confirming the association with IL12B variants (p = 0.003). The cumulative risk for carrying increased number of CD risk haplotypes from genes in this pathway rises to an odds ratio of 4.3 for carrying 5 risk haplotypes. We have previously demonstrated an association between this cohort and IL23R haplotypes. Pairwise analyses suggest that there is statistical interaction between variants in IL17A and IL23R (p = 0.047) and between variants in IL17RA and IL23R (p = 0.036). Furthermore, a significant association between CD and the widely replicated IL23R variants is only seen in the presence of IL17A or IL17RA variants. These data support the investigation of pathways implicated in CD pathogenesis in order to identify further susceptibility genes and also suggest that important gene-gene interaction is present in CD susceptibility.
Introduction
The novel interleukin IL23 was identified as recently as 2000(1). IL23 is a heterodimer cytokine composed of a p40 subunit (shared with IL12) and the unique p19 subunit encoded by the IL23 gene. Recent genetic findings have confirmed previous immunological data that have implicated the IL23/IL17 pathway in inflammatory bowel disease (IBD) pathogenesis. In 2006, a number of studies identified the pivotal role of IL23 in driving intestinal inflammation via inflammatory mediators such as IL17(2). The IL23 receptor (IL23R) is expressed on activated myeloid and T cells and IL23 is essential for maintaining the Th17 response(3). IL17, the pivotal Th17 cytokine, is found at increased levels in IBD(4), and in a model of bacteria-induced T cell dependant colitis, IL23 was implicated in driving both IFNγ and IL17 thereby suggesting that IL23 plays a pivotal role in the development of mucosal inflammation(2).
Further evidence of the key role of IL23 in mucosal inflammation was provided in 2006 when an association between genetic variants in the IL23 receptor (IL23R) gene and IBD in Caucasians was identified by The North American IBD Genetics Consortium in a genome-wide association study (GWAS)(5). Importantly this association with Crohn’s disease (CD) has been widely replicated in diverse Caucasian populations(6-10). The original association with a single non-synonymous single nucleotide polymorphism (SNP) has been supplemented with the finding that variants within the gene have a much greater contribution to disease susceptibility when haplotype-based analyses are performed(11). A recently published meta-analysis of three CD GWASs not only confirmed the original association with IL23R, but also identified that genetic variants within IL12B, a downstream member of the IL23 pathway, are also implicated in Crohn’s disease pathogenesis, further highlighting the contribution of this pathway to the development of disease(12). These data suggest that it would be worthwhile to further examine genes within the IL23/IL17 pathway in CD susceptibility. CD is a genetically complex disease where there are gene/environmental interactions and possibly gene/gene interactions (epistasis).There is increasing evidence from other genetically complex diseases, such as schizophrenia, that investigation of extended pathways known to contain susceptibility genes may reveal novel susceptibility variants(13)
The aim of this study was to examine the role of genes in the IL23/Th17 pathway in CD susceptibility and to investigate for any evidence of epistasic interaction between these genes.
MATERIALS AND METHODS
Subjects
Following informed consent and approval by the Cedars-Sinai Medical Center Institutional Review Board, subjects were recruited at the Cedars-Sinai Medical Center Inflammatory Bowel Disease center. Diagnosis of CD was confirmed using a combination of standard endoscopic, histological, and radiographic features (14). Controls were mainly spouses and acquaintances of CD subjects, and there was no difference in mean age between CD subjects and controls. In keeping with our previously published work, Ashkenazi Jewish ethnicity was assigned when at least one grandparent was of Ashkenazi Jewish origin (15, 16).
Selection of Candidate Genes and SNPs
Genes were included for investigation if they were known to play a role in the IL23/IL17 pathway(17). Specifically, the following ten genes were chosen: IL12B, IL17A, IL17RA, IL17RC, IL17RD, IL17F, IL17C, IL17D, IL12RB1 and IL12RB2. Data from previous investigation of IL23R were also available from a prior study(11). SNPs were selected using “Tagger” (18) and data from the International HapMap Project (19, 20). SNPs that “tagged” major Caucasian haplotypes and that were compatible with Illumina technology were genotyped. Since we were interested in major genetic effects rather than rare alleles, the goal of “tagging” was to find a set of tagSNPs in linkage disequilibrium with all SNPs in the HapMap data with a minor allele frequency ≥ 5%; in some cases this goal was not completely met due to the limitations of the Illumina technology. We also included non-synonymous SNPs with a minor allele frequency greater than 3% and, when available, additional markers listed on the SeattleSNPs website (http://pga.mbt.washington.edu/welcome.html). This redundancy was added to compensate for marker failure in the large-scale genotyping.
Genotyping
DNA was isolated from Epstein Barr virus transformed lymphoblastoid cell lines using proteinase K digestion, organic extraction, and ethanol precipitation (21). Single nucleotide markers (SNPs) were genotyped using the oligonucleotide ligation assay, Illumina Golden Gate technology (22) (Illumina, San Diego, CA). Details of all SNPs studied are given in Table 1 and supplementary table 1.
Table 1.
Association of IL17-IL23 pathway-related haplotypes with CD
| Gene | Percent Controls with Haplotype Present |
Percent CD with Haplotype Present |
p empirical* |
Odds Ratio |
95% Confidence Interval |
Estimate of Population Attributable Risk or of Prevented Fraction |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n=257 | n=763 | |||||||||||||
|
IL17A 1) all subjects |
rs2275913 | rs3819025 | rs10484879 | rs1974226 | ||||||||||
| H1 | G | G | C | G | 68.1% | 67.8% | ||||||||
| H2 | A | G | A | G | 37.0% | 41.3% | ||||||||
| H3 | G | G | C | A | 32.7% | 37.4% | ||||||||
| protective | H4 | A | G | C | G | 20.5% | 13.5% | 0.010 | 0.020 | 0.61 | 0.42,0.88 | − 0.08 | ||
| H5 | G | A | C | G | 11.0% | 9.8% | ||||||||
| 2) Ashkenazi Jewish subjects | rs2275913 | rs3819025 | rs10484879 | rs1974226 | ||||||||||
| H1 | G | G | C | G | 58.8% | 68.8% | ||||||||
| protective | H2 | A | G | A | G | 56.9% | 40.1% | 0.017 | 0.018 | 0.51 | 0.28,0.92 | −0.28 | ||
| H3 | G | G | C | A | 33.3% | 39.2% | ||||||||
| H4 | A | G | C | G | 5.9% | 9.9% | ||||||||
| H5 | G | A | C | G | 13.7% | 11.2% | ||||||||
| 3) non-Jewish subjects | rs2275913 | rs3819025 | rs10484879 | rs1974226 | ||||||||||
| H1 | G | G | C | G | 70.4% | 67.0% | ||||||||
| risk | H2 | A | G | A | G | 32.0% | 42.1% | 0.012 | 0.009 | 1.54 | 1.12,2.26 | + 0.15 | ||
| H3 | G | G | C | A | 32.5% | 36.1% | ||||||||
| protective | H4 | A | G | C | G | 24.1% | 16.0% | 0.009 | 0.018 | 0.59 | 0.39,0.89 | − 0.10 | ||
| H5 | G | A | C | G | 10.3% | 8.9% | ||||||||
| IL17RA (all subjects) | ||||||||||||||
| 1) Block 1 | rs5748864 | rs6518660 | ||||||||||||
| H1 | G | A | 92.5% | 95.0% | ||||||||||
| H2 | A | G | 27.2% | 24.1% | ||||||||||
| protective | H3 | A | A | 15.8% | 7.5% | 0.000 | 0.001 | 0.43 | 0.28,0.67 | − 0.09 | ||||
| 2) Block 2 | rs721930 | rs2241046 | rs2241049 | rs879574 | rs879577 | rs882643 | ||||||||
| H1 | C | A | A | T | A | G | 40.6% | 38.8% | ||||||
| H2 | G | A | G | T | G | G | 31.5% | 37.0% | ||||||
| H3 | C | G | A | T | G | G | 36.6% | 35.0% | ||||||
| risk | H4 | C | A | A | A | G | G | 18.9% | 27.0% | 0.006 | 0.007 | 1.59 | 1.12,2.26 | + 0.10 |
| H5 | C | A | G | T | G | C | 27.2% | 24.1% | ||||||
| IL17RD (all subjects) | ||||||||||||||
| 1) Block 1 | rs6809523 | rs2129821 | rs17057718 | rs6780995 | rs747089 | rs6810042 | ||||||||
| H1 | A | T | C | A | C | G | 64.7% | 71.2% | ||||||
| protective | H2 | A | T | C | A | C | A | 50.2% | 39.1% | 0.002 | 0.001 | 0.64 | 0.48,0.85 | −0.18 |
| H3 | G | T | T | G | T | G | 30.5% | 31.5% | ||||||
| H4 | G | G | C | G | C | G | 17.7% | 19.8% | ||||||
| 2) Block 2 | rs12495640 | rs6788981 | rs7374667 | |||||||||||
| H1 | T | C | T | 49.0% | 54.8% | |||||||||
| risk | H2 | T | C | C | 45.4% | 55.0% | 0.009 | 0.006 | 1.47 | 1.1,1.97 | +0.17 | |||
| protective | H3 | C | C | C | 47.4% | 37.8% | 0.008 | 0.007 | 0.68 | 0.51,0.90 | −0.15 | |||
| H4 | C | T | C | 28.1% | 27.6% | |||||||||
| IL12B (all subjects) | ||||||||||||||
| rs3212227 | rs2853694 | |||||||||||||
| protective | H1 | A | A | 77.2% | 68.3% | 0.008 | 0.003 | 0.64 | 0.46,0.87 | − 0.28 | ||||
| H2 | A | C | 47.2% | 54.0% | ||||||||||
| H3 | C | C | 39.0% | 36.8% | ||||||||||
| IL12RB1 (all subjects) | ||||||||||||||
| rs375947 | rs436857 | |||||||||||||
| risk | H1 | A | G | 83.5% | 90.2% | 0.007 | 0.003 | 1.82 | 1.21,2.73 | +0.40 | ||||
| H2 | G | A | 37.0% | 35.5% | ||||||||||
| H3 | G | G | 24.4% | 24.8% | ||||||||||
|
IL12RB2 1) all subjects |
rs1495964 | rs1908632 | rs11209063 | |||||||||||
| H1 | T | T | T | 79.0% | 78.5% | |||||||||
| H2 | C | T | T | 33.9% | 31.9% | |||||||||
| risk | H3 | C | G | T | 21.4% | 30.3% | 0.010 | 0.002 | 1.6 | 1.14,2.24 | +0.11 | |||
| protective | H4 | C | G | G | 24.9% | 16.6% | 0.010 | 0.05 | 0.6 | 0.43,0.84 | −0.10 | |||
| 2) Ashkenazi Jewish subjects | rs1495964 | rs1908632 | rs11209063 | |||||||||||
| risk | H1 | T | T | T | 64.2% | 78.6% | 0.017 | 0.017 | 2.04 | 1.1,3.8 | +0.4 | |||
| H2 | C | T | T | 32.1% | 26.5% | |||||||||
| H3 | C | G | T | 22.6% | 39.8% | 0.043 | 0.023 | 2.25 | 1.14,4.46 | +0.22 | ||||
| protective | H4 | C | G | G | 45.3% | 19.9% | 0.003 | 0.001 | 0.3 | 0.16,0.55 | −0.32 | |||
| 3) non- Jewish subjects |
rs1495964 | rs1908632 | rs11209063 | |||||||||||
| H1 | T | T | T | 82.5% | 79.4% | |||||||||
| H2 | C | T | T | 35.4% | 34.9% | |||||||||
| H3 | C | G | T | 20.4% | 23.0% | |||||||||
| H4 | C | G | G | 19.4% | 16.1% | |||||||||
Haplotypes with frequency > 5% are shown
Variants are reported as the nucleotide on the forward strand of the NCBI Genome Build 36 and dbSNP v 126.
p empirical obtained by 1000 permutations of CD status with respect to haplotype (see Methods).
Statistical Analyses
HAPLOVIEW was used to determine the structure of haplotype blocks and to test for association between haplotypes and CD(23). The association with CD and the presence of a haplotype was tested using the chi-square test, with estimation of the odds ratio and the 95% confidence interval for the haplotype effect. In order to assess the impact of multiple testing on the results, corrected P values were calculated by performing 1,000 random permutations of disease status for each tested haplotype and the proportion of permuted tests for which the value of the chi-square statistic was greater than for the actual or observed test. Results with significance were defined by p < 0.05 by permutation test. Individual haplotypes were reconstructed using PHASE (v2) by assigning each haplotype with maximum probability(24). Sixty-seven percent of haplotype assignments had probabilities of 100% and 98.5% had probabilities greater than 90%.
Mantel-Haenszel and logistic regression methods were used to determine the interaction between haplotypes in different genes (25). In these analyses the results reported are for Jewish and non-Jewish subjects combined due to the fact that in the majority of cases there were not significant differences between Jewish and non-Jewish subjects (Table 1). The exceptions to this were: (1) that an IL17A “risk” haplotype specific to the non-Jewish population was identified and used for subsequent gene-gene interaction studies; and (2) that an IL12RB2 haplotype was observed specific to the Ashkenazi Jewish population and is reported below. Population attributable risk (PAR) or prevented fraction (PF) was estimated by assuming: (1) the frequency of a particular haplotype in the controls reflected the population frequency of that haplotype; and (2) the odds ratio for the association of a given haplotype reflected the relative risk of that haplotype for Crohns disease (26, 27). Thus, PAR = Phap (OR-1)/[Phap (OR-1)+1], when OR >1, or PF = Phap(1-OR), when OR <1, where Phap = frequency of a risk or preventive haplotype in the control group.
Haplotypes with frequency ≥ 5% are numbered in order of frequency (H1, H2, and so on). Gene-gene interactions were tested by using an interaction test in logistic regression models (28).
RESULTS
We genotyped 763 CD subjects and 254 controls for SNPs in IL12B, IL17A, IL17RA, IL17RC, IL17RD, IL17F, IL17C, IL17D, IL12RB1 and IL12RB2 (for full list of SNPs please see table 1 and supplementary table 1). In addition to the associations demonstrated here with CD and genetic variation within the IL23/IL17 pathway, we have previously published an association between this CD cohort and haplotypes within the gene encoding the IL23 receptor (IL23R)(11) (it is important to stress that these IL23R results are included in this paper not as independent replication of this locus but in order to perform the ‘pathway’ analyses and to assess for gene-gene interaction). These previous analyses suggested that IL23R variants make a substantial contribution to CD susceptibility with a PAR of approximately 20%(11).
We found no association between CD and genetic variation in IL17RC (6 SNPs), IL17F (6 SNPs), IL17C (1 SNP) and IL17D (4 SNPs) (supplementary table 1.).
We found both risk and protective CD associated haplotypes in IL17A, IL17RA, IL17RD, IL12B, IL12RB1 and IL12RB2 and CD (table 1). In addition there was borderline association between an individual IL17RD SNP (rs768713, OR 0.80 (95% confidence intervals 0.65 – 0.98) and CD, an association that did not contribute to any of the haplotypes listed in table 1.
The IL17A haplotypes demonstrate ethnic differences as the H2 haplotype (rs2275913, rs3819025, rs10484879, rs1974226) is a protective haplotype in Ashkenazi Jewish CD (OR 0.51 (0.28 – 0.92)), whereas it is a risk haplotype in the non-Jewish CD (OR 1.54 (1.12 – 2.26)). There is an analogous but converse finding in IL12RB2, where the protective H4 (rs1495964, rs1908632, rs11209063) haplotypic effect is seen in Ashkenazi Jews but not in non-Jewish cases (Table 1).
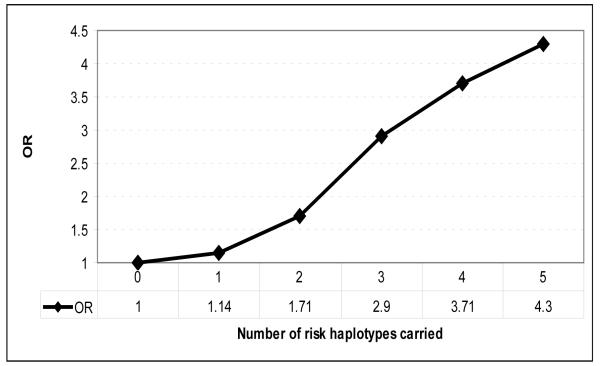
The cumulative risk of carrying an increased number of risk haplotypes from IL23R, IL17A, IL17RA, IL12RB1 and IL17RD in this cohort demonstrates an additive risk for CD as the number of risk haplotypes increases (Figure 1). Assuming an odds ratio of 1 for carriage of no risk haplotypes, the risk increases to an odds ratio of 4.3 for carriage of 5 risk haplotypes. Whilst limited power means that the individual analyses confidence intervals are wide and cross 1, the proportion trend test for this analyses reveals a highly significant result with a p value = 1.7 × 10−7 (Figure 1).
Figure 1. Increasing odds ratios for CD with increasing carriage of risk haplotypes.
Odds ratio for CD for the presence of 0, 1, 2, 3, 4 or 5 risk haplotypes from IL23R, IL17A, IL17RA, IL12RB1 and IL17RD. Proportion trend test p value = 1.7 × 10−7. For haplotype assignments, see Table 1.
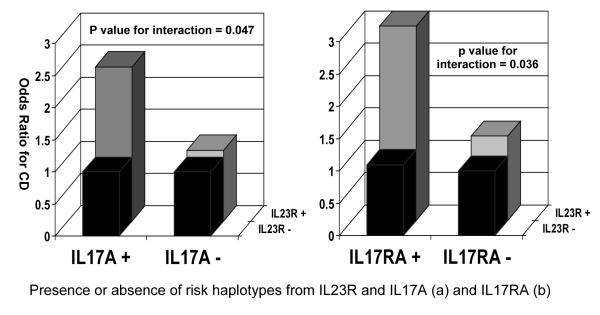
Further evidence of the synergistic effect of the genes in this pathway in the development of CD can be seen in table 2 a, b and c and figure 2 when the haplotypes are analyzed in pairs. These data should be contrasted with the association of Il23/IL17 pathway haplotypes with CD when analyzed individually (Table 1). In non-Jewish patients with CD, a significant increase in CD risk is seen for an IL23R risk haplotype, Block 2 Haplotype 1 or Block 3 Haplotype 1, only when the risk haplotype for IL17A, Haplotype 2, is also present (Table 2a, p for Mantel-Haenszel test = 0.0017). A formal statistical interaction between these two risk haplotypes is significant, despite the modest size of sample available for this analysis (Table 2a, p for interaction=0.047). By a similar analysis, a significant increase in CD risk is seen for an IL23R risk haplotype only when the IL17RA risk haplotype is also present (Table 2b, p for Mantel-Haenszel test = 0.0003, p interaction also significant at p=0.036). In addition a significant decrease in CD risk is seen for an IL23R protective haplotype, Block 2 Haplotype 2 or Block 3 Haplotype 3, only when the protective haplotype of IL12RB2, haplotype 4, is also present (Table 2c, p for Mantel-Haenszel < 0.0001). A significant statistical interaction between these two protective haplotypes was also observed (p interaction = 0.001). These data suggest that the widely replicated IL23R risk associations with CD may be limited to individuals who also ‘carry’ other genetic variants within the IL17/IL23R pathway.
Table 2.
Synergistic Interaction Between IL23R and IL17A, and IL23R and IL17RA
| a) IL23R and IL17A | |||||||
|---|---|---|---|---|---|---|---|
| Interaction between IL23R risk haplotypes and IL17A risk haplotype in non-Jewish subjects. | |||||||
| Presence of IL23R Block 2 H1 or IL23R Block 3 H1 |
Presence of IL17A H2 |
CD | Control | Odds Ratio |
95% Confidence Interval |
Mantel- Haenszel P value |
Interaction P value |
| No | No | 90 | 52 | 1 | 0.0017 | 0.047 | |
| No | Yes | 52 | 30 | 1.0 | 0.6-1.8 | ||
| Yes | No | 166 | 84 | 1.1 | 0.7-1.8 | ||
| Yes | Yes | 133 | 32 | 2.4 | 1.4-4.0 | ||
| b) IL23R and IL17RA | |||||||
|---|---|---|---|---|---|---|---|
| Interaction between IL23R risk haplotypes and IL17RA risk haplotype in all subjects. | |||||||
| Presence of IL23R Block 2 H1 or IL23R Block 3 H1 |
Presence of IL17RA H4 |
CD | Control | Odds Ratio |
95% Confidence Interval |
Mantel- Haenszel P value |
Interaction P value |
| No | No | 175 | 78 | 1 | 0.0003 | 0.036 | |
| No | Yes | 65 | 27 | 1.1 | 0.6-1.8 | ||
| Yes | No | 370 | 126 | 1.3 | 0.9-1.8 | ||
| Yes | Yes | 138 | 20 | 3.0 | 1.8-5.2 | ||
| c) IL23R and IL12RB2 | |||||||
|---|---|---|---|---|---|---|---|
| Interaction between IL23R risk haplotypes and IL12RB risk haplotype in all subjects. | |||||||
| IL12RB2 Haplotype 4 |
Presence of IL23R Block 2 H2 |
CD | Control | OR | 95% Confidence Interval |
Mantel- Haenszel P value |
Interaction P value |
| No | No | 294 | 55 | 1 | 0.001 | ||
| No | Yes | 329 | 132 | 0.47 | 0.33 - 0.66 | ||
| Yes | No | 45 | 32 | 0.26 | 0.15 - 0.45 | <0.0001 | |
| Yes | Yes | 76 | 29 | 0.49 | 0.29 - 0.82 | ||
Figure 2. Summary of genetic interaction between (a) IL23R and IL17 haplotypes and (b) IL23R and IL17RA haplotypes.
Pair-wise analysis demonstrate association between IL23R block 2 HI or block 3 H1 and IL17A (p = 0.047 for interaction) (a) and IL17RA (p = 0.036 for interaction) (b) haplotypes demonstrating that risk haplotypes from both genes are required for disease susceptibility.
DISCUSSION
Previously published data using this cohort of subjects have demonstrated that for CD there are both “risk” and “protective” haplotypes in IL23R(11). These haplotypic analyses substantially increase the estimate of the population attributable risk for the IL23R gene to the order of ~20% and reinforce the importance of IL23R in CD susceptibility. The significant genetic associations and high population attributable risks reported in this paper support the hypothesis that, in addition to IL23R, other genes in the IL17-IL23 pathway are major contributors to CD susceptibility. In addition to IL23R, associations were also observed between CD and common haplotypes in IL17A, IL17RA, IL17RD, IL12B, IL12RB1 and IL12RB2 (Table 2). A “risk” haplotype, conferring a greater susceptibility to CD, and a “protective” haplotype, conferring a reduced susceptibility to CD, were observed with IL17A and with IL17RA, with a population attributable risk (PAR) for each of the order of ~10%, and a “risk” haplotype was observed with IL12RB1 (PAR ~40%). Furthermore, “risk” haplotypes of IL23R and IL17A and of IL23R and IL17RA synergistically interacted to increase CD risk (Table 2 and figure 2). The analysis herein using logistic regression modeling suggested that the increased risk for IL23R SNPs observed previously and for IL23R “risk” haplotypes in this report required the presence of additional “risk” haplotypes from IL17A or IL17RA. We have also demonstrated that specific haplotypes in genes can have differing effects on disease susceptibility in different ethnic groups, as an IL17A haplotype (H2) appears to be a risk haplotype in the non-Jewish subjects and a protective haplotype in the Jewish subjects. These findings substantiate similar findings that we have published in the TNFSF15 gene(29) and emphasize the need for appropriate accounting of ethnicity within genetic studies.
Further support for the importance of the IL17-IL23 pathway-related variants in CD susceptibility was the observation of an increasing odds ratio for CD as the “risk” haplotypes for these genes were combined. While the OR for each “risk” haplotype alone is modest, of the order of 1.4-1.6, the combined OR for the identified haplotypes is substantial, being greater than 4 (Figure 1).
A limitation of this analysis is the relatively limited power as the number of subjects in the sub-groups becomes modest with the increase in the number of risk and protective haplotypes to be analyzed in combination. The main observation of this study, namely that genes in the IL23-IL17 pathway contribute to genetic susceptibility for CD, is robust and is supported by data from the GWAS meta-analysis(12); however, many of the details on interactions and the calculations of population attributable risks should be considered provisional estimates or hypotheses at this time until confirmation in other cohorts of subjects have been reported. The impact of the number of haplotypes studied on the “multiple testing problem,” that positive results may appear by chance and not by the presence of a true association, was considered for the analyses of this paper by comparing the significance of results with ~1,000 random permutations of the CD phenotype with the haplotype assignments.
The technological advances that have facilitated high-throughput and cost-effective genotyping and the increased understanding of the genetic structure of the human genome through the HapMap project have greatly accelerated gene-finding for non-Mendelian/complex genetic traits. However it is becomingly increasingly clear that identifying individual genes is only the first step in unraveling the genetic contribution to diseases such as CD. The observations in this study suggest that further genetic analysis of the pathway related to the original “hit” may uncover still more genetic variants that contribute substantially to susceptibility for the particular phenotype under study. The data also suggest that more complex analyses of related genes may need to be performed if subtle genetic interactions are to be revealed. These types of analyses may also reveal hitherto unknown genetic associations that only ‘reveal’ themselves in these types of analyses.
In summary we have identified novel genetic variants within the IL23/IL17 pathway that contribute to CD susceptibility. Further work to consolidate these findings in other populations and to elucidate the ‘functional’ consequences for the IL23/IL17 pathway of these genetic variants are warranted.
Supplementary Material
Acknowledgments
Financial support: NIH/NIDDK grant P01 DK046763; Cedars-Sinai Medical Center Inflammatory Bowel Disease Research Funds; The Feinstein Family Chair in IBD (SRT); The Cedars-Sinai Board of Governors’ Chair in Medical Genetics (JIR). Genotyping was supported in part by M01-RR00425 to the Cedars-Sinai GCRC genotyping core and by DK62413 (KDT).
References
- 1.Oppmann B, Lesley R, Blom B, et al. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity. 2000;13:715–725. doi: 10.1016/s1074-7613(00)00070-4. [DOI] [PubMed] [Google Scholar]
- 2.Yen D, Cheung J, Scheerens H, et al. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. J Clin Invest. 2006;116:1310–1316. doi: 10.1172/JCI21404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mangan PR, Harrington LE, O’Quinn DB, et al. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 2006;441:231–234. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- 4.Fujino S, Andoh A, Bamba S, et al. Increased expression of interleukin 17 in inflammatory bowel disease. Gut. 2003;52:65–70. doi: 10.1136/gut.52.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duerr RH, Taylor KD, Brant SR, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buning C, Schmidt HH, Molnar T, et al. Heterozygosity for IL23R p.Arg381Gln confers a protective effect not only against Crohn’s disease but also ulcerative colitis. Aliment Pharmacol Ther. 2007;26:1025–1033. doi: 10.1111/j.1365-2036.2007.03446.x. [DOI] [PubMed] [Google Scholar]
- 7.Glas J, Seiderer J, Wetzke M, et al. rs1004819 is the main disease-associated IL23R variant in German Crohn’s disease patients: combined analysis of IL23R, CARD15, and OCTN1/2 variants. PLoS ONE. 2007;2:e819. doi: 10.1371/journal.pone.0000819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberts RL, Gearry RB, Hollis-Moffatt JE, et al. IL23R R381Q and ATG16L1 T300A are strongly associated with Crohn’s disease in a study of New Zealand Caucasians with inflammatory bowel disease. Am J Gastroenterol. 2007;102:2754–2761. doi: 10.1111/j.1572-0241.2007.01525.x. [DOI] [PubMed] [Google Scholar]
- 9.Dubinsky MC, Wang D, Picornell Y, et al. IL-23 receptor (IL-23R) gene protects against pediatric Crohn’s disease. Inflamm Bowel Dis. 2007;13:511–515. doi: 10.1002/ibd.20126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baptista ML, Amarante H, Picheth G, et al. CARD15 and IL23R influences Crohn’s disease susceptibility but not disease phenotype in a Brazilian population. Inflamm Bowel Dis. 2008 doi: 10.1002/ibd.20372. [DOI] [PubMed] [Google Scholar]
- 11.Taylor KD, Targan SR, Mei L, et al. IL23R haplotypes provide a large population attributable risk for Crohn’s disease. Inflamm Bowel Dis. 2008;14:1185–1191. doi: 10.1002/ibd.20478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barrett JC, Hansoul S, Nicolae DL, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hennah W, Tomppo L, Hiekkalinna T, et al. Families with the risk allele of DISC1 reveal a link between schizophrenia and another component of the same molecular pathway, NDE1. Hum Mol Genet. 2007;16:453–462. doi: 10.1093/hmg/ddl462. [DOI] [PubMed] [Google Scholar]
- 14.Mow WS, Vasiliauskas EA, Lin YC, et al. Association of antibody responses to microbial antigens and complications of small bowel Crohn’s disease. Gastroenterology. 2004;126:414–424. doi: 10.1053/j.gastro.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 15.Roth MP, Petersen GM, McElree C, et al. Familial empiric risk estimates of inflammatory bowel disease in Ashkenazi Jews. Gastroenterology. 1989;96:1016–1020. doi: 10.1016/0016-5085(89)91618-1. [DOI] [PubMed] [Google Scholar]
- 16.Yang H, McElree C, Roth MP, et al. Familial empirical risks for inflammatory bowel disease: differences between Jews and non-Jews. Gut. 1993;34:517–524. doi: 10.1136/gut.34.4.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weaver CT, Hatton RD, Mangan PR, et al. IL-17 family cytokines and the expanding diversity of effector T cell lineages. Annu Rev Immunol. 2007;25:821–852. doi: 10.1146/annurev.immunol.25.022106.141557. [DOI] [PubMed] [Google Scholar]
- 18.de Bakker PI, Yelensky R, Pe’er I, et al. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 19.International HapMap Consortium The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 20.de Bakker P. 2004.
- 21.Sambrook J, Fritsch E, Maniatis T. Molecular Cloning. Cold Spring Harbor Laboratory; New York: 1989. [Google Scholar]
- 22.Shen R, Fan JB, Campbell D, et al. High-throughput SNP genotyping on universal bead arrays. Mutat Res. 2005;573:70–82. doi: 10.1016/j.mrfmmm.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 23.Barrett JC, Fry B, Maller J, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 24.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuritz SJ, Landis JR, Koch GG. A general overview of Mantel-Haenszel methods. Ann Rev Public Health. 1988;9:123–160. doi: 10.1146/annurev.pu.09.050188.001011. [DOI] [PubMed] [Google Scholar]
- 26.Sugimura K, Taylor KD, Lin YC, et al. A novel NOD2/CARD15 haplotype conferring risk for Crohn disease in Ashkenazi Jews. Am J Hum Genet. 2003;72:509–518. doi: 10.1086/367848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kleinbaum DG, Kupper LL, Morgenstern H. Epidemiologic Research. Lifetime Learning Publications; Belmont, CA: 1982. [Google Scholar]
- 28.Thornton-Wells TA, Moore JH, Haines JL. Genetics, statistics and human disease: analytical retooling for complexity. Trends Genet. 2004;20:640–647. doi: 10.1016/j.tig.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 29.Picornell Y, Mei L, Taylor K, et al. TNFSF15 is an ethnic-specific IBD gene. Inflamm Bowel Dis. 2007;13:1333–1338. doi: 10.1002/ibd.20223. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.