Summary
Protein families with functionally diverse members can illuminate the structural determinants of protein function and the process by which protein structure and function evolve. To identify the key amino acid changes that differentiate one family member from another, most studies have taken a “horizontal” approach, swapping candidate residues between present-day family members. This approach has often been stymied, however, by the fact that shifts in function often require multiple interacting mutations; chimeric proteins are often non-functional, either because one lineage has amassed mutations that are incompatible with key residues that conferred a new function on other lineages, or because it lacks mutations required to support those key residues. These difficulties can be overcome by using a vertical strategy, which reconstructs ancestral genes and uses them as the appropriate background in which to study the effects of historical mutations on functional diversification. In this review, we discuss the advantages of the vertical strategy and highlight several exemplary studies that have used ancestral gene reconstruction to reveal the molecular underpinnings of protein structure, function, and evolution.
Introduction
Biochemists would like to know how protein sequence determines structure and function; molecular evolutionary biologists are interested in the processes that generated the diverse structures and functions of extant proteins. Answering either question requires some knowledge of the distribution of structures and functions through the multidimensional “space” of possible protein sequences [1,2]. Characterizing that distribution is extremely difficult, however, because of the vast number of possible sequences and the time required to experimentally generate and study them: even high-throughput methods for generating and screening mutant libraries explore tiny regions around defined starting points within sequence space.
One solution to this problem is to analyze the evolutionary record. Evolution has been a massive experiment in the diversification and optimization of protein structure-function relations, conducted in countless parallel lineages over vast periods of time. The outcomes of that experiment are preserved in the sequences, structures, and functions of modern-day protein families. Evolutionary analysis of these families therefore has the potential to provide key insights into the nature of protein sequence space and the determinants of protein structure and function.
Horizontal vs. vertical analysis of protein families
How can protein families best be studied? A major goal for biochemists and evolutionary biologists alike is to identify the necessary and sufficient subset of residues that cause functional differences between family members. One strategy is to identify candidate amino acid differences between divergent family members using sequence-based or structural analysis [3–6], and then test the functional role of these residues by swapping them between family members using site-directed mutagenesis. This “horizontal” approach often identifies residues that are important to one function, because changing them results in an impaired or nonfunctional protein [7–9], but it rarely identifies the set of residues sufficient to switch the function of one protein to that of another. The enolases, for example, are a well-studied family whose members share a common fold and enzymatic mechanism but catalyze diverse reactions [10,11]. Despite many attempts, few have succeeded in altering one enolase to catalyze the reaction of another [12], and even these have generated enzymes with considerably lower efficiency than their natural counterparts [13,14], indicating that not all residues important for function have been identified.
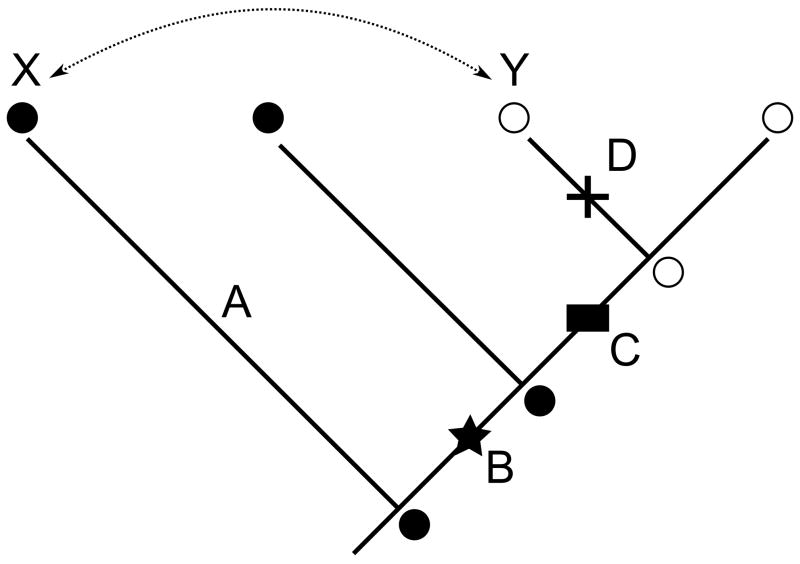
The reason studies of this type fall short is that they ignore history. Protein function evolved as mutations accumulated through time—vertically—in ancestral protein lineages, whereas horizontal comparisons of modern proteins involve only the tips of the evolutionary tree. The horizontal approach suffers from two major problems. First, it is inefficient, because many sequence differences irrelevant to the functional difference may have accumulated during intervals in which they functions of interest did not change (Fig. 1).
Figure 1.
Dissecting the sequence determinants of function within a family that has an ancestral function (filled circle) and a derived function (open circle). The functional change was caused by a subset of the sequence changes along branch C (black box). In the scenario shown here, permissive mutations on branch B (star) were required to allow the protein to tolerate the function-switching mutations; restrictive mutations incompatible with the ancestral function accumulated on branch D (cross). Swapping residues between modern proteins (arrow) is inefficient because the sequences differ by all mutations along A, B, C, and D. Further, protein X does not have the permissive mutations and cannot take on the derived function. Protein Y has restrictive mutations that do not allow it tolerate the ancestral function.
Second, lineage-specific sequence changes may lead to epistasis [15–19], interdependence between mutations that cause a single change to have different effects in different protein family members. If epistasis occurs, studies in a modern protein background may not reveal the effect of mutations on other family members or the ancestral proteins in which new functions actually evolved. Two varieties of epistatic mutations along evolutionary trajectories are particularly relevant. Permissive mutations introduce amino acids required for a protein to tolerate key function-switching mutations. These mutations may, for example, increase protein stability to buffer the protein against destabilizing functional residues [15,20]. Conversely, restrictive mutations introduce residues that are incompatible with the functions of other family members, because they produce steric clashes, for examples [21]. Swapping putative function-switching residues into proteins from lineages in which restrictive mutations occurred—or permissive mutations did not occur—will result in a nonfunctional or impaired protein, even if the residues tested indeed were the historical cause of the new function.
An explicitly phylogenetic approach to studies of functional diversity within protein families could address these issues. A vertical strategy would focus on mutations that occurred along the branch in the family tree on which functional diversification occurred. This strategy would be more efficient, because only those mutations that occurred during a limited period of evolutionary time need be investigated as candidates (Fig. 1). Moreover, by using the protein background in which the sequence changes actually occurred, this approach could avoid the effect of epistatic interactions that can confound experimental tests of the functional importance of those candidates. A vertical strategy would even allow the restrictive and permissive epistatic mutations to be specifically identified [21,22].
Resurrecting ancient proteins
The difficulty, of course, is that studying evolution along a branch requires access to the nodes on either end of the branch. The endpoints of internal branches are ancestral proteins that, by definition, no longer exist. But ancestral sequence reconstruction (ASR), a recently developed strategy for studying molecular evolution [23,24], can circumvent this problem. In 1965, Pauling and Zuckerkandl speculated that it would one day be possible to use the sequences of modern proteins to infer the sequences of ancestral proteins, which could be synthesized and studied experimentally [25]. Decades later, ASR has become a mature technique, which has been used to study many protein families, including GFP-like proteins [26,27], steroid receptors [21,28,22,29], opsins [30–32], and others [33–37]. ASR first infers ancestral sequences from an alignment of extant protein sequences, given the phylogeny that describes their historical relationships and a statistical model of amino acid substitution that describes the relative probability of replacing each amino acid with any other amino acid. The maximum likelihood sequence at any ancestral node on the phylogeny is the sequence with the highest probability of generating all of the sequence data in modern-day proteins [38]. Once the ancestral protein sequence is known, a DNA molecule coding for it is synthesized, allowing the ancestral protein to be expressed and characterized experimentally [23]. ASR also allows the functional impact of sequence changes that happened in the deep past to be studied by introducing historical mutations into the ancestral background, recapitulating mutational trajectories that occurred during evolution. State-of-the-art studies in the field acknowledge that reconstructed ancestors are approximations of historical reality; these studies carefully explore the robustness of their functional inferences to uncertainty about the reconstructed ancestors by experimentally characterizing alternate plausible reconstructions [23,26].
In the sections that follow, we discuss how ASR has been used to gain important insights into the underlying determinants of protein structure, function and evolution. Using several case studies, we demonstrate the effectiveness of ASR studies to quantitatively dissect the interactions that determine function, reveal multiple amino acids that underlie function, and determine the role of epistasis in shaping protein evolution. We conclude with a discussion of the expanding role that ASR can play in understanding the molecular determinants of protein function and evolution.
Opsins: quantifying functional interactions
The benefits of using the ancestral background for studying the effects of function-switching mutations were demonstrated recently in an elegant study of the opsins, a family of G-protein coupled receptors that absorb light in the vertebrate visual system. All opsins use the same covalently attached chromophore, but each opsin has a distinct wavelength of maximum absorption (λmax). Years of work have shown that the sequence determinants of λmax are complex, making comparative studies in modern opsins difficult to interpret. For example, human red and green opsins have λmax of 563 and 531 nm, respectively. Switching three amino acids in the red opsin to their states in their green paralog is sufficient to yield a green-absorbing pigment. The reverse, however, is not true: inserting the three “red” amino acids into the green opsin yields an intermediate opsin; several additional mutations are required to achieve the red phenotype [39]. Clearly, the background modulates the effect of individual substitutions on function. Yokoyama and colleagues estimate that some 60% of the opsin mutations reported in the literature show evidence of interaction with other mutations [30].
Yokoyama and colleagues dissected these interactions using ASR. They reasoned that the ancestral sequence provided the appropriate background for determining the effects of key mutations that occurred during opsin evolution. They began by resurrecting the ancestor of the red and green opsin genes and used this as a background for mutagenesis to identify the historical importance of key mutations [30,31]. When reconstituted in cultured cells, the ancestral pigment absorbed maximally in the red. They next identified five historical amino acid changes that were conserved in one state in red opsins and another state in the green opsins. When these five residues were introduced together into the ancestral background, they fully recapitulated the shift in λmax from red to green [31]. Yokoyama and colleagues then introduced each mutation singly and in sets of two and three and measured λmax of each variant. This approach allowed them to statistically partition the main, background-independent effects of each mutation from the effects of among-mutation interactions. Twenty-seven percent of the total shift in λmax was the result of epsitatic interactions rather than the direct effects of the individual mutations. Yokoyama and colleagues then fit a quantitative model to their results and found, remarkably, that it could predict from the states at these five sites alone the λmax of a wide variety of extant opsins to within 5 nm. Thus, where many studies in modern proteins yielded contradictory results about the functional importance of key mutations, a single study in the ancestral protein yielded results universally applicable to the family as a whole.
GFP-like proteins: when many amino acids determine function
Yokoyama’s work illustrates how reconstructed ancestral sequences provide the proper background for testing the functional impact of key mutations. ASR also provides an efficient way to identify those residues in the first place, as illustrated by the work of Mikhail Matz’s laboratory on GFP-like proteins from scleratinian (reef-building) corals. These proteins fluoresce at wavelengths determined by their amino acid sequence. Corals within the suborder Faviina are particularly diverse in the color of their fluorescence. By using ASR to characterize ancient sequences throughout the family, Matz’s group found that the GFP-like protein in the ancestral Faviina fluoresced in the green, followed by diversification into a variety of other colors [26,27]. Matz and colleagues then sought to identify the mutations responsible for the evolution of red fluorescence from this green ancestor in the great star coral Montastrea cavernosa. They found that 37 amino acid changes occurred between the GFP of the Faviina ancestor and that of M. cavernosa (compared to 108 differences between M. cavernosa and its closest modern-day green relative). Exhaustive characterization of all 237 —137 billion— mutational combinations would be intractable, so Matz and colleagues generated a library of variants in which each protein contained approximately half of the 37 residues in the ancestral state, half in the derived state. They then assessed the fluorescence of a large number of red and green clones from this library and statistically analyzed the association between the state at each site and the wavelength the protein emitted. This approach allowed Matz and colleagues to identify the set of historical mutations likely to contribute to the derived phenotype, including those that contribute only in some backgrounds due to epistasis. They found that 12 of the 37 mutations were significantly associated with red fluorescence. When this set was introduced into the ancestral green background, they yielded a red-emitting protein indistinguishable from the modern protein.
This set of key historical substitutions contained some residues previously identified as being important for red fluorescence based on structural considerations [40]; Q65H, for example, is required for red fluorescence because the histidine is incorporated into the red fluorophore. Yet it also revealed mutations whose importance would have been difficult to predict from structural observations. Only five of the mutations they identified were in the vicinity of the fluorophore; the remaining seven were spread widely throughout the three-dimensional structure and would have been almost impossible to identify using a horizontal approach. Despite their distance from the fluorophore, these mutations interacted strongly to bring about the derived phenotype in the ancestral background.
Steroid receptors: epistasis in the evolution of ligand specificity
ASR can be combined with structural biology to reveal the mechanisms by which interacting residues lead to complex functions. Our own group’s work investigating the evolution of the mineralcorticoid and glucocorticoid recptors (MR and GR) provides an example. MR and GR are nuclear transcription factors that directly regulate gene expression in a ligand-dependent fashion. MR and GR arose by duplication of a single ancestral receptor deep in the vertebrates and then diverged to bind different ligands and regulate different processes. MR is activated by aldosterone to regulate osmolarity; it is also activated by cortisol, albeit to a lesser extent. GR regulates the stress response and is activated only by cortisol. Despite a wealth of functional and structural information, identifying the sequence differences that underlie the functional difference between extant GR and MR using a horizontal approach been challenging. For example, a structural comparison of human MR and GR suggested that two sequence changes (S106P and L111Q) were likely to be important determinants of ligand specificity. When tested experimentally, however, swapping these residues between hGR and hMR yielded receptors that could not activate at all [41].
By resurrecting key ancestral proteins in MR/GR evolution, characterizing the effect of historical mutations on their functions in various combinations, and determining their crystal structures, our group was able to determine the molecular basis of the difference in MR/GR function. We found that the ancestor of all MRs and GRs (AncCR) was MR-like, with sensitivity to both aldosterone and cortisol [28]. By resurrecting successive ancestors in the GR lineage (Fig. 2), we found that cortisol specificity arose during a 40 million year period between AncGR1 (GR in the ancestor of all jawed vertebrates, which had the ancestral, MR-like phenotype) and AncGR2 (GR2 in the ancestor of bony vertebrates, which was cortisol specific). Thirty-seven amino acid changes occurred along this branch, but only five have been conserved in one state in the MRs and in another in the GRs, suggesting a key role in maintaining their different functions. We introduced these five mutations singly and in pairs into AncGR1. None of the single mutations enhanced cortisol specificity, but the combination of S106P and L111Q switched AncGR1’s preference to cortisol over aldosterone by radically reducing mineralocorticoid sensitivity. Strong epistasis was apparent: L111Q has no apparent functional effect when introduced alone, and S106P dramatically reduces activation by all ligands, but together they recapitulate a large portion of the functional switch from AncGR1 to AncGR2.
Figure 2.
Permissive and restrictive mutations shaped GR evolution. GRs and MRs result from an ancient gene duplication (arrow). ancCR was activated by aldosterone and cortisol (filled square, filled triangle). Cortisol specificity (open square, filled triangle) evolved between ancGR1 and ancGR2. The mutations that led to the functional change (black box) were preceded by permissive mutations (star) and followed by restrictive mutations (X). MRs do not have the permissive mutations and cannot tolerate the mutations in the black box. Human and fish GRs have restrictive mutations that do not allow reversion of the mutations in the black box to the ancestral state.
To identify the mechanism underlying this effect, we determined the X-ray crystal structures of ancestral receptors before and after the functional switch [21,22] (Figure 3A). Pro-106 causes a kink that remodels one side of the ligand-binding pocket, dramatically shifting the receptor’s helix 7 and destabilizing interactions with all ligands. Gln-111 is on the repositioned helix; in its new location, the polar side chain forms a hydrogen bond to a hydroxyl group unique to cortisol, recovering binding in a cortisol-specific manner. The cause of the interaction between these two mutations is therefore conformational: the effect of L111Q on function is determined by its spatial location, which depends on whether or not S106P has occurred.
Figure 3.
Structural basis for the evolution of cortisol specificity. A) Structure of AncCR (white, 2Q1H) with bound aldosterone (orange) overlaid on the structure of AncGR2 (blue, 3GN8) with bound DEX (purple), a cortisol analog. Mutation S106P disrupts a helix cap and introduces a kink at the N-terminus of helix 7, repositioning helix 7 so that Gln-111 of AncGR2 can form a cortisol-specific hydrogen bond. B) Schematic representation of restrictive mutations G114Q and L197M. The derived Gln-Met pair in AncGR2 can be tolerated in the derived conformation (right) but clash when helix 7 is in its ancestral position (left). The hydrogen-bond of Gln-111 with cortisol (purple) is shown in red.
Why do these mutations, which switch the function of the ancestral receptor, radically impair function when they are introduced into the modern MR or are reversed in the modern GR [22,41]? We found that some of the other 32 mutations that occurred between AncGR1 and AncGR2 have strong modulating effects. Specifically, three additional mutations (L29M, F98I and S212Δ) fine-tuned the derived function, eliminating all remnants of the response to mineralocorticoids and yielding a fully cortisol-specific receptor. These mutations further destabilize the receptor and cannot be tolerated, however, unless they are preceded by two permissive mutations (N26T and Q105L), which have virtually no effect on function when introduced alone. A third stabilizing permissive mutation (Y72R), which occurred earlier— between AncCR and AncGR1—is also essential for the function-switching mutations to be tolerated. Without these permissive mutations, the key mutations that historically produced a cortisol-specific receptor yield a receptor that does not activate transcription at all. These findings reveal why it has been so difficult to convert a modern MR into a GR-like protein using horizontal approaches: the MR lacks the permissive mutations that occurred in the GR lineage, so it cannot tolerate the destabilizing effects of the mutations that switched the GR’s functions.
We also identified restrictive mutations that occurred later in the GR lineage, which are incompatible with the MR-like conformation. Specifically, five of the mutations that accumulated in the evolving GR after the functional switch are incompatible with the ancestral conformation because they produce a steric clash or eliminate favorable interactions necessary to support the ancestral position of helix 7 (Figure 3B). For example, in the ancestral state Gly-114 sits directly across from Leu-197 between helix 7 and helix 10 in the receptor. In AncGR2 and its descendants, the Gly-Leu pair is replaced with the longer side chains of Gln-Met; this pair can be accommodated in AncGR2 and its descendants because of the shift in helix 7, but it produces a steric clash if the helix is returned to the ancestral conformation. These results reveal that the modern GRs cannot be converted to the MR-like structure and function because of restrictive mutations that occurred hundreds of millions of years ago in the GR lineage.
Implications
These ASR projects make clear that epistasis is common along evolutionary trajectories, and this fact has significant implications for how we study protein structure and function. Because mutations may have different effects in different sequence backgrounds, the only way to understand the historical relevance of a mutation is to test it in the ancestral background. Ancestral sequences are also necessary to identify the primary determinants of functional differences between extant proteins, because the accumulation of permissive and restrictive mutations may yield spurious results if key sequence differences are experimentally analyzed in modern backgrounds.
The prominent role of epistasis has profound implications for the processes by which proteins evolve. In some instances, such as the evolution of contemporary antibiotic resistance, protein evolution appears to proceed by pure functional hill climbing, with each new mutation leading to an incremental increase in function under the influence of selection [42]. But studies of evolution in more ancient proteins reveal a much more complex picture in which permissive mutations of no apparent effect transiently open paths to new functional states, while restrictive mutations close them [21,43,22,44]. Which paths are open at any moment may therefore depend on which neutral (or nearly neutral) mutations happen to be present in an evolving population. The evolution of major changes in protein structure and function may be possible or inevitable given one set of prior historical mutations, but nearly impossible given other equally probable events. The molecular basis of function in modern-day proteins may thus be far from optimal, instead representing the outcome of an historical process in which chance plays a major role.
The discovery of permissive and restrictive epistatic mutations using ASR raises additional questions, some of which may be answerable using ASR. How large is the set of potentially permissive mutations that could allow a protein to tolerate a specific change in structure and function? Are there general physical chemical properties that characterize permissive mutations? Once restrictive mutations have occurred in a lineage, will selection to reacquire the ancestral function tend to produce proteins with new structure-function relations that differ from those in the ancestor? One prospective approach to these questions is to combine ASR studies with directed evolution [45,46], to explore some of the many “might-have-been” trajectories that were not taken by natural evolution.
A final benefit of ASR is that it provides a bridge between mechanistic biochemistry and evolutionary biology, fields that have been largely separate, despite the long-standing existence of fascinating questions at their interface [47–50]. As the case studies we have reviewed here show, a detailed understanding of the historical processes and mechanisms by which proteins acquired their functions can help us understand how and why proteins work as they do today. Further, because evolution represents an exploration of protein sequence space by independent lineages over long periods of time, a reconstruction of the historical trajectories of protein evolution has the potential to shed some light on the distribution of functions and structures through protein space. By enabling rigorous biochemical assessments of ancient proteins, ASR promises new insights into the physical-chemical determinants that have shaped protein evolution and the historical determinants of protein architecture.
Acknowledgments
Supported by National Science Foundation IOB-0546906 and National Institutes of Health R01-GM081592, F32-GM074398, and F32-GM090650. J.W.T. is an Early Career Scientist of the Howard Hughes Medical Institute. M.J.H. is supported by National Research Service Fellowship 1F32GM989650.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Michael J. Harms, Email: harms@uoregon.edu.
Joseph W. Thornton, Email: joet@uoregon.edu.
References
- 1.Smith J. Natural Selection and the Concept of a Protein Space. Nature. 1970;225:563–564. doi: 10.1038/225563a0. [DOI] [PubMed] [Google Scholar]
- 2.Romero PA, Arnold FH. Exploring protein fitness landscapes by directed evolution. Nat Rev Mol Cell Biol. 2009;10:866–876. doi: 10.1038/nrm2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buske FA, Their R, Gillam EMJ, Bodén M. In silico characterization of protein chimeras: Relating sequence and function within the same fold. Proteins: Struct Funct Bioinf. 2009;77:111–120. doi: 10.1002/prot.22422. [DOI] [PubMed] [Google Scholar]
- 4.Chakrabarti S, Lanczycki CJ. Analysis and prediction of functionally important sites in proteins. Protein Sci. 2007;16:4–13. doi: 10.1110/ps.062506407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Capra JA, Singh M. Characterization and Prediction of Residues Determining Protein Functional Specificity. Bioinformatics. 2008 doi: 10.1093/bioinformatics/btn214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donald JE, Shakhnovich EI. SDR: a database of predicted specificity-determining residues in proteins. Nucl Acids Res. 2009;37:D191–194. doi: 10.1093/nar/gkn716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Datta AK. Comparative sequence analysis in the sialyltransferase protein family: analysis of motifs. Curr Drug Targets. 2009;10:483–498. doi: 10.2174/138945009788488422. [DOI] [PubMed] [Google Scholar]
- 8.Desai TA, Rodionov DA, Gelfand MS, Alm EJ, Rao CV. Engineering transcription factors with novel DNA-binding specificity using comparative genomics. Nucleic Acids Res. 2009;37:2493–2503. doi: 10.1093/nar/gkp079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maita N, Nyirenda J, Igura M, Kamishikiryo J, Kohda D. Comparative structural biology of eubacterial and archaeal oligosaccharyltransferases. J Biol Chem. 2010;285:4941–4950. doi: 10.1074/jbc.M109.081752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neidhart DJ, Kenyon GL, Gerlt JA, Petsko GA. Mandelate racemase and muconate lactonizing enzyme are mechanistically distinct and structurally homologous. Nature. 1990;347:692–694. doi: 10.1038/347692a0. [DOI] [PubMed] [Google Scholar]
- 11.Hoffman M. On the road to mandelate … racemase. Science. 1991;251:31–32. doi: 10.1126/science.1986411. [DOI] [PubMed] [Google Scholar]
- 12.Gerlt JA, Babbitt PC. Enzyme (re)design: lessons from natural evolution and computation. Current Opinion in Chemical Biology. 2009;13:10–18. doi: 10.1016/j.cbpa.2009.01.014. The authors review attempts to perform “horizontal” comparisons between divergent enolases. They gather examples that demonstrate challenges that face the approach. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiang H, Luo L, Taylor KL, Dunaway-Mariano D. Interchange of Catalytic Activity within the 2-Enoyl-Coenzyme A Hydratase/Isomerase Superfamily Based on a Common Active Site Template. Biochemistry. 1999;38:7638–7652. doi: 10.1021/bi9901432. [DOI] [PubMed] [Google Scholar]
- 14.Ijima Y, Matoishi K, Terao Y, Doi N, Yanagawa H, Ohta H. Inversion of enantioselectivity of asymmetric biocatalytic decarboxylation by site-directed mutagenesis based on the reaction mechanism. Chem Commun. 2005 doi: 10.1039/b416398b. [DOI] [PubMed] [Google Scholar]
- 15.Bershtein S, Segal M, Bekerman R, Tokuriki N, Tawfik DS. Robustness-epistasis link shapes the fitness landscape of a randomly drifting protein. Nature. 2006;444:929–932. doi: 10.1038/nature05385. [DOI] [PubMed] [Google Scholar]
- 16.Hayashi Y, Aita T, Toyota H, Husimi Y, Urabe I, Yomo T. Experimental Rugged Fitness Landscape in Protein Sequence Space. PLoS ONE. 2006;1:e96. doi: 10.1371/journal.pone.0000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phillips PC. Epistasis - the essential role of gene interactions in the structure and evolution of genetic systems. Nat Rev Genet. 2008;9:855–867. doi: 10.1038/nrg2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parera M, Perez-Alvarez N, Clotet B, Martínez MA. Epistasis among Deleterious Mutations in the HIV-1 Protease. J Mol Biol. 2009;392:243–250. doi: 10.1016/j.jmb.2009.07.015. [DOI] [PubMed] [Google Scholar]
- 19.O’Maille PE, Malone A, Dellas N, Andes Hess B, Smentek L, Sheehan I, Greenhagen BT, Chappell J, Manning G, Noel JP. Quantitative exploration of the catalytic landscape separating divergent plant sesquiterpene synthases. Nat Chem Biol. 2008;4:617–623. doi: 10.1038/nchembio.113. The authors created a library with all possible combinations of nine mutations between two enzymes, and then characterized the products of each protein. The mutants exhibited extreme epistasis: 1) no single amino acid correlated with relative product output, and 2) that the effect of the mutation depended on the state of the other eight residues. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bloom JD, Labthavikul ST, Otey CR, Arnold FH. Protein stability promotes evolvability. Proc Natl Acad Sci USA. 2006;103:5869–5874. doi: 10.1073/pnas.0510098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bridgham JT, Ortlund EA, Thornton JW. An epistatic ratchet constrains the direction of glucocorticoid receptor evolution. Nature. 2009;461:515–519. doi: 10.1038/nature08249. By combining ancestral sequence reconstruction, phylogenetic inference, and structural biology, our group was able to identify mutations that shaped the evolution of steroid receptors. We identified a set of mutations that closed a previously accessible evolutionary path, revealing one reason why horizontal comparisons of steroid receptors have proven difficult to interpret. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ortlund EA, Bridgham JT, Redinbo MR, Thornton JW. Crystal Structure of an Ancient Protein: Evolution by Conformational Epistasis. Science. 2007;317:1544–1548. doi: 10.1126/science.1142819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thornton JW. Resurrecting ancient genes: experimental analysis of extinct molecules. Nat Rev Genet. 2004;5:366–375. doi: 10.1038/nrg1324. [DOI] [PubMed] [Google Scholar]
- 24.Liberles DA. Ancestral sequence reconstruction. Oxford University Press; USA: 2007. [Google Scholar]
- 25.Pauling L, Zuckerkandl E. Chemical Paleogenetics: Molecular “Restoration Studies” of Extinct Forms of Life. Acta Chem Scand. 1963;17:S9–S16. [Google Scholar]
- 26.Ugalde JA, Chang BSW, Matz MV. Evolution of Coral Pigments Recreated. Science. 2004;305:1433. doi: 10.1126/science.1099597. [DOI] [PubMed] [Google Scholar]
- 27.Field SF, Matz MV. Retracing Evolution of Red Fluorescence in GFP-Like Proteins from Faviina Corals. Mol Biol Evol. 2010;27:225–233. doi: 10.1093/molbev/msp230. Starting from an ancestral protein, the authors used a novel screening method to identify epistatic interactions that led to the evolution of the red phenotype in GFP-like proteins from coral. Their approach revealed that many of the mutations necessary for the phenotype have no effect on color unless introduced in combination with other mutations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bridgham JT, Carroll SM, Thornton JW. Evolution of Hormone-Receptor Complexity by Molecular Exploitation. Science. 2006;312:97–101. doi: 10.1126/science.1123348. [DOI] [PubMed] [Google Scholar]
- 29.Carroll SM, Bridgham JT, Thornton JW. Evolution of Hormone Signaling in Elasmobranchs by Exploitation of Promiscuous Receptors. Mol Biol Evol. 2008;25:2643–2652. doi: 10.1093/molbev/msn204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yokoyama S, Yang H, Starmer WT. Molecular Basis of Spectral Tuning in the Red- and Green-Sensitive (M/LWS) Pigments in Vertebrates. Genetics. 2008;179:2037–2043. doi: 10.1534/genetics.108.090449. They used the reconstructed ancestor of modern red and green opsins to study the effects of five mutations on opsin absorbance spectra. By doing studies in this background, they were able to quantitatively explain the variation in modern red and green opsins, something that has been unattainable using modern proteins as backgrounds for mutagenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yokoyama S, Radlwimmer FB. The Molecular Genetics and Evolution of Red and Green Color Vision in Vertebrates. Genetics. 2001;158:1697–1710. doi: 10.1093/genetics/158.4.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yokoyama S, Tada T, Zhang H, Britt L. Elucidation of phenotypic adaptations: Molecular analyses of dim-light vision proteins in vertebrates. Proc Natl Acad Sci USA. 2008;105:13480–13485. doi: 10.1073/pnas.0802426105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaiser SM, Malik HS, Emerman M. Restriction of an Extinct Retrovirus by the Human TRIM5{alpha} Antiviral Protein. Science. 2007;316:1756–1758. doi: 10.1126/science.1140579. [DOI] [PubMed] [Google Scholar]
- 34.Kuang D, Yao Y, MacLean D, Wang M, Hampson DR, Chang BSW. Ancestral reconstruction of the ligand-binding pocket of Family C G protein-coupled receptors. Proc Natl Acad Sci USA. 2006;103:14050–14055. doi: 10.1073/pnas.0604717103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gaucher EA, Thomson JM, Burgan MF, Benner SA. Inferring the palaeoenvironment of ancient bacteria on the basis of resurrected proteins. Nature. 2003;425:285–288. doi: 10.1038/nature01977. [DOI] [PubMed] [Google Scholar]
- 36.Gaucher EA, Govindarajan S, Ganesh OK. Palaeotemperature trend for Precambrian life inferred from resurrected proteins. Nature. 2008;451:704–707. doi: 10.1038/nature06510. [DOI] [PubMed] [Google Scholar]
- 37.Thomson JM, Gaucher EA, Burgan MF, De Kee DW, Li T, Aris JP, Benner SA. Resurrecting ancestral alcohol dehydrogenases from yeast. Nat Genet. 2005;37:630–635. doi: 10.1038/ng1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Z, Kumar S, Nei M. A New Method of Inference of Ancestral Nucleotide and Amino Acid Sequences. Genetics. 1995;141:1641–1650. doi: 10.1093/genetics/141.4.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Asenjo AB, Rim J, Oprian DD. Molecular determinants of human red/green color discrimination. Neuron. 1994;12:1131–1138. doi: 10.1016/0896-6273(94)90320-4. [DOI] [PubMed] [Google Scholar]
- 40.Field S, Bulina M, Kelmanson I, Bielawski J, Matz M. Adaptive Evolution of Multicolored Fluorescent Proteins in Reef-Building Corals. J Mol Evol. 2006;62:332–339. doi: 10.1007/s00239-005-0129-9. [DOI] [PubMed] [Google Scholar]
- 41.Li Y, Suino K, Daugherty J, Xu HE. Structural and Biochemical Mechanisms for the Specificity of Hormone Binding and Coactivator Assembly by Mineralocorticoid Receptor. Mol Cell. 2005;19:367–380. doi: 10.1016/j.molcel.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 42.Weinreich DM, Delaney NF, DePristo MA, Hartl DL. Darwinian Evolution Can Follow Only Very Few Mutational Paths to Fitter Proteins. Science. 2006;312:111–114. doi: 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- 43.Field SF, Matz MV. Retracing evolution of red fluorescence in GFP-like proteins from Faviina corals. J Mol Evol. 2009 doi: 10.1093/molbev/msp230.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wagner A. Neutralism and selectionism: a network-based reconciliation. Nat Rev Genet. 2008;9:965–974. doi: 10.1038/nrg2473. A thoughtful review that explores how molecular epistasis may shape evolutionary trajectories, particularly focusing on its relationship to the neutralist/selectionist debate. [DOI] [PubMed] [Google Scholar]
- 45.Bloom J, Arnold F. In the light of directed evolution: Pathways of adaptive protein evolution. Proc Natl Acad Sci USA. 2009;106:9995–10000. doi: 10.1073/pnas.0901522106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tokuriki N, Tawfik DS. Protein Dynamism and Evolvability. Science. 2009;324:203–207. doi: 10.1126/science.1169375. [DOI] [PubMed] [Google Scholar]
- 47.Anfinsen C. Molecular Basis of Evolution. John Wiley & Sons; 1959. [Google Scholar]
- 48.Blundell TL, Wood SP. Is the evolution of insulin Darwinian or due to selectively neutral mutation? Nature. 1975;257:197–203. doi: 10.1038/257197a0. [DOI] [PubMed] [Google Scholar]
- 49.Perutz MF. Species adaptation in a protein molecule. Mol Biol Evol. 1983;1:1–28. doi: 10.1093/oxfordjournals.molbev.a040299. [DOI] [PubMed] [Google Scholar]
- 50.Serrano L, Day AG, Fersht AR. Step-wise mutation of barnase to binase. A procedure for engineering increased stability of proteins and an experimental analysis of the evolution of protein stability. J Mol Biol. 1993;233:305–12. doi: 10.1006/jmbi.1993.1508. [DOI] [PubMed] [Google Scholar]