Summary of recent advances
Recent technologies have allowed high resolution genome-wide binding profiles of numerous transcription factor and other proteins. A widespread observation has emerged from studies in diverse mammalian systems: most binding events are located at great distances from gene promoters. It is becoming apparent that the traditional one-dimensional view of gene regulation via the proximal cis regulatory elements is over-simplified. True proximity and functional relevance can be revealed by studying the three-dimensional structure of the genome packaged inside the nucleus. Thus the spatial architecture of the genome has attracted a lot of interest and has intensified its significance in modern cell biology. Here we discuss current methods, concepts, and controversies in this rapidly evolving field.
Introduction
The 3.4 billion base pairs of the human genome are packaged in hierarchical structures in a cell nucleus of about 10 micron diameter. The different levels of chromatin compaction may impact numerous nuclear events such as transcription and replication. Moreover, regulation of these processes may be influenced by the three dimensional organization of the chromatin in the nuclear space. The spatial configuration of chromosomes results in inter- and intra-chromosomal interactions that bring distant genomic elements to close proximity. The nucleus is also highly compartmentalized with defined nuclear bodies like the nucleoli, nuclear speckles, PML bodies, and Cajal bodies. Here functional domains can be formed by enriched foci or compartmentalization of nuclear proteins. Despite these non-random spatial features, it is still not clear what the principles of nuclear organization are and how they are related to function.
In the past, nuclear architecture could be studied mostly by DNA or RNA fluorescence in situ hybridization (FISH) that determines the location of a genomic locus relatively to another locus or a nuclear landmark such as the nuclear periphery. Recently developed molecular approaches, chromosome conformation capture (3C) and other closely related assays, utilize crosslinking, enzymatic digestion, ligation, and amplification procedures to probe the network of spatial interactions. 3C assesses the relative contact frequency between specific pairs of genomic elements of interest. High throughput applications [1–3**] allow increased coverage and quantitative information that are necessary to understand the long-range contacts between genomic elements and the whole genome in the native state. For example, the 3C on Chip (4C) technique [4] measures the interaction frequency of a genomic point of interest (bait) with the whole genome. Now it has become possible to map the entire interaction network (all-against-all) using a novel assay, Hi-C, albeit with a limited resolution [3**].
These recent genome-wide studies demonstrated that there exist at least two tiers of environments for any given locus. A genomic locus is in contact with adjacent elements, up to a few million bases along the linear DNA, at a frequency that is distinctively higher than that for more distant genomic loci on the same chromosome or on other chromosomes [3**–5] (Figure 1). The different levels of environment have varying biophysical properties, and may have differential impact on function. In this review we will discuss the nuclear architecture at these two orders of spatial organization. The first part will focus on the long range contacts between genes and distant cis regulatory elements, within a few Mb range. The second will address the organization of the nuclear space on the order of chromosome folding structures that result in intra- and inter-chromosomal contacts. We will also discuss recent findings regarding the features of long-range contacts, nuclear sub-environments, and their function in regulating nuclear processes in mammals. Particular attention will be given to current controversies in the field and how they may relate to technical variations in imaging methods. Finally, we will discuss how new molecular approaches could be used in parallel with previous methods to address important questions in this rapidly evolving arena.
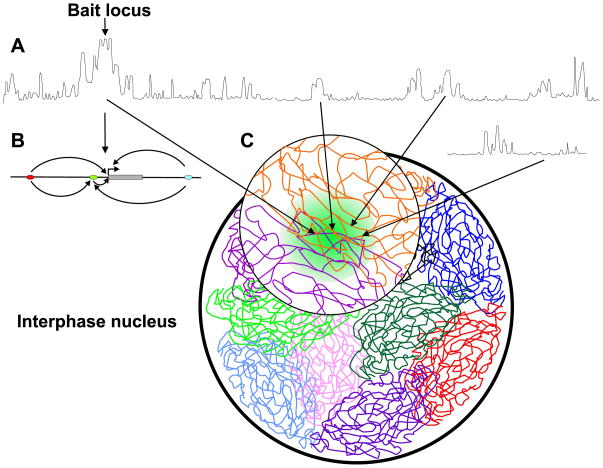
Figure 1.
Microarray profile of long range interactions by 4C. (A) Long-range contacts of the bait across the same chromosome (top profile) and with loci on another chromosome (bottom profile). The peaks represent long-range contacts and their magnitudes indicate the interaction frequency. The dominant peak cluster centered at the bait reflects the high interaction frequency of the locus with the nearby sequences within a few million bp distance. Most of the reported distant regulatory elements are found within this region (panel B). (B) Finer-scale spatial interactions within the bait region. Regulatory sites (ellipses) are in long-range contacts (arrows) with other regulatory sites and genes (gray rectangle), generating a complex network of direct and indirect contacts. (C) The chromosomes are folded in the interphase nucleus into chromosome territories. Chromosomal loci are non-randomly engaged in long-range contacts with other distant loci from the same chromosomes and with loci from other chromosomes creating spatial micro-environments.
Long-range interactions by looping
A fundamental feature of mammalian genome organization is that regulatory elements such as insulators, enhancers, silencers, and locus control regions (LCRs) are predominantly located in great distances, up to hundreds of kilobases, away from the regulated genes. These elements, as for the well studied beta globin LCR, are in some cases critical for the correct spatial and temporal expression of target genes [6]. Mechanisms involved in long-range transcription regulation remain elusive, with little conclusive evidence to support either of the leading models: looping and spreading [7]. In the spreading model, a signal is initiated at an enhancer element and is transmitted along the chromatin fiber to the target gene promoter. In the looping model, the enhancer contacts the promoter through their associated proteins, with the intervening DNA looping out.
A breakthrough for probing in vivo interaction between distal cis regulatory elements came when the chromosome conformation capture (3C) approach was applied to mammalian systems [8]. This and later studies showed that the beta-globin enhancer and other regulatory elements reside in close proximity to the active beta-globin gene. The intrinsic ability of the beta-globin LCR to upregulate the expression of nearby genes was demonstrated by knocking the LCR into an unrelated housekeeping gene cluster. The LCR was found to induce the transcription of nearby genes. Interestingly, the LCR looped to the upregulated genes, suggesting that long-range contact between regulatory element and a gene can determine the gene transcriptional activity [9**]. In the past years, many studies reported long-range interactions between distal genomic elements, starting to address the functional relevance of these interactions to gene activity. Despite the recent focus on long-range chromatin interactions, the spreading mechanism may prevail in some cases, such as in the activation of the human epsilon-globin gene by the LCR [10].
Proteins that mediate looping
If looping of DNA elements is important, what maintains the loops together? Given the inherent flexibility of the chromatin fiber, some chance encounters are expected for a locus. For example, 3C studies show that a region of interest contacts its neighboring genomic sequences, separated up to hundreds of kb, regardless of any functional relevance. Therefore, a specific long-range contact is characterized by an interaction frequency much greater than this random expectation [11]. Such a loop is most likely mediated by proteins associated with the contact points. The identity of these proteins and their regulatory roles would shed light on potential functions of the loops that they maintain. Indeed many proteins were recently found to occupy the base of the loops, and fall into different functional categories such as transcription factors (TFs) [12–16], nuclear receptors (NRs) [17–19], insulators, Polycomb [20;21], chromatin remodelers, and architectural proteins (additional papers reviewed in [22]).
For the extensively studied beta-globin locus, EKLF, FOG-1, GATA-1, GATA-2 [23;24*;25], the insulator protein CTCF [26] and the architectural protein SATB1 [27] have been shown to be involved in the loop structure. CTCF, thought to be important for loop structures, is bound across the mammalian genome at cohesin binding sites, suggesting that cohesin also has a role in the long-range structures [28–31]. It has been suggested that the TF binding at a promoter and a distant enhancer reflects the specificity observed in such interactions [32].
In addition to transcription regulators, it has been proposed that the transcriptional machinery may mediate the loop structure between distant regulatory elements and the transcribed gene. Nascent transcripts and active RNA polymerase II (Pol II) were found to aggregate in discrete foci within the nucleus termed transcription factories. In this model, transcriptionally active Pol II influences genome organization [33] as genes would migrate to the immobilized factories to be transcribed. However, the chromatin contacts between the LCR and the beta-globin gene in erythroid cells is maintained when transcription initiation and/or elongation is inhibited and Pol II binding to these elements is decreased [5;34]. Therefore, unlike TFs, Pol II may not have a direct role in maintaining the chromatin loops. The above results do not rule out the possibility that Pol II may participate in establishing the loops.
Functional significance of loops
Numerous studies have investigated the functional correlates of the long range loops, especially with respect to gene expression. Polycomb group (PcG) proteins are epigenetic chromatin modifiers involved in gene repression over many cell generations. The human GATA-4 locus is organized in a multi-loop structure in undifferentiated embryonic carcinoma cells. The contact regions were found to be enriched for PcG proteins and DNA hypermethylation when the locus is repressed. After receiving differentiation signals, activation of GATA-4 transcription was accompanied by DNA hypomethylation and loss of PcG proteins. Knocking down the EZH2 Polycomb protein or reduction of DNA methylation relaxed the contacts and increased expression, demonstrating a high correlation between the PcG-mediated repressive loop structure and expression [35]. Similarly, EZH2 also mediates a 35 kb repressive loop between the tumor suppressor genes INK4b and INK4a [36]. In light of the decrease of interaction frequency with the GATA-4 locus, it will be interesting to look for newly formed contacts that are correlated with gene activity. A switch of the contact partners between the active and the inactive state was demonstrated in erythrocytes, where the Kit promoter transitions from interacting with the upstream enhancer to contacting a downstream element as its transcription is silenced during differentiation [24*]. Another study showed that GATA-1 and Ikaros repress the gamma-globin gene during erythrogenesis when erythrocytes switch globin expression from the gamma to the beta gene, concurrent with conformational changes in the globin locus [13].
To better understand how the long-range loops relate to gene activity, several groups have studied the effects of knocking down the relevant TF. In many cases there was a change in the locus topology together with inhibition of transcription. Unfortunately these findings do not address whether the loop is the cause or the consequence of the transcriptional process, because the approach does not uncouple the structural effects from those of the TF action on the promoter region. Nonetheless, evidence exists that the interaction with a distant regulatory element enhances the level of transcription. During erythroid differentiation, the beta globin gene is already expressed and harbors active chromatin marks, prior to long-range contact with the distant LCR and elevated transcription [37]. In addition, the deletion of the globin LCR in mice does not completely shut down beta globin and active chromatin marks still remain [38].
Loops and dynamics
An outstanding question has generated a great deal of interest and controversy: How dynamic are the chromatin loops? NRs are particularly suitable for this problem because gene regulation can be rapidly initiated and controlled by the ligand. In addition, long-range interaction between NR binding sites and target genes is probably important, because the majority of their genomic binding sites are located at great distances from genes, and some regulated genes do not have any binding sites nearby [39–41].
A recent high throughput method, ChIA-PET, has established extensive hormone-dependent chromatin loops between multiple estrogen receptor (ER) alpha binding sites and gene promoters [42*]. However, it will be informative to look beyond the interactions between NR binding sites. In a recent study, when interaction partners of a Glucocorticoid Receptor (GR)-induced promoter were examined, the promoter was found to loop out to the promoter of another GR-regulated gene. Surprisingly, the authors did not find a GR binding site at the other side of the loop by ChIP. Transient and very short-lived protein binding events may not be preserved by the widely used formaldehyde crosslinking [43]. In addition, the long range interaction was set prior to GR activation [17]. It is possible that different NRs act via different dynamic mechanisms, but clearly more studies are required to address this important issue of long range interaction dynamics.
The controversy has intensified by a number of reports on various dynamic aspects of NR actions. Gene activation can be cyclic for some NRs such as ERα and vitamin D receptor (VDR) [44–46]. The ligand can be released in vivo in an hourly cycle, giving rise to transcriptional pulses [47]. As for long range interactions, transient loops between ER binding sites have been reported [19;48] as well as VDR binding sites interacting in a cyclic manner which correlates with gene transcription, chromatin modification, and Pol II activity [49**]. These data do not necessarily imply that the two ends of the loop are brought together by an active mechanism. Rather, dynamic formation of a loop may arise from stabilization of random collisions that exist in the cell population which results in a higher contact frequency.
Further studies for loops
Recent studies have revealed a more complex relationship between regulatory elements than previously thought [50;51]. Genomic regulatory sites are often identified as DNaseI hypersensitive sites (DHSs), regardless of the proteins occupying them. Indeed, multiple DHSs form contacts with the globin LCR in a developmental stage-dependent manner [8;52]. In light of the observed topological complexity, it will be worthwhile to identify all DHSs [53] and rigorously examine their contacts, rather than focusing on a few specific protein binding sites. DHSs could serve as useful reference points for 3C or the high throughput Chromosome Conformation Capture Carbon Copy (5C) technology [1;2], and will generate valuable insight on the nature of various loop structures.
Chromosome territories
In addition to the non-random and transcriptionally relevant chromatin folding in the few million base range outlined in previous sections, a locus will non-randomly associate with more distant loci on the same chromosome or on other chromosomes at lower frequency (Figure 1). A significant component of the non-random organization of the nucleus involves the heterogeneous spatial distribution of chromosomes [54;55]. Imaging-based studies of nuclear architecture have determined that individual chromosomes are not scattered over the whole nuclear space but are restrained in discrete regions termed chromosome territories (CT) [56]. CTs do not have physical barriers, and the chromatin fibers of each CT intermingle with those of neighboring chromosomes [57] thereby increasing the complexity of potential spatial neighbors for a given locus. In spherical nuclei, CTs have preferred radial positions, in that gene-rich chromosomes tend to be internally located and gene-poor chromosomes are closer to the nuclear periphery [58]. Aside from this general feature, loci organization within a CT [59] and the organization of CTs relative to each other are cell type-specific [60], suggestive of functional relevance. A recent cutting-edge work has brought new insights on the biophysical principles of CT folding. The authors developed a new technology, Hi-C, which is a high-throughput version of 3C that allows the capture of large scale chromatin interactions of the entire genome. Remarkably, the chromatin conformation of the human genome in the megabase scale was consistent with a model based on fractal globular polymers. The fractal globules enable dense packing, and are knot-free, allowing the flexibility to easily fold and unfold chromosomes during nuclear processes such as gene expression and cell cycle [3**].
Inter-chromosomal associations: Functional specificity?
Several studies have presented evidence for the intriguing phenomenon of non-random association between functionally related genes that are located very far or on different chromosomes. Regarding the extensively characterized globin genes in erythroid cells, imaging studies have shown that beta-globin co-localizes with other distant erythroid-specific genes [61–63]. However, frequent co-localization of active erythroid-specific genes does not establish the existence of functionally specialized nuclear environments. Some groups propose that the interactions occur at sites enriched with active Pol II termed transcription factories [33], while others find the associations to co-localize with nuclear splicing bodies [63].
Overcoming the limited scope in interaction partners that are probed by imaging methods, two recent studies characterized the spatial neighborhood of the beta-globin locus by genome-wide 4C approaches, with somewhat conflicting results. Simonis et al. found that the beta-globin contact loci, although interacting with many active genes, were not enriched for erythroid-specific genes [4]. On the other hand, Schoenfelder et al. probed Pol II associated beta- and alpha-globin environments and reported that the contact loci are enriched for other active and erythroid-specific genes [64*]. The authors also observed that erythroid-specific TF Klf1 is enriched in a subset of transcription factories, and mediates preferential associations of Klf1-regulated genes, suggesting that regulatory factors create specialized nuclear environments. Although they focused solely on actively transcribed globins, this does not explain the discrepant results from the two studies because the globin genes are highly transcribed and engaged with factories in about 90% of these cells [61]. Significant differences might have come from the very different approaches employed for statistical data analysis (W. de Laat, personal communication), as the former study found predominantly intra-chromosomal contacts of beta-globin while the latter reported mostly inter-chromosomal interactions.
Interestingly, inter-chromosomal co-localization of inactive genes has also been reported. The imprinting control region (ICR) of the maternally expressed H19 gene was found to engage in allele-specific inter-chromosomal interactions with other imprinted genes [65;66]. A recent 4C study showed that imprinted loci across the whole genome frequently associate each other and that the CTCF binding site within the ICR has a global effect on the spatial proximity between imprinted loci and their epigenetic state [67*]. However Sandhu et al. did not account for the gene density or the transcriptional status of imprinting-independent genes in the contacts. Therefore, it is difficult to determine whether these hubs regulate imprinted genes or are a consequence of more general features of nuclear architecture like gene density and activity.
Some studies reported remarkable findings on the functional and dynamic specificity of large-scale inter-chromosomal interactions in response to external stimuli. Co-localization of the IFN-beta gene with three distant NF-kappaB bound loci was shown to be dynamic and concomitant with transcriptional activation by a viral infection [68]. In another study, ER induced genes were found to co-localize frequently after estradiol treatment [69]. Despite these interesting observations, it remains critically important to investigate whether such dynamic phenomena are a general mechanism of gene regulation using comprehensive approaches.
Nuclear ‘hubs’ that can be defined by distinct factors and genomic elements, like the nucleoli, are an appealing concept for coordinated regulation of specific genes. However, a clearer picture will emerge from probing the whole environment of the loci of interest. Another point to consider, as we expand our understanding, involves the nature of the cell types under study. For example, it is unclear how much of the extensive knowledge about erythroid cells, a highly specialized cell type dedicated to globin production, translates into other cell types of physiological importance. For a cell type that needs to be versatile in response to various stimuli, the nuclear organization may be more complicated.
Nuclear bodies and landmarks for genome organization
Nuclear bodies, such as nucleoli, PML, and Cajal bodies, are dynamic environments enriched for specific protein factors and genomic loci. Complete mapping of genomic regions in proximity to these bodies could be useful in building three dimensional maps of nuclear architecture. A disadvantage in this approach is that the number of nuclear bodies is highly variable, depending on the cell type, transcriptional activity, the cell cycle stage, and also varies in the cell population. Moreover, the association of the genome loci with the nuclear bodies may vary.
The nuclear envelope is also a well-defined nuclear landmark with associated genomic regions. Although nuclear lamina associated loci are generally thought to be gene-poor and repressed [70] several different approaches, including gene tethering to the periphery, have revealed a more complex relationship [71–74]. These studies underscore the fact that the nuclear periphery is not an exclusively silent environment.
The nuclear envelope has also served in recent years as a convenient reference for mapping the nuclear architecture in a more quantitative fashion [75]. In addition to mapping individual chromosomes along the center-periphery axis, attempts have been made to correlate radial positioning to gene expression. However, the positioning in the radial axis is poorly associated with gene activity [76]. Another study examined the radial position of 11 cancer-related genes during early mammary tumor formation and development [77]. Although each gene had a preferred radial position, alterations of positioning patterns during differentiation and tumorigenesis were unrelated to transcription.
Taken together, it seems that there is no clear correlation between gene activity and the distance from the nuclear periphery. This is in agreement with the fact that sites of active transcription are evenly distributed throughout the nucleus with no preference towards the nuclear center [78]. In addition, there may be other repressive environments in the nucleus. Therefore, although the radial position of a gene relative to the periphery serves as a convenient reference, it is not clearly linked to the functional status [75].
‘C’ assays and DNA FISH: Which is validating what?
Since the early studies that introduced 3C and 4C methodologies, DNA FISH has been used to ‘validate’ a selected sample of interacting pairs to see if they are indeed in close physical proximity in the nucleus. Here we discuss some inherent differences between the two assays and how they affect the apparent degree of correlation.
3C-based assays obtain information from DNA-DNA interactions that are summed over millions of cells. At any given moment, certain contacts may be abundant and exist in the majority of cells, while other interactions may be transient or present in a small subpopulation of cells. Although direct translation of the 4C/5C signals to the contact frequency scale is very difficult, it is generally thought that high and low signals roughly correspond to frequent and rare interactions, respectively. The dramatic shift of the signal strength within versus outside of the bait neighborhood in 4C data exemplifies the wide spectrum of contact frequency.
DNA FISH produces information at the level of single cells with the spatial resolution dictated by the optical limit and the probe size. In a widely used protocol, each locus is represented by BAC probes that typically span a few hundred kilobases and therefore may cover a larger region than the relevant contact locus. Also, the probe-annealing step requires denaturing the nuclear DNA in a high temperature which may perturb the native positions of genomic loci [79]. Although the contact frequency is directly estimated by the fraction of cells containing co-localizing loci, the assay is more prone to sampling error because the number of nuclei that are examined ranges in the hundreds at best.
Aside from these general features, it is noteworthy that DNA FISH assays are implemented in various ways. These include the three methods that we describe here. In the first method, one simply collects images from several focal planes through the nuclei and scores two loci as co-localizing if their FISH signals overlap in the same z plane [80]. In the second method, cryoFISH is performed and a thin physical section through the nuclei is examined to count those that have co-localizing FISH signals [4;57]. In these two methods, one obtains an estimate of the contact frequency for the pair of interest by counting the cells that have ‘co-localizing’ FISH signals. Typically the percent of nuclei showing co-localization is reported for each pair of loci. Co-localization of a pair of loci is determined based on an overlap between the two FISH signals, which is influenced by the particular image acquisition and thresholding of the intensities for the fluorescence channels. In an alternative method (Hakim, Sung, Hager, unpublished data), all the focal planes from DNA FISH images are utilized to calculate the three dimensional position of each signal. Effects from imaging and thresholding can be minimized if the position of a FISH signal is determined by its brightest pixel (the center) rather than its edges. Then the center-to-center distance in 3D is calculated in an automated fashion for a large number of cells. Unlike the previous methods, one obtains the information on the distribution of 3D distances, and can also report the contact frequency by introducing a distance cut-off for co-localization. Here it has to be taken into account that even overlapping loci may well have a significant (center-to-center) distance between them. Interestingly, it has been observed that the distance distribution itself has some correlation with interaction measurements from 4C or 5C [Job Dekker, personal communication; Hakim, Sung, Hager, unpublished]. It seems possible that spatially proximal loci represent those that contact each other in a dynamic fashion.
Because the apparent ‘validation’ rate is directly affected by the limited spatial resolution, small sample size, and variable quantification methods for DNA FISH, it can be difficult to establish a high concordance with 3C-based assays. Employment of objective automated quantification method will not only increase the number of cells examined but also ensure a standardized protocol for treating and reporting DNA FISH data accurately.
Challenges and new opportunities
Imaging approaches like FISH and immunofluorescence, that have been instrumental in making interesting observations, are nevertheless descriptive and limited in the number of loci that can be studied simultaneously. They do not provide comprehensive information about the spatial environment for a given locus. Similarly, although nuclear bodies are known to associate with particular loci in certain conditions, imaging results do not convey the full spectrum of genomic contacts for these nuclear bodies.
Current high-throughput technologies allow higher-resolution quantitative studies of DNA-DNA interactions across the genome. Many variations of the 3C approach and 3C combined with protein interactions are now available [1;42*;64*]. The 4C (3C on chip, circular 3C) approaches identify all the genomic regions in contact with a locus of interest, while the 5C assesses all the interactions spanning a large locus of interest. Although rather high-resolution, 5C analysis is limited to a confined genomic region and does not cover the entire genome. A major advance in genomic technologies for studying nuclear architecture has recently been achieved by the Hi-C method [3**]. It is based on selective purification of spatially proximal ligation products followed by massively parallel sequencing, which reveals chromatin interactions across the whole genome. The assay resolution is currently about 1Mb, and may potentially approach that of 4C, in the range of 10 kb, as the Hi-C library is sequenced much deeper. The ChIA-PET approach is a powerful combination of ChIP and 3C, developed for long range interactions mediated by a protein of interest [42*].
The different versions of the high-throughput methods provide data on different aspects of nuclear organization, and are therefore complementary. For example, ChIA-PET provides information for interactions between genomic elements that are in contact with a specific protein, but does not seem to capture inter-chromosomal interactions efficiently. 4C and Hi-C detect long-range interactions regardless of whether they bind to a specific protein.
Conclusion
Recent investigations of the functional roles of nuclear organization have led to many interesting discoveries as well as new questions. While imaging methods like DNA FISH have been extremely useful, great advances have been made for high resolution molecular assays that dramatically improve quantification of genome-wide interaction patterns. Combining the genome-wide chromatin configuration methodologies with existing genomic tools will be instrumental for obtaining comprehensive insights on principles of genome organization. In parallel, it will be necessary to standardize and increase the throughput of imaging approaches for a more accurate picture at the single cell level. In addition to FISH and other fixed cell assays, live cell imaging approaches will be essential in addressing dynamic aspects of chromatin interactions. Lastly, correct interpretation of data remains an important issue in this fast-evolving, technology-driven field. Sound statistical analyses and computational modeling can facilitate almost all aspects of the relevant techniques, not only by minimizing sources of discrepancies but also by uncovering novel insights. Interdisciplinary collaborations including researchers with mathematical and biophysical expertise will significantly enhance many studies in the future. Armed with these exciting new approaches, the field has high hopes for witnessing fruitful contributions toward a clear relationship between nuclear architecture and function.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Simonis M, Kooren J, de Laat W. An evaluation of 3C-based methods to capture DNA interactions. Nat Methods. 2007;4:895–901. doi: 10.1038/nmeth1114. [DOI] [PubMed] [Google Scholar]
- 2.Ohlsson R, Gondor A. The 4C technique: the ‘Rosetta stone’ for genome biology in 3D? Curr Opin Cell Biol. 2007;19:321–325. doi: 10.1016/j.ceb.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 3**.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. This article describes the Hi-C assay that probes all the chromatin long-range contacts in the cell nucleus. This molecular approach confirmed the organization of chromosomes in territories and further suggests that chromatin conformation at the megabase scale is consistent with a fractal globule, a knot-free, polymer conformation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat Genet. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 5.Palstra RJ, Simonis M, Klous P, Brasset E, Eijkelkamp B, de Laat W. Maintenance of long-range DNA interactions after inhibition of ongoing RNA polymerase II transcription. PLoS One. 2008;3:e1661. doi: 10.1371/journal.pone.0001661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Q, Peterson KR, Fang X, Stamatoyannopoulos G. Locus control regions. Blood. 2002;100:3077–3086. doi: 10.1182/blood-2002-04-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ptashne M. Gene regulation by proteins acting nearby and at a distance. Nature. 1986;322(6081):697–701. doi: 10.1038/322697a0. [DOI] [PubMed] [Google Scholar]
- 8.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 9**.Noordermeer D, Branco MR, Splinter E, Klous P, van Ijcken W, Swagemakers S, Koutsourakis M, van der Spek P, Pombo A, de Laat W. Transcription and chromatin organization of a housekeeping gene cluster containing an integrated beta-globin locus control region. PLoS Genet. 2008;4:e1000016. doi: 10.1371/journal.pgen.1000016. This article demonstrates the correlation between nuclear repositioning and long-range interactions to gene expression. Integration of the human beta-globin LCR into an unrelated mouse gene-rich region changed the locus position relatively to the chromosome territory but did not have a major effect on the association with transcription factories or gene expression. Interestingly, the integrated LCR formed direct long-range interactions with two upregulated genes in the locus and propose that LCR-gene contacts via chromatin looping determine which genes are transcriptionally enhanced. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu X, Ling J, Zhang L, Pi W, Wu M, Tuan D. A facilitated tracking and transcription mechanism of long-range enhancer function. Nucleic Acids Res. 2007;35:5532–5544. doi: 10.1093/nar/gkm595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dekker J. The three ‘C’ s of chromosome conformation capture: controls, controls, controls. Nat Methods. 2006;3:17–21. doi: 10.1038/nmeth823. [DOI] [PubMed] [Google Scholar]
- 12.Chavanas S, Adoue V, Mechin MC, Ying S, Dong S, Duplan H, Charveron M, Takahara H, Serre G, Simon M. Long-range enhancer associated with chromatin looping allows AP-1 regulation of the peptidylarginine deiminase 3 gene in differentiated keratinocyte. PLoS One. 2008;3:e3408. doi: 10.1371/journal.pone.0003408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bottardi S, Ross J, Bourgoin V, Fotouhi-Ardakani N, Affar eB, Trudel M, Milot E. Ikaros and GATA-1 combinatorial effect is required for silencing of human gamma-globin genes. Mol Cell Biol. 2009;29:1526–1537. doi: 10.1128/MCB.01523-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keys JR, Tallack MR, Zhan Y, Papathanasiou P, Goodnow CC, Gaensler KM, Crossley M, Dekker J, Perkins AC. A mechanism for Ikaros regulation of human globin gene switching. Br J Haematol. 2008;141:398–406. doi: 10.1111/j.1365-2141.2008.07065.x. [DOI] [PubMed] [Google Scholar]
- 15.Du MJ, Lv X, Hao DL, Zhao GW, Wu XS, Wu F, Liu DP, Liang CC. MafK/NF-E2 p18 is required for beta-globin genes activation by mediating the proximity of LCR and active beta-globin genes in MEL cell line. Int J Biochem Cell Biol. 2008;40:1481–1493. doi: 10.1016/j.biocel.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 16.Liu Z, Garrard WT. Long-range interactions between three transcriptional enhancers, active Vkappa gene promoters, and a 3′ boundary sequence spanning 46 kilobases. Mol Cell Biol. 2005;25:3220–3231. doi: 10.1128/MCB.25.8.3220-3231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hakim O, John S, Ling JQ, Biddie SC, Hoffman AR, Hager GL. Glucocorticoid receptor activation of the Ciz1-Lcn2 locus by long range interactions. J Biol Chem. 2009;284:6048–6052. doi: 10.1074/jbc.C800212200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gothard LQ, Hibbard JC, Seyfred MA. Estrogen-mediated induction of rat prolactin gene transcription requires the formation of a chromatin loop between the distal enhancer and proximal promoter regions. Mol Endocrinol. 1996;10:185–195. doi: 10.1210/mend.10.2.8825558. [DOI] [PubMed] [Google Scholar]
- 19.Perillo B, Ombra MN, Bertoni A, Cuozzo C, Sacchetti S, Sasso A, Chiariotti L, Malorni A, Abbondanza C, Avvedimento EV. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science. 2008;319:202–206. doi: 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- 20.Lanzuolo C, Roure V, Dekker J, Bantignies F, Orlando V. Polycomb response elements mediate the formation of chromosome higher-order structures in the bithorax complex. Nat Cell Biol. 2007;9:1167–1174. doi: 10.1038/ncb1637. [DOI] [PubMed] [Google Scholar]
- 21.Li T, Hu JF, Qiu X, Ling J, Chen H, Wang S, Hou A, Vu TH, Hoffman AR. CTCF regulates allelic expression of Igf2 by orchestrating a promoter-polycomb repressive complex 2 intrachromosomal loop. Mol Cell Biol. 2008;28:6473–6482. doi: 10.1128/MCB.00204-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sexton T, Bantignies F, Cavalli G. Genomic interactions: chromatin loops and gene meeting points in transcriptional regulation. Semin Cell Dev Biol. 2009;20:849–855. doi: 10.1016/j.semcdb.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 23.Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA. Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 24*.Jing H, Vakoc CR, Ying L, Mandat S, Wang H, Zheng X, Blobel GA. Exchange of GATA factors mediates transitions in looped chromatin organization at a developmentally regulated gene locus. Mol Cell. 2008;29:232–242. doi: 10.1016/j.molcel.2007.11.020. This article shows the exchange of GATA transcription factors binding and long-range contacts in the Kit locus in different transcription states throughout erythrogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drissen R, Palstra RJ, Gillemans N, Splinter E, Grosveld F, Philipsen S, de Laat W. The active spatial organization of the beta-globin locus requires the transcription factor EKLF. Genes Dev. 2004;18:2485–2490. doi: 10.1101/gad.317004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, Grosveld F, Galjart N, de Laat W. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006;20:2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L, Di LJ, Lv X, Zheng W, Xue Z, Guo ZC, Liu DP, Liang CC. Inter-MAR association contributes to transcriptionally active looping events in human beta-globin gene cluster. PLoS One. 2009;4:e4629. doi: 10.1371/journal.pone.0004629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, Cobb BS, Yokomori K, Dillon N, Aragon L, Fisher AG, Merkenschlager M. Cohesins Functionally Associate with CTCF on Mammalian Chromosome Arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 29.Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, Yahata K, Imamoto F, Aburatani H, Nakao M, Imamoto N, Maeshima K, Shirahige K, Peters JM. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 30.Stedman W, Kang H, Lin S, Kissil JL, Bartolomei MS, Lieberman PM. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 2008;27:654–666. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, Aebersold R, Ranish JA, Krumm A. CTCF physically links cohesin to chromatin. Proc Natl Acad Sci USA. 2008;105:8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nolis IK, McKay DJ, Mantouvalou E, Lomvardas S, Merika M, Thanos D. Transcription factors mediate long-range enhancer-promoter interactions. Proc Natl Acad Sci USA. 2009;106:20222–20227. doi: 10.1073/pnas.0902454106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sutherland H, Bickmore WA. Transcription factories: gene expression in unions? Nat Rev Genet. 2009;10:457–466. doi: 10.1038/nrg2592. [DOI] [PubMed] [Google Scholar]
- 34.Mitchell JA, Fraser P. Transcription factories are nuclear subcompartments that remain in the absence of transcription. Genes Dev. 2008;22:20–25. doi: 10.1101/gad.454008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tiwari VK, McGarvey KM, Licchesi JD, Ohm JE, Herman JG, Schubeler D, Baylin SB. PcG proteins, DNA methylation, and gene repression by chromatin looping. PLoS Biol. 2008;6:2911–2927. doi: 10.1371/journal.pbio.0060306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kheradmand KS, Solaimani KP, Farahbakhshian E, Pourfarzad F, von Lindern M, Verrijzer CP. EZH2-dependent chromatin looping controls INK4a and INK4b, but not ARF, during human progenitor cell differentiation and cellular senescence. Epigenetics Chromatin. 2009;2:16. doi: 10.1186/1756-8935-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kooren J, Palstra RJ, Klous P, Splinter E, von Lindern M, Grosveld F, de Laat W. Beta-globin active chromatin Hub formation in differentiating erythroid cells and in p45 NF-E2 knock-out mice. J Biol Chem. 2007;282:16544–16552. doi: 10.1074/jbc.M701159200. [DOI] [PubMed] [Google Scholar]
- 38.Schubeler D, Groudine M, Bender MA. The murine beta-globin locus control region regulates the rate of transcription but not the hyperacetylation of histones at the active genes. Proc Natl Acad Sci USA. 2001;98:11432–11437. doi: 10.1073/pnas.201394698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao H, Falt S, Sandelin A, Gustafsson JA, Dahlman-Wright K. Genome-wide identification of estrogen receptor alpha-binding sites in mouse liver. Mol Endocrinol. 2008;22:10–22. doi: 10.1210/me.2007-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.John S, Sabo PJ, Johnson TA, Sung MH, Biddie SC, Lightman SL, Voss TC, Davis SR, Meltzer PS, Stamatoyannopoulos JA, Hager GL. Interaction of the glucocorticoid receptor with the global chromatin landscape. Mol Cell. 2008;29:611–624. doi: 10.1016/j.molcel.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 41.Nielsen R, Pedersen TA, Hagenbeek D, Moulos P, Siersbaek R, Megens E, Denissov S, Borgesen M, Francoijs KJ, Mandrup S, Stunnenberg HG. Genome-wide profiling of PPARgamma:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev. 2008;22:2953–2967. doi: 10.1101/gad.501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42*.Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, Chew EG, Huang PY, Welboren WJ, Han Y, Ooi HS, Ariyaratne PN, Vega VB, Luo Y, Tan PY, Choy PY, Wansa KD, Zhao B, Lim KS, Leow SC, Yow JS, Joseph R, Li H, Desai KV, Thomsen JS, Lee YK, Karuturi RK, Herve T, Bourque G, Stunnenberg HG, Ruan X, Cacheux-Rataboul V, Sung WK, Liu ET, Wei CL, Cheung E, Ruan Y. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. This article reports the global chromatin interactions network bound by oestrogen receptor alpha (ER-alpha), using the chromatin interaction analysis by paired-end tag sequencing (ChIA-PET) assay. Gene promoters communicate with multiple distant ER-alpha-binding sites by long-range interactions. These contacts were shown to be dependent on ER-alpha activation and correlated with gene expression suggesting that ER-alpha mediates chromatin looping to bring genes together for coordinated gene expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmiedeberg L, Skene P, Deaton A, Bird A. A temporal threshold for formaldehyde crosslinking and fixation. PLoS One. 2009;4:e4636. doi: 10.1371/journal.pone.0004636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim S, Shevde NK, Pike JW. 1,25-Dihydroxyvitamin D3 stimulates cyclic vitamin D receptor/retinoid X receptor DNA-binding, co-activator recruitment, and histone acetylation in intact osteoblasts. J Bone Miner Res. 2005;20:305–317. doi: 10.1359/JBMR.041112. [DOI] [PubMed] [Google Scholar]
- 45.Banwell CM, MacCartney DP, Guy M, Miles AE, Uskokovic MR, Mansi J, Stewart PM, O’Neill LP, Turner BM, Colston KW, Campbell MJ. Altered nuclear receptor corepressor expression attenuates vitamin D receptor signaling in breast cancer cells. Clin cancer Res. 2006;12:2004–2013. doi: 10.1158/1078-0432.CCR-05-1218. [DOI] [PubMed] [Google Scholar]
- 46.Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 47.Stavreva DA, Wiench M, John S, Conway-Campbell BL, McKenna MA, Pooley JR, Johnson TA, Voss TC, Lightman SL, Hager GL. Ultradian hormone stimulation induces glucocorticoid receptor-mediated pulses of gene transcription. Nat cell Biol. 2009;11:1093–1102. doi: 10.1038/ncb1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cullen KE, Kladde MP, Seyfred MA. Interaction between transcription regulatory regions of prolactin chromatin. Science. 1993;261:203–206. doi: 10.1126/science.8327891. [DOI] [PubMed] [Google Scholar]
- 49**.Saramaki A, Diermeier S, Kellner R, Laitinen H, Vaisanen S, Carlberg C. Cyclical chromatin looping and transcription factor association on the regulatory regions of the p21 (CDKN1A) gene in response to 1alpha,25-dihydroxyvitamin D3. J Biol Chem. 2009;284:8073–8082. doi: 10.1074/jbc.M808090200. This work reports a dynamic long-range contact between the vitamin D receptor element (VDREs) and the transcription start site (TSS) of the p21 gene. The cyclic (45–60 min) chromatin loop was correlated with VDR and RNA polymerase II binding, chromatin epigenetic modifications and transcription. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, Lobanenkov V, Reik W, Ohlsson R. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci USA. 2006;103:10684–10689. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qiu X, Vu TH, Lu Q, Ling JQ, Li T, Hou A, Wang SK, Chen HL, Hu JF, Hoffman AR. A complex deoxyribonucleic acid looping configuration associated with the silencing of the maternal Igf2 allele. Mol Endocrinol. 2008;22:1476–1488. doi: 10.1210/me.2007-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Palstra RJ, Tolhuis B, Splinter E, Nijmeijer R, Grosveld F, de Laat W. The beta-globin nuclear compartment in development and erythroid differentiation. Nat Genet. 2003;35:190–194. doi: 10.1038/ng1244. [DOI] [PubMed] [Google Scholar]
- 53.Sabo PJ, Kuehn MS, Thurman R, Johnson BE, Johnson EM, Cao H, Yu M, Rosenzweig E, Goldy J, Haydock A, Weaver M, Shafer A, Lee K, Neri F, Humbert R, Singer MA, Richmond TA, Dorschner MO, McArthur M, Hawrylycz M, Green RD, Navas PA, Noble WS, Stamatoyannopoulos JA. Genome-scale mapping of DNase I sensitivity in vivo using tiling DNA microarrays. Nat Methods. 2006;3:511–518. doi: 10.1038/nmeth890. [DOI] [PubMed] [Google Scholar]
- 54.Simonis M, de Laat W. FISH-eyed and genome-wide views on the spatial organisation of gene expression. Biochim Biophys Acta. 2008;1783:2052–2060. doi: 10.1016/j.bbamcr.2008.07.020. [DOI] [PubMed] [Google Scholar]
- 55.Misteli T. Concepts in nuclear architecture. Bioessays. 2005;27:477–487. doi: 10.1002/bies.20226. [DOI] [PubMed] [Google Scholar]
- 56.Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 57.Branco MR, Pombo A. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol. 2006;4:e138. doi: 10.1371/journal.pbio.0040138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reddy TE, Pauli F, Sprouse RO, Neff NF, Newberry KM, Garabedian MJ, Myers RM. Genomic determination of the glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome Res. 2009;19:2163–2171. doi: 10.1101/gr.097022.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hepperger C, Mannes A, Merz J, Peters J, Dietzel S. Three-dimensional positioning of genes in mouse cell nuclei. Chromosoma. 2008;117:535–551. doi: 10.1007/s00412-008-0168-2. [DOI] [PubMed] [Google Scholar]
- 60.Marella NV, Seifert B, Nagarajan P, Sinha S, Berezney R. Chromosomal rearrangements during human epidermal keratinocyte differentiation. J Cell Physiol. 2009;221:139–146. doi: 10.1002/jcp.21855. [DOI] [PubMed] [Google Scholar]
- 61.Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, Fraser P. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- 62.Negrini M, Silini A, Kozak C, Tsujimoto Y, Croce CM. Molecular analysis of MBCL-2: Structure and expression of the murine gene homologous to the human gene involved in follicular lymphoma. Cell. 1987;49:455–63. doi: 10.1016/0092-8674(87)90448-x. [DOI] [PubMed] [Google Scholar]
- 63.Brown JM, Green J, das Neves RP, Wallace HA, Smith AJ, Hughes J, Gray N, Taylor S, Wood WG, Higgs DR, Iborra FJ, Buckle VJ. Association between active genes occurs at nuclear speckles and is modulated by chromatin environment. J Cell Biol. 2008;182:1083–1097. doi: 10.1083/jcb.200803174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64*.Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, Eskiw CH, Luo Y, Wei CL, Ruan Y, Bieker JJ, Fraser P. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet. 2010;42:53–61. doi: 10.1038/ng.496. This work developed the e4C to show that the transcriptionally active alpha- and beta-globin genes associated with other active genes in erythroid cells. These Klf1 mediated inter- and intra-chromosomal associations take place in transcription factories that are enriched with Klf1 transcription factor and harbor Klf1 regulated genes. The authors suggest that expression of co-regulated genes take place in specialized nuclear sub compartments enriched with regulatory and transcription factors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW, Cherry AM, Hoffman AR. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- 66.Zhao Z, Tavoosidana G, Sjolinder M, Gondor A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, Pant V, Tiwari V, Kurukuti S, Ohlsson R. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated i. Nat Genet. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 67*.Sandhu KS, Shi C, Sjolinder M, Zhao Z, Gondor A, Liu L, Tiwari VK, Guibert S, Emilsson L, Imreh MP, Ohlsson R. Nonallelic transvection of multiple imprinted loci is organized by the H19 imprinting control region during germline development. Genes Dev. 2009;23:2598–2603. doi: 10.1101/gad.552109. This report describes the spatial clustering of imprinted genes, located on several chromosomes. Remarkably, the authors report that one genomic site, the CTCF-binding sites within the H19 imprinting control region (ICR) determines the spatial proximity between imprinted loci and their epigenetic state. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Apostolou E, Thanos D. Virus Infection Induces NF-kappaB-dependent interchromosomal associations mediating monoallelic IFN-beta gene expression. Cell. 2008;134:85–96. doi: 10.1016/j.cell.2008.05.052. [DOI] [PubMed] [Google Scholar]
- 69.Hu Q, Kwon YS, Nunez E, Cardamone MD, Hutt KR, Ohgi KA, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG, Fu XD. Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules. Proc Natl Acad Sci USA. 2008;105:19199–19204. doi: 10.1073/pnas.0810634105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, van Steensel B. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 71.Brown CR, Silver PA. Transcriptional regulation at the nuclear pore complex. Curr Opin Genet Dev. 2007;17:100–106. doi: 10.1016/j.gde.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 72.Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–247. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- 73.Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, Perry P, Ylstra B, Chubb JR, Bickmore WA. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 2008;4:e1000039. doi: 10.1371/journal.pgen.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kumaran RI, Spector DL. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J Cell Biol. 2008;180:51–65. doi: 10.1083/jcb.200706060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Takizawa T, Meaburn KJ, Misteli T. The meaning of gene positioning. Cell. 2008;135:9–13. doi: 10.1016/j.cell.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Williams RR, Azuara V, Perry P, Sauer S, Dvorkina M, Jorgensen H, Roix J, McQueen P, Misteli T, Merkenschlager M, Fisher AG. Neural induction promotes large-scale chromatin reorganisation of the Mash1 locus. J Cell Sci. 2006;119:132–140. doi: 10.1242/jcs.02727. [DOI] [PubMed] [Google Scholar]
- 77.Meaburn KJ, Misteli T. Locus-specific and activity-independent gene repositioning during early tumorigenesis. J Cell Biol. 2008;180:39–50. doi: 10.1083/jcb.200708204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Elbi CC, Misteli T, Hager GL. Recruitment of the Dioxin Receptor to Active Transcription Sites. Mol Biol Cell. 2002;13:2001–2015. doi: 10.1091/mboc.13.6.mk0602002001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Muller WG, Rieder D, Karpova TS, John S, Trajanoski Z, McNally JG. Organization of chromatin and histone modifications at a transcription site. J Cell Biol. 2007;177:957–967. doi: 10.1083/jcb.200703157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Takizawa T, Gudla PR, Guo L, Lockett S, Misteli T. Allele-specific nuclear positioning of the monoallelically expressed astrocyte marker GFAP. Genes Dev. 2008;22:489–498. doi: 10.1101/gad.1634608. [DOI] [PMC free article] [PubMed] [Google Scholar]