Summary
Protein interactions give rise to networks that control cell fate in health and disease; selective means to probe these interactions are therefore of wide interest. We discuss here Evolutionary Tracing (ET), a comparative method to identify protein functional sites and to guide experiments that selectively block, recode, or mimic their amino acid determinants. These studies suggest, in principle, a scalable approach to perturb individual links in protein networks.
Keywords: Evolutionary Trace, protein-protein interactions, allosteric pathway, functional annotation, protein design, network medicine
INTRODUCTION
Protein interactions are an emerging frontier for therapy because they underlie all aspects of cellular activity [1]. They organize cellular components into complexes, macromolecular machines, cellular pathways and biological networks that sustain development, growth and homeostasis. Upon disruption, deregulated interactions can lead to amyloidosis, to cancer, or to many other ailments [2].
Unfortunately, such disruptions are common and diverse. A survey of deleterious protein mutations recently suggested 65 diseases likely caused by a gain, or loss, of specific protein-protein interactions (PPI) [3]. Moreover, in a complex disorder such as ataxia, the same disease may arise in different individuals from defects in different interconnected proteins [4]. Therapies directed to a single specific protein may thus fail. This realization, plus the slow rate of new drug development relative to the rapid expansion of biological knowledge, make a case for a network approach to medicine [5], namely, discovering the components of a disease process; elucidating their interactions; diagnosing those at fault; and developing flexible therapeutic tools to counter their abnormal interaction. This review focuses on the last step in this process: approaches to understand the molecular details of protein functional sites in order to gain control over them [6].
ANALYSIS AND PREDICTION OF FUNCTIONAL SITES
A first step to manipulate a protein interaction is to characterize the amino acids that control it and which, together, define a functional site. Many different approaches try to detect various types of sites: for catalysis [7], for binding small ligands [8], for macromolecules [9], or sites and amino acids that control functional specificity [10]. Nearly all these approaches search a protein structure for features typical of a functional signature. This includes geometric searches for ligand binding pockets and clefts [11,12], chemical titration models to identify catalytic residues [13]; and energy calculations to identify ligand binding, or “hot spot”, residues [14–16]. Other approaches focus on evolutionary conservation such as Consurf [17], siteFiNDER3 [18], INTREPID [19], and Joint Evolutionary Trace (JET) [20], often related to Evolutionary Trace (ET) [21–24].
In turn these features can be combined to increase the accuracy of searches. DISCERN, for example, identifies catalytic residues from their structural clustering and conservation [25], and many other methods search for surface cavities that exhibit sequence conservation in order to detect potential small ligand sites [8,26]. The PPI-pred server focuses on protein-protein interfaces [27]; it is a machine learning method that optimizes predictions by combining structural information, surface features, evolutionary information, and residue composition. FINDSITE is a threading based method for ligand binding site prediction [28].
To compare these many different functional site prediction methods is desirable but not straightforward. Ultimately, this is an experimental task that must be tailored to the problem at hand. Arguably, residues beyond a catalytic site can define its activity [29] and thus could be part of it; conversely, not all PPI residues are energetically important to an interaction [30] so that the elements of an interface may not all contribute to it equally, or even significantly. Moreover, there are distinct approaches to define an interface. They agree broadly but can differ in details: some approaches are based on proximity between chains [31], and others are based on surface buried away from the solvent [32] or on pure geometry [33]. More problematic, gold standard mutational studies are themselves limited in the number of substitutions and assays that probe the biological role of sequence position. Finally, while some amino acids are important for structure rather than function [34,35], it is clear that the former are a main cause of deficits in the context of human genetic variation [36]. These observations suggest that stringent tests of functional site and residues predictions might focus on whether they are necessary and sufficient for function: (a) can they guide protein function redesign experiments efficiently; and (b) can they predict protein function based on residue similarities?
REDESIGN OF PROTEIN INTERACTIONS
Functional site redesign strategies are distinct from larger transfer of sequence segments that form modular protein chimeras [37]. Rather redesign means to target, or graft, the amino acids of a functional epitope to modulate function [38,39]. Often the focus of these experiments is on controlling the character of an interaction.
Some studies manipulate a protein to raise its affinity [40]. In calmodulin, affinity with CAM-dependent protein kinase II was increased 900-fold [41]. The method requires a reliable structure as a starting point and knowledge of the actual site meant for redesign. The strategy is then to increase the hydrophobic surface area of the binding site, and assess whether this is likely to increase binding energy while maintaining a stable structure as determined by energy, and protein models, from Rosetta [42] or CHARMM [43]. Binding affinity may increase as much as 10-fold by replacing a single polar residue with a hydrophobic one, or small hydrophobic residues by larger ones [44]. Strikingly, MHC peptide binding to its T Cell receptor improved nearly 100-fold by several single point mutations that each alone improved affinity six-fold or less [45]. Thus increasing the overall hydrophobicity of the interface synergistically improved affinity.
Other studies aim to redesign specificity, either at protein-ligand binding sites [46] or at protein-protein interfaces [47–50]. Thus, some enzymes have been rationally redesigned to be active with a particular substrate [51,52]. Typically, this entails mutations that: increase electrostatics complementarity to the desired ligand (positive design); decrease the fit to undesired ones (negative design); and maintain stability in the original structure [53]. A challenging example of negative design developed target-specific peptides for 20 bZIP families, this despite the high similarity in sequence and structure of these proteins [54]. The method, CLASSY, systematically explored positive and negative designs. To gain efficiency, the group combined linear programming to optimize energy calculations and a cluster expansion to convert the structure-based problem to a sequence-based one. In 40 of the 48 cases, the new peptide bound the intended target while losing binding to undesired competitor targets.
Of note, these protein redesign methods are typically not closely linked to functional site predictions. Instead they exploit a priori knowledge of a known complex to focus on contact residues and compute potential gains in affinity or specificity based on energetics, taking the functional site for granted. Comparative methods such as the Evolutionary Trace, however, frequently link both together.
EVOLUTIONARY TRACING
The Evolutionary Trace aims to guide experiment to the amino acids involved directly in protein function [21]. It does so by ranking the impact of each sequence position on evolutionary divergence, as illustrated in Figure 1. Conceptually, ET mimics experimental mutational scanning. Whereas, in the laboratory, a sequence residue is “important” when its mutation changes the response of an assay, here ET assumes a residue is (more or less) important when its variations correlate with (greater or lesser) evolutionary divergences [21]. Unlike conservation-based methods, ET information depends crucially on the evolutionary branching pattern, every split being interpreted as a functional divergence.
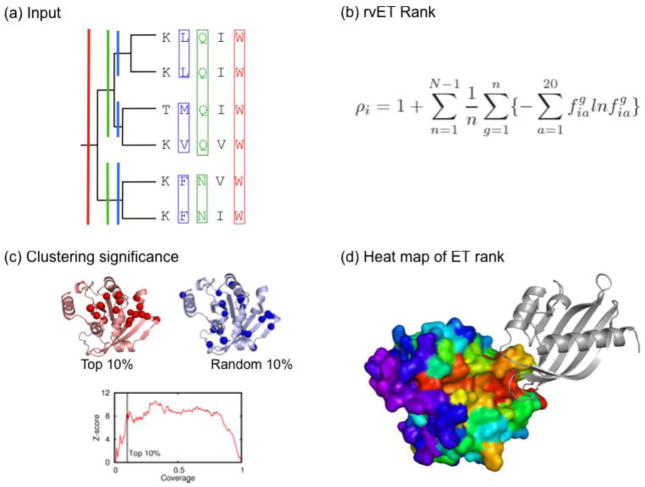
Figure 1. The Evolutionary Trace Method.
(a) A fragment of a multiple sequence alignment and the evolutionary tree of a protein family are displayed. ET considers, for each sequence position, every branch and sub branch from root to tip. The variation pattern of a rank 1 residue is complete invariance, since this correlates perfectly with the whole tree viewed as a single branch (red). The variation pattern at rank 2, when the tree is split into its first two branches, is variation between these two branches but invariance within each one (green). The relevant pattern of variation at rank 3 is, likewise, invariance within all first three branches but variation between the two nearest ones (blue). Thus, every position gets a rank–the earliest tree node after which it varies no further [21]. (b) A drawback is that sequence errors, gaps, or insertions arise ever more frequently as sequence space grows. The heuristic requirement for absolute invariance within branches can be relaxed by summing entropy terms over the sub-branches (g), weighted by the nodes (n) at which they occur in a tree with N leaves. This more robust hybrid phylogenetic-entropy approach called rvET for real-value ET [22] produces a non-integer rank of evolutionary importance (ρi) for each residue (i) making up the query protein. The frequency of amino acid a in the MSA column for residue i, in sub-branch g, is . This more robust hybrid phylogenetic-entropy approach produces non-integer rank of evolutionary importance (ρi) and is called rvET for real-value ET [22]. (c) When trace residues are mapped onto the structure, they typically cluster (red) whereas randomly picked amino acids do not (blue) [55]. Large clustering Z-scores imply the analysis is reliable and the ET site likely to be functionally relevant. (d) The resolution of functional site discovery is a parameter under operator control, it modifies the percentile rank coverage, or the threshold on a heat map of the structure where evolutionary importance decreases from red to blue, illustrated by a trace of the GDP-bound form of the Ras-family GTPase Ran bound to the ribbon form of nuclear transport factor 2 (PDB:1a2K, [93]). Public ET servers are available [23,24].
Critically, top-ranked ET residues are far from being random: they typically cluster together spatially in the protein structure, and these clusters map out functional sites. The structural clustering (for example at, but not restricted to, the 30th percentile-rank) was statistically significant in nearly all of the 46 proteins tested with diverse functions [55]. And this could then can be quantified statistically on a large scale, and in closed form, by a clustering z-score: the distance between the observed clustering pattern and the one expected by chance, expressed in units of standard deviation [56]. In turn, the surface clusters of top-ranked residues overlapped known functional sites significantly more than expected by chance, as seen retrospectively in 79 diverse proteins [57], or in prospective case studies [58,59].
The clustering and biological relevance of evolutionary important residues are tightly interconnected and general features of sequence, structure and function. First, structurally, the better top-ranked residues clustered in decoy models of protein folds, the closer these models were to the native fold [57], an observation that was independently verified [60]. Likewise, functionally, the more top-ranked residues clustered in the structure, the better these clusters predicted functional sites–a correlation tested in over 50 diverse proteins [61]. Although the structural resolution was not sufficient to guide protein folding usefully, it does provide a feedback mechanism to improve functional site predictions [62]. Together, these studies show that ET predictions are non-random; their reliability is quantified through a measure of confidence, the clustering z-score; they identify functional sites in retrospective controls; and they are widely applicable to the structural proteome.
APPLICATIONS TO PROTEIN REDESIGN
Besides these retrospective controls, laboratory studies extensively tested whether ET information could guide experiments to perturb protein interactions. A simple test was to selectively separate functions in multifunctional proteins by targeting point mutations to top-ranked amino acids [59,63–65]. In one instance, the Ku heterodimer, ET-guided experiments that produced in months many more separation of function mutants than a multi-year experimental screen in yeast, and which showed that the site mediating double-strand break DNA repair and telomere maintenance segregated to opposite ends of the Ku structure, thereby clarifying how these antagonistic functions could coexist in one complex [66].
In a second type of perturbation, manufactured peptides copy the molecular determinants of a binding site and then compete with or substitute for a native interaction [67]. In one case, a helical peptide engineered to mimic the most important residues of a new binding site suggested by ET disrupted function [68]. Although peptides have non-traditional pharmaceutical profiles, much work aims to increase their delivery and stability [69] and, clearly, they can be effective at disrupting critical pathways, such as Notch signaling [70]. Moreover, the preponderance of protein-peptide interactions, estimated to form 15–40% of all interactions within a cell [71], makes this approach a rich potential source of new molecular perturbing agents.
A third type of network perturbation rewires an interaction to a different function [6]. Since ET explicitly points out which residues are important and how they vary from branch to branch of the evolutionary tree, this provides, in principle, a protein family-specific cipher to recode function among protein homologs by swapping their cognate top-ranked residues [72]. This hypothesis has been extensively tested and led, in vitro, to swapped activity or binding [73,74], and, in vivo, to adapt a frog proneural transcription factor to a fly environment, and vice versa [75].
These different examples of functional perturbation demonstrate that bona fide predictions could be repeatedly validated in a variety of different experimental systems and laboratories. Of note, all these examples focused so far on functional residues at, or near a protein surface. A fourth type of network perturbation can also target the internal components of a protein structure.
G PROTEIN SIGNALING APPLICATIONS
About 30% of current drugs target G protein-coupled receptors (GPCRs) [76] or their associated protein network. ET was created specifically to study this pathway, which underlies smell, taste, vision, pain and much of endocrine and autonomic pharmacology. One goal is to identify and then rationally modify the molecular basis of signaling to identify novel possible therapeutic targets. Thus, following the same type of protein redesigns as above, separation of function mutations in the receptor [77], Gα [59], and Regulator of G protein Signaling (RGS) [74] validated prior predictions of interaction sites for export, receptor coupling [58], and downstream effector activation [65], respectively. Functional rewiring was demonstrated in the RGS case by switching top-ranked cognate residues among homologs [74]. And a peptide designed to mimic the top-ranked residues of a novel site in G protein receptor kinases impaired GPCR phosphorylation, confirming a role in this interaction [68]. All these studies target top-ranked ET residues at, or near, the surface of the protein.
However, evolutionary analysis can also inform the internal mechanisms of a protein. A trace of visual rhodopsins versus the broader set of rhodopsin-like GPCRs identified two separate structural subdomains buried in the core of the seven-helical transmembrane bundle [78]. The first one, unique to rhodopsins, was a putative ligand-specific binding site. The other one, common to all GPCRs, was a putative evolutionary conserved allosteric pathway that, over a distance, transforms ligand binding into effector activation. As predicted, point mutations to these sites then respectively impaired ligand binding or caused constitutive activity [78]. Moreover, three mutations in the allosteric pathway near the G protein coupling site blocked G protein signaling but kept the β-arrestin signaling intact [79]. These studies highlight the existence of functional modules within the structure and how they may be exploited to effectively sever just one of the two signaling branches efferent from activated receptors. More recently, related work investigated the correlations between sequence positions within a protein family and found similar structure-function partitioning of the protein into groups of residue positions referred to as “protein sectors” [80]. This analysis was extended to the S1A serine proteases and found mutations of the individual “protein sectors” lead to different effects focused either on catalytic power or thermal stability.
To test further how well such evolutionary modules guide protein engineering, and to understand the origin of ligand-biased signaling, whereby different ligands signal via G proteins or β-arrestin to different extents, a study swapped top-ranked ET residues from the putative common allosteric pathway between two antagonistic psychoactive receptors, those for serotonin and dopamine [81], Figure 2. All single point mutants were expressed and bound normally to a bioamine antagonist. Strikingly, all of them also exhibited altered binding or signaling, by either dopamine or serotonin, and these effects were mostly separable or even paradoxical. Notably, four mutants significantly enhanced serotonin response without increasing serotonin binding. And two of these four mutants had decreased dopamine signaling, even though dopamine affinity was as good or better than in the wild type receptor.
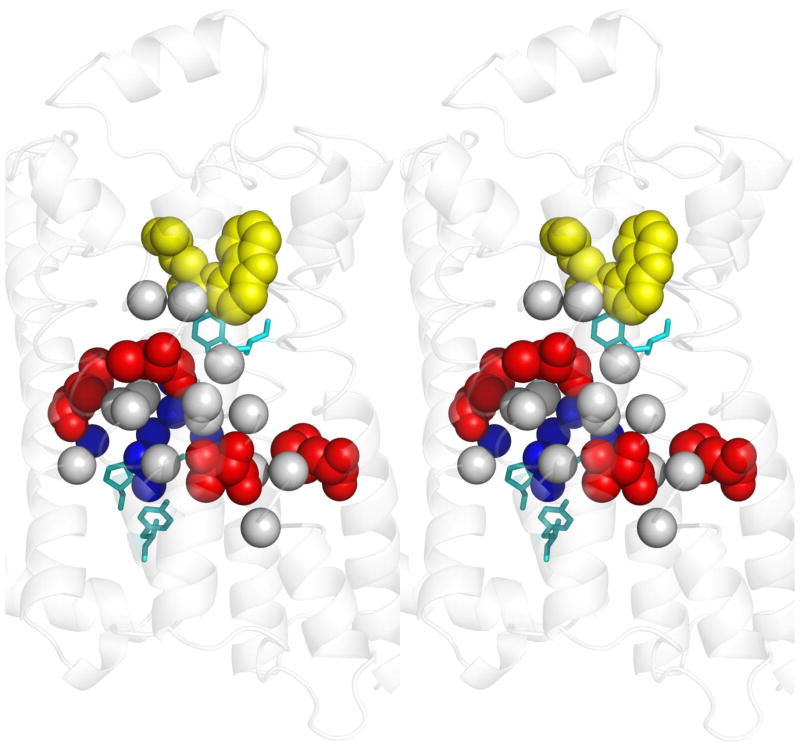
Figure 2. Determinants of allosteric specificity in psychoactive receptors.
A stereo view of the β2-adrenergic receptor structure bound to carazolol (yellow) (PDB:2rh1 [94]) provides a homology model for bioamine receptors, including the D2R and 5HT2AR dopamine and serotonin receptors. Four top-ranked amino acids (red), that are part of a putative allosteric pathway identified by ET, were swapped into D2R from their cognates in 5HT2AR and this conferred significant serotonin responsiveness to the mutants even though none had increased affinity to serotonin (red). Two of the four also had decreased dopamine responsiveness with either no change or with a paradoxical increase in dopamine affinity. Although distant from the ligand binding site, these residues are closely associated to other top-ranked ET residues in the putative allosteric pathway that are invariant between D2DR and 5HT2AR (Cα atoms of residues within 5 Å, gray). Together, these top-ranked residues link the toggle switch (top, cyan residue) and the NPxxY motif (lower cyan residues) that are generic GPCR mediators of activation; and they also surround a pocket of structural waters (blue spheres). Like rekeying the tumblers of a lock, the exchange of these residues shifted the sensitivity of the allosteric pathway from one ligand to another, independently of changes to binding affinity. These allosteric specificity determinants are consistent with the pharmacology of ligand-biased signaling [81].
This independent reprogramming of binding and of signaling from dopamine to serotonin highlights allosteric specificity, namely, the pathway itself can determine which bound ligands signal, separate of binding site affinity. Moreover, the key determinants of the allosteric pathway can be traced evolutionarily and then recoded, one top-ranked ET residues at a time–much as the tumblers of a lock are rekeyed. Presumably, during evolution, single mutations constantly change ligand affinity, effector biases, or the wiring between them, and thus probe alternative wiring at GPCR network nodes. In practice, many of the key residues surround structural waters, as shown in Figure 2, suggesting a potential site where a drug could influence ligand-biased signaling [81].
FROM DETERMINANTS TO LARGE SCALE FUNCTION PREDICTION
Case studies such as these are informative, but they cannot prove that a method is broadly applicable. To do so would require that functional determinants be identified and shown to be predictive of function–on a proteomic scale. A simple example is the Serine-Histidine-Aspartate catalytic triad, a three amino-acid structural motif often sufficient to identify proteases [82]. More generally, methods to annotate the unknown function of the novel structures produced by Structural Genomics [83] follow this logic and transfer annotations between proteins based on local sequence and structure similarities [84,85].
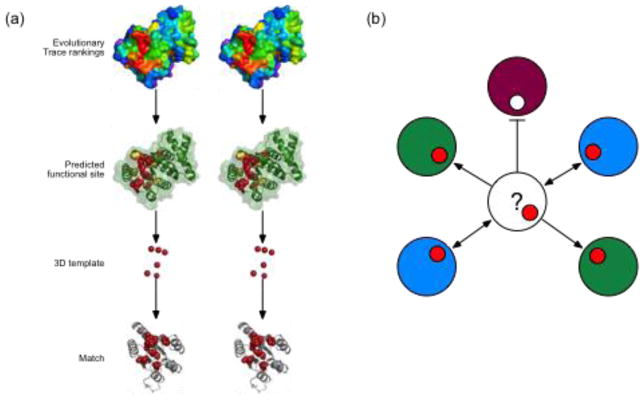
Likewise, an Evolutionary Trace Annotation (ETA) server was developed to predict the function of novel protein structures, as illustrated in Figure 3. Starting with a query structure, ETA traces it, identifies the largest surface cluster of top-ranked residues and picks from those a structural motifs, or 3D template. The template can then be used to search all PDB structures for similarities that suggest a common function. While geometric matches within 2 Å root mean square deviation are often random, the specificity rises to over 90% once these matches are also filtered for (i) the importance of the matched site [86], (ii) reciprocity, so the 3D-template of the match matches back to the query [87], and (iii) plurality, so that multiple matches point to the same function more than to any other [88]. This approach is scalable the structural proteome to annotate over 1200 structural genomics enzyme up to three Enzyme Classification digits with 92% accuracy [89], or non-enzymes using the Gene Ontology functional classification [29]. These annotations suggest that only six top-ranked amino acids are sufficient to identify function. Moreover, in enzymes, simply substituting other residues that may be more directly associated with catalysis lowered rather than raised accuracy, showing that the definition of a necessary set of residues to define function is complex [29]. Overall, these studies complement case controls to confirm the proteome-wide possibility of picking functional determinants from evolutionary comparisons.
Figure 3. Structure function prediction based on evolutionary 3D templates.
(a) Evolutionary Trace rankings of residue importance for Mycobacterium tuberculosis v1626 (PDB 1sd5, chain A) are represented as a heat map of the structure’s surface (red, most important; purple, least important). Based on these rankings, the most important residues are mapped onto the structure (green ribbons) to identify a solvent-accessible cluster of 10 ET residues (red and yellow spheres). The Cα coordinates of the top six residues (red) are used as the template and searched against a database of annotated target structures. (b) A conceptual diagram of ETA heuristic filtering shows proteins as large circles, with color representing functions and templates matches as smaller circles. ETA first discards matched sites if they are not themselves evolutionary important. Red matches indicate importance and pass the filter (arrows), while white matches are unimportant and do not pass this filter (flat-headed line). ETA then examines match reciprocity, with one-way (single-headed arrows) matches rejected, and reciprocal matches (double-handed arrows) accepted. Finally, ETA requires that a predicted function achieve a vote plurality; here, after all filters are used, the two proteins with the “blue” function represent the majority of the matches. ETA would therefore predict that the query protein (question mark) has the blue function. Public ETA servers are available at http://mammoth.bcm.tmc.edu [89].
CONCLUDING REMARKS
Predictive algorithms must fulfill specific objective criteria: (a) to produce results that are non-random; (b) to match retrospective controls; (c) to also match prospective controls, i.e. make genuine predictions that are then experimentally validated; (d) and to be scalable to a well-defined domain of application. A fifth requirement is, since in biology a single method is unlikely to be unfailingly predictive, (e) to quantify prediction confidence to distinguish favorable cases from others that are less so. As seen above, the Evolutionary Trace fulfills these conditions.
A biological result is that ET servers offer a reliable approach to focus protein redesign studies to the most relevant parts of a protein [23,24,89]. The elucidation of allosteric specificity determinants linked to ligand-based signaling in GPCRs is an example. From a theoretical perspective, ET also points to general proteomic rules and to fundamental evolutionary patterns: Sequence residues are ranked by evolution; the important ones cluster; these clusters indicate functional sites; clustering quality correlated with functional site prediction quality; and variations at top-ranked residues generally control functional specificity.
It is helpful to understand the origin of this diverse information, which seems at odds with what might be expected from simple conservation analysis. First, it is important to stress that ET does not rely on conservation but rather on variation. The variations linked to small divergences are ranked poorly, those linked to major divergences are ranked superiorly. In effect, the tree divergences provide virtual functional “assays”. Since a tree with N sequences has N−1 nodes, ET analysis benefits from vastly more functional assays than an experimental laboratory. Moreover, the sequence variations under study are all informative since they occur in living species, and thus are evolutionary successful. Thus, comparative analysis with ET benefits from a wealth of relevant biological information. Second, the clustering of top-ranked is also informative, as it links functional impact to the structural continuity of evolutionary selection forces. The tree thus literally acts as a cipher to deconvolute evolutionary variations in sequence and function and, with a structure, enables high-resolution definition of functional sites.
An open question remains the ultimate domains of application of these techniques, tested thus far in structurally well-defined proteins. For example, many important interactions couple protein folding with protein binding in intrinsically disordered regions [90]. Whether disordered proteins can fit into the same evolutionary framework is not demonstrated. Even more challenging would be to fit in exotic experiments that demonstrate the remarkable change that a single residue mutation can bring about on both structure and function [91]. Also with the advent of personal genomes [92], improved interpretations of sequence variations from an evolutionary perspective would be desirable.
In the near term, since comparative analysis does not rely on energetics, these distinct and experimentally validated approaches should be complementary. This suggests that as protein networks become better defined, computational tools may reliably design protein variants and peptide tools that guide systematic perturbations in order to assess the mechanisms, and screen for therapeutics, aimed at networks.
Acknowledgments
We thank Matthew Ward and Serkan Erdin for contributing Figure 3. O.L. gratefully acknowledges support by grants from the NIH, GM079656 and GM066099, and from the NSF, DBI-0547695 and CCF 0905536. A.D.W, was supported by training fellowships from the National Library of Medicine to the Keck Center for Interdisciplinary Bioscience Training of the Gulf Coast Consortia (NLM grant 5T15LM07093).
Abbreviations
- PPI
Protein-protein interaction
- ET
Evolutionary Trace
- MSA
Multiple Sequence Alignment
- PDB
Protein Databank
- GPCR
G protein-coupled receptor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 2.Zanzoni A, Soler-Lopez M, Aloy P. A network medicine approach to human disease. FEBS Lett. 2009;583:1759–1765. doi: 10.1016/j.febslet.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Schuster-Bockler B, Bateman A. Protein interactions in human genetic diseases. Genome Biol. 2008;9:R9. doi: 10.1186/gb-2008-9-1-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lim J, Hao T, Shaw C, Patel AJ, Szabo G, Rual JF, Fisk CJ, Li N, Smolyar A, Hill DE, et al. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125:801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 5.Pawson T, Linding R. Network medicine. FEBS Lett. 2008;582:1266–1270. doi: 10.1016/j.febslet.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 6.van der Sloot AM, Tur V, Szegezdi E, Mullally MM, Cool RH, Samali A, Serrano L, Quax WJ. Designed tumor necrosis factor-related apoptosis-inducing ligand variants initiating apoptosis exclusively via the DR5 receptor. Proc Natl Acad Sci U S A. 2006;103:8634–8639. doi: 10.1073/pnas.0510187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tong W, Wei Y, Murga LF, Ondrechen MJ, Williams RJ. Partial order optimum likelihood (POOL): maximum likelihood prediction of protein active site residues using 3D Structure and sequence properties. PLoS Comput Biol. 2009;5:e1000266. doi: 10.1371/journal.pcbi.1000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Capra JA, Laskowski RA, Thornton JM, Singh M, Funkhouser TA. Predicting protein ligand binding sites by combining evolutionary sequence conservation and 3D structure. PLoS Comput Biol. 2009;5:e1000585. doi: 10.1371/journal.pcbi.1000585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ofran Y, Rost B. Protein-protein interaction hotspots carved into sequences. PLoS Comput Biol. 2007;3:e119. doi: 10.1371/journal.pcbi.0030119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chakrabarti S, Bryant SH, Panchenko AR. Functional specificity lies within the properties and evolutionary changes of amino acids. J Mol Biol. 2007;373:801–810. doi: 10.1016/j.jmb.2007.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glaser F, Morris RJ, Najmanovich RJ, Laskowski RA, Thornton JM. A method for localizing ligand binding pockets in protein structures. Proteins. 2006;62:479–488. doi: 10.1002/prot.20769. [DOI] [PubMed] [Google Scholar]
- 12.Davis IW, Baker D. RosettaLigand docking with full ligand and receptor flexibility. J Mol Biol. 2009;385:381–392. doi: 10.1016/j.jmb.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 13.Ondrechen MJ, Clifton JG, Ringe D. THEMATICS: a simple computational predictor of enzyme function from structure. Proc Natl Acad Sci U S A. 2001;98:12473–12478. doi: 10.1073/pnas.211436698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pettit FK, Bare E, Tsai A, Bowie JU. HotPatch: a statistical approach to finding biologically relevant features on protein surfaces. J Mol Biol. 2007;369:863–879. doi: 10.1016/j.jmb.2007.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shazman S, Celniker G, Haber O, Glaser F, Mandel-Gutfreund Y. Patch Finder Plus (PFplus): a web server for extracting and displaying positive electrostatic patches on protein surfaces. Nucleic Acids Res. 2007;35:W526–530. doi: 10.1093/nar/gkm401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tuncbag N, Gursoy A, Keskin O. Identification of computational hot spots in protein interfaces: combining solvent accessibility and inter-residue potentials improves the accuracy. Bioinformatics. 2009;25:1513–1520. doi: 10.1093/bioinformatics/btp240. [DOI] [PubMed] [Google Scholar]
- 17.Glaser F, Pupko T, Paz I, Bell RE, Bechor-Shental D, Martz E, Ben-Tal N. ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics. 2003;19:163–164. doi: 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
- 18.Innis CA. siteFiNDER|3D: a web-based tool for predicting the location of functional sites in proteins. Nucleic Acids Res. 2007;35:W489–494. doi: 10.1093/nar/gkm422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sankararaman S, Sjolander K. INTREPID--INformation-theoretic TREe traversal for Protein functional site IDentification. Bioinformatics. 2008;24:2445–2452. doi: 10.1093/bioinformatics/btn474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Engelen S, Trojan LA, Sacquin-Mora S, Lavery R, Carbone A. Joint evolutionary trees: a large-scale method to predict protein interfaces based on sequence sampling. PLoS Comput Biol. 2009;5:e1000267. doi: 10.1371/journal.pcbi.1000267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lichtarge O, Bourne HR, Cohen FE. An evolutionary trace method defines binding surfaces common to protein families. J Mol Biol. 1996;257:342–358. doi: 10.1006/jmbi.1996.0167. [DOI] [PubMed] [Google Scholar]
- 22.Mihalek I, Res I, Lichtarge O. A family of evolution-entropy hybrid methods for ranking protein residues by importance. J Mol Biol. 2004;336:1265–1282. doi: 10.1016/j.jmb.2003.12.078. [DOI] [PubMed] [Google Scholar]
- 23.Mihalek I, Res I, Lichtarge O. Evolutionary trace report_maker: a new type of service for comparative analysis of proteins. Bioinformatics. 2006;22:1656–1657. doi: 10.1093/bioinformatics/btl157. [DOI] [PubMed] [Google Scholar]
- 24.Morgan DH, Kristensen DM, Mittelman D, Lichtarge O. ET viewer: an application for predicting and visualizing functional sites in protein structures. Bioinformatics. 2006;22:2049–2050. doi: 10.1093/bioinformatics/btl285. [DOI] [PubMed] [Google Scholar]
- 25.Sankararaman S, Sha F, Kirsch JF, Jordan MI, Sjolander K. Active site prediction using evolutionary and structural information. Bioinformatics. 26:617–624. doi: 10.1093/bioinformatics/btq008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang B, Schroeder M. LIGSITEcsc: predicting ligand binding sites using the Connolly surface and degree of conservation. BMC Struct Biol. 2006;6:19. doi: 10.1186/1472-6807-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bradford JR, Westhead DR. Improved prediction of protein-protein binding sites using a support vector machines approach. Bioinformatics. 2005;21:1487–1494. doi: 10.1093/bioinformatics/bti242. [DOI] [PubMed] [Google Scholar]
- 28.Skolnick J, Brylinski M. FINDSITE: a combined evolution/structure-based approach to protein function prediction. Brief Bioinform. 2009;10:378–391. doi: 10.1093/bib/bbp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29•.Erdin S, Ward RM, Venner E, Lichtarge O. Evolutionary trace annotation of protein function in the structural proteome. J Mol Biol. 2010;396:1451–1473. doi: 10.1016/j.jmb.2009.12.037. Reliable prediction of function in enzymes and non-enzymes alike, based on local structural similarities of just six evolutionary important amino acids. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 31.Tsai CJ, Lin SL, Wolfson HJ, Nussinov R. A dataset of protein-protein interfaces generated with a sequence-order-independent comparison technique. J Mol Biol. 1996;260:604–620. doi: 10.1006/jmbi.1996.0424. [DOI] [PubMed] [Google Scholar]
- 32.Jones S, Thornton JM. Principles of protein-protein interactions. Proc Natl Acad Sci U S A. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loriot S, Cazals F. Modeling Macro-Molecular Interfaces with Intervor. Bioinformatics. doi: 10.1093/bioinformatics/btq052. [DOI] [PubMed] [Google Scholar]
- 34.Wang K, Horst JA, Cheng G, Nickle DC, Samudrala R. Protein meta-functional signatures from combining sequence, structure, evolution, and amino acid property information. PLoS Comput Biol. 2008;4:e1000181. doi: 10.1371/journal.pcbi.1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chelliah V, Chen L, Blundell TL, Lovell SC. Distinguishing structural and functional restraints in evolution in order to identify interaction sites. J Mol Biol. 2004;342:1487–1504. doi: 10.1016/j.jmb.2004.08.022. [DOI] [PubMed] [Google Scholar]
- 36.Yue P, Moult J. Identification and analysis of deleterious human SNPs. J Mol Biol. 2006;356:1263–1274. doi: 10.1016/j.jmb.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 37.Coward P, Wada HG, Falk MS, Chan SD, Meng F, Akil H, Conklin BR. Controlling signaling with a specifically designed Gi-coupled receptor. Proc Natl Acad Sci U S A. 1998;95:352–357. doi: 10.1073/pnas.95.1.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fazelinia H, Cirino PC, Maranas CD. OptGraft: A computational procedure for transferring a binding site onto an existing protein scaffold. Protein Sci. 2009;18:180–195. doi: 10.1002/pro.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu S, Zhu X, Liang H, Cao A, Chang Z, Lai L. Nonnatural protein-protein interaction-pair design by key residues grafting. Proc Natl Acad Sci U S A. 2007;104:5330–5335. doi: 10.1073/pnas.0606198104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reynolds KA, Thomson JM, Corbett KD, Bethel CR, Berger JM, Kirsch JF, Bonomo RA, Handel TM. Structural and computational characterization of the SHV-1 beta-lactamase-beta-lactamase inhibitor protein interface. J Biol Chem. 2006;281:26745–26753. doi: 10.1074/jbc.M603878200. [DOI] [PubMed] [Google Scholar]
- 41.Yosef E, Politi R, Choi MH, Shifman JM. Computational design of calmodulin mutants with up to 900-fold increase in binding specificity. J Mol Biol. 2009;385:1470–1480. doi: 10.1016/j.jmb.2008.09.053. [DOI] [PubMed] [Google Scholar]
- 42.Rohl CA, Strauss CE, Misura KM, Baker D. Protein structure prediction using Rosetta. Methods Enzymol. 2004;383:66–93. doi: 10.1016/S0076-6879(04)83004-0. [DOI] [PubMed] [Google Scholar]
- 43.Brooks BR, Brooks CL, 3rd, Mackerell AD, Jr, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, et al. CHARMM: the biomolecular simulation program. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sammond DW, Eletr ZM, Purbeck C, Kimple RJ, Siderovski DP, Kuhlman B. Structure-based protocol for identifying mutations that enhance protein-protein binding affinities. J Mol Biol. 2007;371:1392–1404. doi: 10.1016/j.jmb.2007.05.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45•.Haidar JN, Pierce B, Yu Y, Tong W, Li M, Weng Z. Structure-based design of a T-cell receptor leads to nearly 100-fold improvement in binding affinity for pepMHC. Proteins. 2009;74:948–960. doi: 10.1002/prot.22203. Mutations predicted to enhance packing and electrostatics at a protein-peptide interface. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boas FE, Harbury PB. Design of protein-ligand binding based on the molecular-mechanics energy model. J Mol Biol. 2008;380:415–424. doi: 10.1016/j.jmb.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Joachimiak LA, Kortemme T, Stoddard BL, Baker D. Computational design of a new hydrogen bond network and at least a 300-fold specificity switch at a protein-protein interface. J Mol Biol. 2006;361:195–208. doi: 10.1016/j.jmb.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 48.Kortemme T, Joachimiak LA, Bullock AN, Schuler AD, Stoddard BL, Baker D. Computational redesign of protein-protein interaction specificity. Nat Struct Mol Biol. 2004;11:371–379. doi: 10.1038/nsmb749. [DOI] [PubMed] [Google Scholar]
- 49.Potapov V, Reichmann D, Abramovich R, Filchtinski D, Zohar N, Ben Halevy D, Edelman M, Sobolev V, Schreiber G. Computational redesign of a protein-protein interface for high affinity and binding specificity using modular architecture and naturally occurring template fragments. J Mol Biol. 2008;384:109–119. doi: 10.1016/j.jmb.2008.08.078. [DOI] [PubMed] [Google Scholar]
- 50.Lee J, Natarajan M, Nashine VC, Socolich M, Vo T, Russ WP, Benkovic SJ, Ranganathan R. Surface sites for engineering allosteric control in proteins. Science. 2008;322:438–442. doi: 10.1126/science.1159052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51•.Chen CY, Georgiev I, Anderson AC, Donald BR. Computational structure-based redesign of enzyme activity. Proc Natl Acad Sci U S A. 2009;106:3764–3769. doi: 10.1073/pnas.0900266106. Structure-based redesign increases specificity to wild type substrates or switches it to a non-cognate substrates against which the wild-type enzyme was previously inactive. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52••.Murphy PM, Bolduc JM, Gallaher JL, Stoddard BL, Baker D. Alteration of enzyme specificity by computational loop remodeling and design. Proc Natl Acad Sci U S A. 2009;106:9215–9220. doi: 10.1073/pnas.0811070106. Precise constrained loop remodeling near the active site changes relative enzyme specificity by 6 orders of magnitude from normal to target substrate. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53•.Bolon DN, Grant RA, Baker TA, Sauer RT. Specificity versus stability in computational protein design. Proc Natl Acad Sci U S A. 2005;102:12724–12729. doi: 10.1073/pnas.0506124102. Strengths and limitations of positive and negative designs to guide and stabilize protein associations into homo- versus heterodimers. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54••.Grigoryan G, Reinke AW, Keating AE. Design of protein-interaction specificity gives selective bZIP-binding peptides. Nature. 2009;458:859–864. doi: 10.1038/nature07885. A step towards the challenging problem of selective inhibition among closely related ancestrally related protein-interactions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Madabushi S, Yao H, Marsh M, Kristensen DM, Philippi A, Sowa ME, Lichtarge O. Structural clusters of evolutionary trace residues are statistically significant and common in proteins. J Mol Biol. 2002;316:139–154. doi: 10.1006/jmbi.2001.5327. [DOI] [PubMed] [Google Scholar]
- 56.Mihalek I, Res I, Yao H, Lichtarge O. Combining inference from evolution and geometric probability in protein structure evaluation. J Mol Biol. 2003;331:263–279. doi: 10.1016/s0022-2836(03)00663-6. [DOI] [PubMed] [Google Scholar]
- 57.Yao H, Kristensen DM, Mihalek I, Sowa ME, Shaw C, Kimmel M, Kavraki L, Lichtarge O. A Sensitive, Accurate, and Scalable Method to Identify Functional Sites in Protein Structures. J Mol Bio. 2003 doi: 10.1016/s0022-2836(02)01336-0. In Press. [DOI] [PubMed] [Google Scholar]
- 58.Lichtarge O, Bourne HR, Cohen FE. Evolutionarily conserved Galphabetagamma binding surfaces support a model of the G protein-receptor complex. Proc Natl Acad Sci U S A. 1996;93:7507–7511. doi: 10.1073/pnas.93.15.7507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Onrust R, Herzmark P, Chi P, Garcia PD, Lichtarge O, Kingsley C, Bourne HR. Receptor and betagamma binding sites in the alpha subunit of the retinal G protein transducin. Science. 1997;275:381–384. doi: 10.1126/science.275.5298.381. [DOI] [PubMed] [Google Scholar]
- 60.Schueler-Furman O, Baker D. Conserved residue clustering and protein structure prediction. Proteins. 2003;52:225–235. doi: 10.1002/prot.10365. [DOI] [PubMed] [Google Scholar]
- 61.Mihalek I, Res I, Lichtarge O. Evolutionary and structural feedback on selection of sequences for comparative analysis of proteins. Proteins. 2006;63:87–99. doi: 10.1002/prot.20866. [DOI] [PubMed] [Google Scholar]
- 62.Yao H, Mihalek I, Lichtarge O. Rank information: a structure-independent measure of evolutionary trace quality that improves identification of protein functional sites. Proteins. 2006;65:111–123. doi: 10.1002/prot.21101. [DOI] [PubMed] [Google Scholar]
- 63.Cushman I, Bowman BR, Sowa ME, Lichtarge O, Quiocho FA, Moore MS. Computational and biochemical identification of a nuclear pore complex binding site on the nuclear transport carrier NTF2. J Mol Biol. 2004;344:303–310. doi: 10.1016/j.jmb.2004.09.043. [DOI] [PubMed] [Google Scholar]
- 64.Rajagopalan L, Patel N, Madabushi S, Goddard JA, Anjan V, Lin F, Shope C, Farrell B, Lichtarge O, Davidson AL, et al. Essential helix interactions in the anion transporter domain of prestin revealed by evolutionary trace analysis. J Neurosci. 2006;26:12727–12734. doi: 10.1523/JNEUROSCI.2734-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sowa ME, He W, Wensel TG, Lichtarge O. A regulator of G protein signaling interaction surface linked to effector specificity. Proc Natl Acad Sci U S A. 2000;97:1483–1488. doi: 10.1073/pnas.030409597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ribes-Zamora A, Mihalek I, Lichtarge O, Bertuch AA. Distinct faces of the Ku heterodimer mediate DNA repair and telomeric functions. Nat Struct Mol Biol. 2007;14:301–307. doi: 10.1038/nsmb1214. [DOI] [PubMed] [Google Scholar]
- 67.Churchill EN, Qvit N, Mochly-Rosen D. Rationally designed peptide regulators of protein kinase C. Trends Endocrinol Metab. 2009;20:25–33. doi: 10.1016/j.tem.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Baameur F, Morgan DH, Yao H, Tran TM, Hammitt RA, Sabui S, McMurray JS, Lichtarge O, Clark RB. Role for the regulator of G-protein signaling homology domain of G protein-coupled receptor kinases 5 and 6 in beta 2-adrenergic receptor and rhodopsin phosphorylation. Mol Pharmacol. 2010;77:405–415. doi: 10.1124/mol.109.058115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Prive GG, Melnick A. Specific peptides for the therapeutic targeting of oncogenes. Curr Opin Genet Dev. 2006;16:71–77. doi: 10.1016/j.gde.2005.12.010. [DOI] [PubMed] [Google Scholar]
- 70.Moellering RE, Cornejo M, Davis TN, Del Bianco C, Aster JC, Blacklow SC, Kung AL, Gilliland DG, Verdine GL, Bradner JE. Direct inhibition of the NOTCH transcription factor complex. Nature. 2009;462:182–188. doi: 10.1038/nature08543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Neduva V, Linding R, Su-Angrand I, Stark A, de Masi F, Gibson TJ, Lewis J, Serrano L, Russell RB. Systematic discovery of new recognition peptides mediating protein interaction networks. PLoS Biol. 2005;3:e405. doi: 10.1371/journal.pbio.0030405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lichtarge O, Yamamoto KR, Cohen FE. Identification of functional surfaces of the zinc binding domains of intracellular receptors. J Mol Biol. 1997;274:325–337. doi: 10.1006/jmbi.1997.1395. [DOI] [PubMed] [Google Scholar]
- 73.Raviscioni M, Gu P, Sattar M, Cooney AJ, Lichtarge O. Correlated evolutionary pressure at interacting transcription factors and DNA response elements can guide the rational engineering of DNA binding specificity. J Mol Biol. 2005;350:402–415. doi: 10.1016/j.jmb.2005.04.054. [DOI] [PubMed] [Google Scholar]
- 74.Sowa ME, He W, Slep KC, Kercher MA, Lichtarge O, Wensel TG. Prediction and confirmation of a site critical for effector regulation of RGS domain activity. Nat Struct Biol. 2001;8:234–237. doi: 10.1038/84974. [DOI] [PubMed] [Google Scholar]
- 75.Quan XJ, Denayer T, Yan J, Jafar-Nejad H, Philippi A, Lichtarge O, Vleminckx K, Hassan BA. Evolution of neural precursor selection: functional divergence of proneural proteins. Development. 2004;131:1679–1689. doi: 10.1242/dev.01055. [DOI] [PubMed] [Google Scholar]
- 76.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 77.Kobayashi H, Ogawa K, Yao R, Lichtarge O, Bouvier M. Functional rescue of beta-adrenoceptor dimerization and trafficking by pharmacological chaperones. Traffic. 2009;10:1019–1033. doi: 10.1111/j.1600-0854.2009.00932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Madabushi S, Gross AK, Philippi A, Meng EC, Wensel TG, Lichtarge O. Evolutionary trace of G protein-coupled receptors reveals clusters of residues that determine global and class-specific functions. J Biol Chem. 2004;279:8126–8132. doi: 10.1074/jbc.M312671200. [DOI] [PubMed] [Google Scholar]
- 79.Shenoy SK, Drake MT, Nelson CD, Houtz DA, Xiao K, Madabushi S, Reiter E, Premont RT, Lichtarge O, Lefkowitz RJ. beta-arrestin-dependent, G protein-independent ERK1/2 activation by the beta2 adrenergic receptor. J Biol Chem. 2006;281:1261–1273. doi: 10.1074/jbc.M506576200. [DOI] [PubMed] [Google Scholar]
- 80.Halabi N, Rivoire O, Leibler S, Ranganathan R. Protein sectors: evolutionary units of three-dimensional structure. Cell. 2009;138:774–786. doi: 10.1016/j.cell.2009.07.038. An approach distinct from ET that also observed functional clusters and subclusters. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81••.Rodriguez GJ, Yao R, Lichtarge O, Wensel TG. Evolution-guided discovery and recoding of allosteric pathway specificity determinants in psychoactive bioamine receptors. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.0914877107. In Press. Evolutionary dissection of a molecular lock and key mechanism: An allosteric pathway is identified and, independent of binding affinity, its amino acid tumblers rekeyed to an alternative ligand. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wallace AC, Laskowski RA, Thornton JM. Derivation of 3D coordinate templates for searching structural databases: application to Ser-His-Asp catalytic triads in the serine proteinases and lipases. Protein Sci. 1996;5:1001–1013. doi: 10.1002/pro.5560050603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nair R, Liu J, Soong TT, Acton TB, Everett JK, Kouranov A, Fiser A, Godzik A, Jaroszewski L, Orengo C, et al. Structural genomics is the largest contributor of novel structural leverage. J Struct Funct Genomics. 2009;10:181–191. doi: 10.1007/s10969-008-9055-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Arakaki AK, Huang Y, Skolnick J. EFICAz2: enzyme function inference by a combined approach enhanced by machine learning. BMC Bioinformatics. 2009;10:107. doi: 10.1186/1471-2105-10-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Laskowski RA, Watson JD, Thornton JM. Protein function prediction using local 3D templates. J Mol Biol. 2005;351:614–626. doi: 10.1016/j.jmb.2005.05.067. [DOI] [PubMed] [Google Scholar]
- 86.Kristensen DM, Chen BY, Fofanov VY, Ward RM, Lisewski AM, Kimmel M, Kavraki LE, Lichtarge O. Recurrent use of evolutionary importance for functional annotation of proteins based on local structural similarity. Protein Sci. 2006;15:1530–1536. doi: 10.1110/ps.062152706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ward RM, Erdin S, Tran TA, Kristensen DM, Lisewski AM, Lichtarge O. De-orphaning the structural proteome through reciprocal comparison of evolutionarily important structural features. PLoS One. 2008;3:e2136. doi: 10.1371/journal.pone.0002136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kristensen DM, Ward RM, Lisewski AM, Erdin S, Chen BY, Fofanov VY, Kimmel M, Kavraki LE, Lichtarge O. Prediction of enzyme function based on 3D templates of evolutionarily important amino acids. BMC Bioinformatics. 2008;9:17. doi: 10.1186/1471-2105-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ward RM, Venner E, Daines B, Murray S, Erdin S, Kristensen DM, Lichtarge O. Evolutionary Trace Annotation Server: automated enzyme function prediction in protein structures using 3D templates. Bioinformatics. 2009;25:1426–1427. doi: 10.1093/bioinformatics/btp160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wright PE, Dyson HJ. Linking folding and binding. Curr Opin Struct Biol. 2009;19:31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91••.Alexander PA, He Y, Chen Y, Orban J, Bryan PN. A minimal sequence code for switching protein structure and function. Proc Natl Acad Sci U S A. 2009;106:21149–21154. doi: 10.1073/pnas.0906408106. A single amino acid change flips a non-natural 4beta+alpha protein that binds IgG into a 3-alpha structure that binds albumin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lupski JR, Reid JG, Gonzaga-Jauregui C, Rio Deiros D, Chen DC, Nazareth L, Bainbridge M, Dinh H, Jing C, Wheeler DA, et al. Whole-Genome Sequencing in a Patient with Charcot-Marie-Tooth Neuropathy. N Engl J Med. 2010 doi: 10.1056/NEJMoa0908094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stewart M, Kent HM, McCoy AJ. Structural basis for molecular recognition between nuclear transport factor 2 (NTF2) and the GDP-bound form of the Ras-family GTPase Ran. J Mol Biol. 1998;277:635–646. doi: 10.1006/jmbi.1997.1602. [DOI] [PubMed] [Google Scholar]
- 94.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, et al. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]