Abstract
The binding of nuclear proteins to chromatin in live cells has been analyzed by kinetic modeling procedures applied to experimental data from fluorescence recovery after photobleaching (FRAP). The kinetic models have yielded a number of important biological predictions about transcription, but concerns have arisen about the accuracy of these predictions. First, different studies using different kinetic models have arrived at very different predictions for the same or similar proteins. Second, some of these divergent predictions have been shown to arise from technical issues rather than biological differences. For confidence and accuracy, gold standards for the measurement of in vivo binding must be established by extensive cross validation using both different experimental methods and different kinetic modeling procedures.
Introduction
Over the past decade, FRAP has become a widely used technique for quantifying the dynamics of proteins in live cells [1,2]. In FRAP (Fig. 1A) a region of the cell is photobleached, and then the rate at which fluorescence recovers there is measured. When examined by FRAP, many nuclear proteins exhibit complete recoveries within seconds [3]. This dynamic behavior suggests that these proteins are transiently bound to chromatin, since stable binding interactions would be characterized by either incomplete or very slow FRAP recoveries.
Figure 1.
(A) FRAP (red circle) of the nucleus is performed by photobleaching a sub-region containing a fluorescently tagged protein (green circles). The bleached molecules (black) move out of the bleach zone (small arrows) and unbleached molecules move in. The rate of fluorescence recovery can be measured by calculating the average fluorescent intensity inside the measurement area (dashed circle) as a function of time, generating a FRAP curve. (B) Kinetic models are used to simulate FRAP experiments. The models are constructed from a set of fixed and variable parameters. The fixed parameters are measurable quantities such as the shape of the nucleus and the shape of the photobleach pattern. The variable parameters are unknown quantities of interest, such as the rates of diffusion and binding for the fluorescently tagged protein. For any combination of parameter values, the kinetic model generates a simulated FRAP curve. The variable parameters can be varied until a good fit to the experimental FRAP curve is obtained. These best-fitting parameters then yield the estimates for the diffusion and binding rates, which are then interpreted to reach biological conclusions.
To extract information about this in vivo binding, FRAP curves must be analyzed quantitatively with kinetic models [4] (Fig. 1B). These models have now been used extensively to measure the in vivo binding of various proteins involved in gene expression.
Here we review how these quantitative analyses of FRAP are done and summarize their conclusions. We show that there are discrepancies among the conclusions from different quantitative FRAP studies and argue that many of these could reflect technical issues. We suggest strategies for resolving these discrepancies, and recommend a combination of continued work and ongoing caution in interpreting kinetic modeling from FRAP data in live cells.
FRAP of nuclear proteins includes information about chromatin binding
Most nuclear proteins recover much more slowly than would be expected if they just diffused through the nucleus. Unconjugated GFP, which has a mass of 27 kD, provides a baseline for purely diffusive behavior. When a 2 µm diameter spot is photobleached, GFP in the nucleus completely recovers in 1.1 sec [5]. This measurement can be used to predict the recovery times for larger diffusing proteins, since with a simple approximation the rate of diffusion goes as the inverted cube root of a protein's mass, presuming a roughly spherical protein [6]. Thus for a 2 µm diameter bleach spot, a 100 kD protein should require 1.6 sec to recover and a 1 MD protein complex should require 3.5 sec.
The recoveries of most nuclear proteins are typically much slower than this, requiring instead at least 15 sec for a 2 µm diameter bleach spot [3]. This retardation compared to pure diffusion has been interpreted in virtually all FRAP studies of nuclear proteins as binding to immobile chromatin structures. Extracting information about this predicted in vivo binding from a FRAP experiment requires a quantitative analysis of the FRAP data using a kinetic model [4].
In vivo predictions are obtained by comparing experimental FRAPs to simulated FRAPs generated with a kinetic model
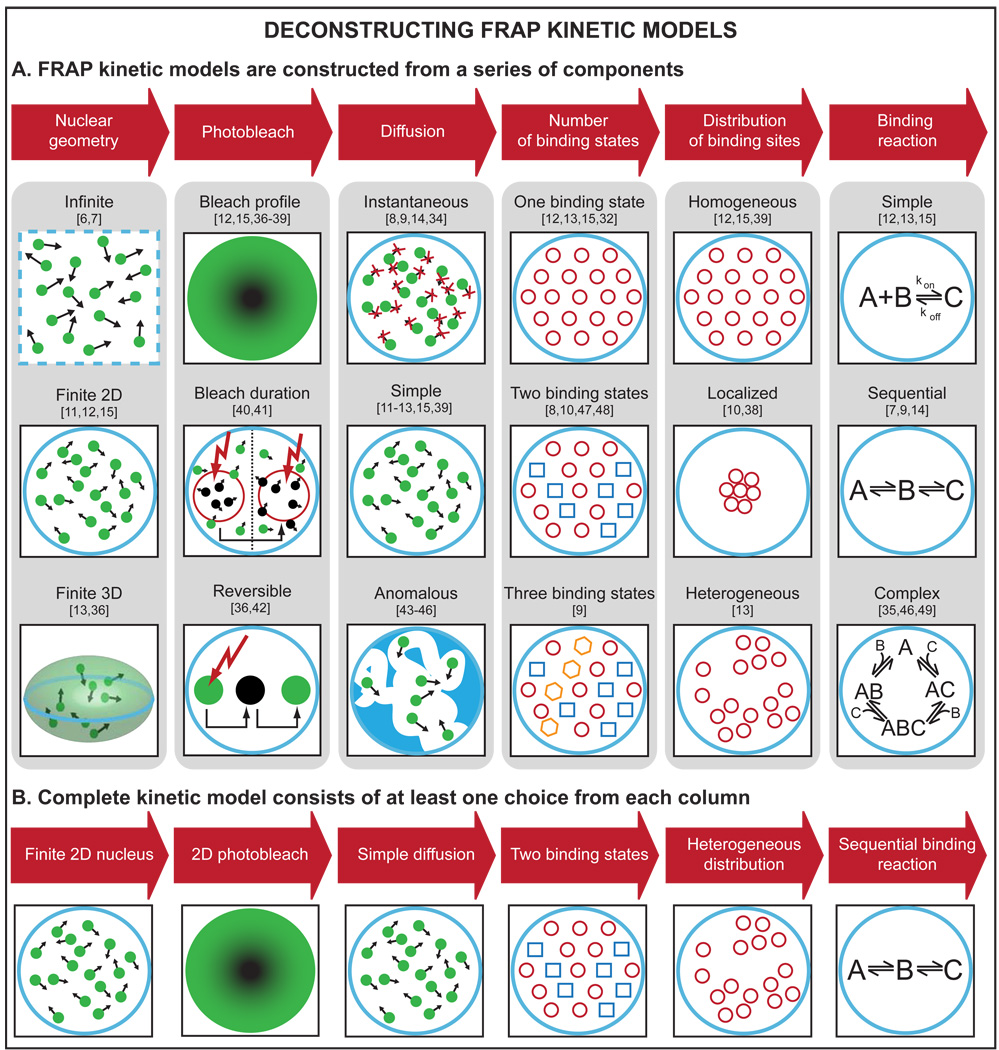
Kinetic models seek to simulate the processes occurring in a FRAP experiment (see Fig. 2 for details and references). All kinetic models have incorporated a photobleach. When applied to FRAPs of nuclear proteins, all kinetic models have also simulated binding at immobile chromatin sites. Many models presume just one binding state, but some have postulated two or three distinct binding states [7–10]. In addition to binding, some kinetic models have also allowed for diffusion of the fluorescent molecule into the bleach spot [6,11–13]. However, other models have presumed that diffusion can be neglected [7–9,14]. This is appropriate if the time to diffuse across the bleach spot is much faster than the time to bind to chromatin [2].
Figure 2.
(A) A FRAP kinetic model is constructed from a series of components (red arrows) that reflect properties of the cell, the photobleach or the fluorescent protein. The images under each component indicate some of the choices that have been made by different published kinetic models. Neither the list in each column nor the references cited for each choice are exhaustive. (B) A complete kinetic model consists of at least one choice from each column (some choices are not mutually exclusive). The number of possible kinetic models is therefore very large (36 = 729 different kinetic models could be constructed from just the list of possibilities shown here).
These basic assumptions are then translated into a mathematical / computational model [6–15] that describes binding by an association and dissociation rate, and diffusion by a diffusion constant (in those cases where diffusion cannot be neglected). Binding and diffusion rates can then be varied to generate a simulated FRAP curve that matches the experimental FRAP data (Fig. 1B), and in this way estimates are obtained for these parameters.
Kinetic models are used to quantify and interpret in vivo binding mechanisms
In addition to the quantitative predictions about diffusion and binding rates, kinetic models also typically lead to biological interpretations about the behavior of the protein that was photobleached.
A good example of the power of kinetic modeling comes from the analysis of polymerase II FRAPs (Table IA). This has generated predictions for the number of kinetically distinct polymerase binding states at a gene, including the fraction sizes of polymerase molecules in each state and their residence times. Knowledge of these kinetic properties leads to important biological conclusions about how the polymerase functions in vivo.
| A. RNA polymerases | |||||
|---|---|---|---|---|---|
| Polymerase | References | Organism (cell line) |
Number of bound states |
Elongation rate |
Nature of assembly/progression |
| I | [7] | Monkey (CM3) |
2 Initiation, Elongation |
5.7 kb/min | Inefficient |
| II | [29] | Hamster (C23) |
1 Elongation |
0.5 kb/min | Inefficient |
| II | [9] | Human (U2OS) |
3 Promoter- binding, Initiation, Elongation |
4.3 kb/min | Inefficient |
| II | [30] | Human (U2OS) |
1 Elongation |
1.9 kb/min | Efficient |
| II | [31] | Fly (UAS) |
2 Elongation, Recycling |
1.5 kb/min | Efficient |
| B. Transcription factors | |||||
|---|---|---|---|---|---|
| Transcription factor |
References | Organism (Cell line) |
Number of bound states |
Residence time |
Nature of binding |
| GR | [6] | Mouse (3617) |
1 | 13 ms | Non-specific |
| Max | [8] | Human (Hela) |
2 | 6 s / 14 s | Non-specific / Specific |
| p53 | [12] | Human (H1299) |
1 | 2.5 s | Non-specific |
| AR | [32] | Human (Hep3B) |
1 | 45 s | Specific |
| C. Histone H1 | |||||
|---|---|---|---|---|---|
| Histone variant |
References | Organism | Number of bound states |
Residence Time |
Nature of binding |
| H1° | [33] | Human (SK-N-SH) |
2 | NA / 52 s | Weak / Tight |
| SUV39H1, H1.1 |
[13] | Rat (NRK) |
≥ 1 | < 170 s | Tight |
| H1° | [8] | Mouse (BALB/c 3T3) |
1 | > 183 s | Tight |
| H1 | [34] | Mouse (NIH 3T3) |
1 | 76 s | Tight |
| H1° | [35] | Mouse (BALB/c 3T3) |
≥ 2 | NA / 100 s | Weak / Tight |
For example, in one kinetic modeling study of pol II three distinct binding states were detected [9]. These were attributed to promoter binding of the polymerase, initiation of the polymerase at the promoter and elongation of the polymerase down the gene. These assignments led to the conclusion that transcription was inefficient, since the fraction of polymerase molecules in the promoter binding state was much larger than in the initiation state which in turn was much larger than in the elongation state. As a consequence, only 1 in 90 polymerases that were recruited to the gene proceeded to elongation. Kinetic modeling also yielded a prediction for the residence time of molecules in the elongation state, which was used to estimate the in vivo elongation rate by dividing the residence time by the number of base pairs in the gene being transcribed.
Thus analysis of polymerase FRAP data has provided information about the different molecular states of the polymerase at a promoter, its efficiency of progressing to elongation, and its in vivo rate of elongation, but how robust are these predictions?
Different kinetic models make different predictions for the same or similar molecules
Table IA summarizes the results of five different kinetic modeling studies of the polymerase. This comparison reveals multiple discrepancies. For example, the number of predicted kinetic states of the polymerase varies from one to three (Table IA). Thus, some studies of the polymerase conclude they detect only elongation, others conclude they detect initiation and elongation and others conclude they detect promoter binding, initiation and elongation. Additionally, some studies predict efficient polymerase assembly, while others predict inefficient assembly (Table IA).
Discrepancies are not limited to the polymerase, but also arise in kinetic modeling of other molecules. The estimated residence times for transcription factors bound to chromatin vary by four orders of magnitude from milliseconds to many seconds (Table IB), and the number of distinct bound states for histone H1 varies from one to two (Table Ic).
The discrepancies in Table I could reflect real biological differences. For example, the different polymerase II studies have examined transcription either throughout the nucleoplasm or at different transgenes, while the transcription factor or H1 studies have looked at either different transcription factors or different H1 variants in different cell lines.
It is also possible, however, that some of the divergent predictions could arise from technical issues. These differences include how the FRAP experiment was performed and what assumptions were made to generate the kinetic model. Since the differences between the kinetic models are much larger than the differences between the FRAP experiments, the kinetic models emerge as the prime suspects to account for technical errors.
Errors in kinetic modeling could arise from many sources
As noted above, to simulate FRAP of a chromatin binding protein, the processes of photobleaching, diffusion and binding must be modeled. For each of these processes, various details remain uncertain. These include whether the 3D bleach is well approximated by a 2D bleach, whether diffusion is simple or anomalous and whether binding can be described by simple chemical kinetics (see Fig. 2 for details and references).
Since there is no agreement yet on the best kinetic model, a number of different, plausible kinetic models have been proposed and applied to FRAP data. Unfortunately, many of these models are capable of fitting the same FRAP curve, even though they can lead to very different biological predictions (Fig. 3A,B).
Figure 3.
As Fig. 2 shows, many different kinetic models can be constructed. It is now recognized that some of these models can fit the same experimental FRAP curve equally well (A). Unfortunately, different fits can lead to very different biological interpretations (B), so it is critical to decide on the correct kinetic model. This can be done by cross validation (C). Under "Current Status", we list the assumptions that have been made for each of the components of a kinetic model (red boxes), and then under "Sample Cross Validation" we suggest how these assumptions can be verified.
For example, it is possible to fit the same FRAP curve presuming completely different roles for diffusion [15]. The different kinetic models derived from these different assumptions about diffusion yield completely different biological predictions, namely either one binding state or two distinct binding states with the potential for large discrepancies in the measured strength of binding.
Cross validation raises questions about the current predictions of kinetic modeling
Since there is no consensus yet on the best kinetic model, there is as yet no gold standard measurement of in vivo binding rates. The results in Table I are the first attempts to measure in vivo binding. We cannot compare them to the in vitro estimates because it is thought that the in vitro experiments fail to capture the complexity of the live cell [3,16].
Therefore the best approach at present is to evaluate different FRAP kinetic modeling procedures by cross validation (Fig. 3C). A direct cross validation has only been performed for a subset of the transcription factor analyses shown in Table IB. The results of this cross validation indicated that the differences reported among three different transcription factor studies were due exclusively to inaccuracies in the kinetic modeling procedures [15].
Specifically, when the three different kinetic modeling procedures were applied to the same transcription factor in the same cell line, three distinctly different sets of predictions were obtained. These disparate predictions for the same transcription factor paralleled those obtained in the original studies for different transcription factors.
As a result, two mistakes were identified in the original published kinetic models. The first was an inaccurate approximation of the photobleach profile and the second was improperly neglecting the role of diffusion. When the kinetic models were corrected to eliminate these errors, the modified FRAP procedures now yielded similar binding estimates not only for the same transcription factor, but also for the three different transcription factors [15]. These results suggest that the predictions of kinetic modeling, including many of those in Table I, should be viewed with caution.
One area of caution is the estimated residence times on chromatin, and as a corollary, the polymerase elongation rate, since this is derived from a residence time estimate. Determining the residence time is difficult because it is not always proportional to the FRAP recovery time. Fluorescent molecules entering the bleach spot may undergo a series of diffusion and binding steps before they reach the center of the bleach spot. If there are ten such steps, for example, then the binding residence time will be about one-tenth the FRAP recovery time. Therefore, the FRAP recovery time sets an upper bound on the residence time, but an accurate kinetic model is required to estimate the actual residence time. An inaccurate kinetic model can result in residence time estimates that are off by three orders of magnitude [15].
The second area of caution is the predicted number of binding states. An inaccurate kinetic model may lead to a poor fit of the experimental FRAP data, which can often be overcome by postulating an additional binding state, since this adds free parameters to the kinetic model. As described above, this occurred in the kinetic modeling of transcription factor FRAPs, where a failure to account for diffusion led to the prediction of an additional, spurious binding state [15]. These artifactual states may then be assigned biologically relevant functions (such as specific-site binding of a transcription factor).
The third area of caution is the predicted size of the bound and free fractions. Inaccurate kinetic models led to significant errors in predicted bound fractions in several different transcription factor studies [15]. If similar errors afflict some of the polymerase studies, then conclusions about the efficiency or inefficiency of polymerase assembly and/or progression may be wrong, since they are often derived from estimates of bound-fraction sizes.
Further cross validations are necessary to improve our confidence in kinetic modeling
Complete confidence in kinetic modeling will only be achieved over time as gold standards for the measurement of in vivo binding become established. This will involve extension of the cross validation approach described above where the same transcription factor in the same cell was measured by different FRAP experiments and kinetic modeling procedures [15]. It will be important to apply this cross validation strategy to the polymerase and histone H1, which also exhibit discrepancies (Table I).
Beyond comparisons of FRAP experimental methods and kinetic modeling procedures, it will be essential to compare completely alternative experimental methods (Fig. 3C). These include fluorescence correlation spectroscopy (FCS) [5,12,17–20], single molecule tracking (SMT) [21,22], competition ChIP [23,24], continuous photobleaching [25,26] and acceptor photobleaching FRAP [27], since kinetic models exist or can be developed for each of these to also provide estimates of in vivo binding rates.
The preceding experimental methods are at least partially orthogonal, and so limitations of one method are not shared by the others. For example, an underlying concern in FRAP is that the intentional photobleach induces damage [28]. This is not an issue in FCS where low light intensities are used. Comparisons of some of these different experimental methods are already underway [15,17,19,20], so rapid progress should be expected.
As these different experimental methods are applied to the same molecule in the same system, flaws in either the experimental method or the kinetic modeling procedures are likely to be uncovered and then corrected. Only then will a consensus gold-standard finally emerge.
Conclusion
While advances in light microscopy and protein tagging have made analysis of live-cell kinetics possible, much work remains to validate the quantitative and qualitative conclusions that have been drawn from the kinetic modeling of FRAP data. The early steps in this validation are already uncovering errors that significantly impact previous conclusions, and this may continue as the field matures and settles on standard procedures that are deemed robust and reliable. This goal will be realized by a concerted effort to address unresolved technical uncertainties that could still confound current analyses. This will be achieved by comparison of the results obtained for the same molecules in the same system with different experimental methods and different kinetic modeling procedures. This effort will be rewarded with the development of accurate tools to probe binding processes in live cells.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
- 1.Houtsmuller AB. Fluorescence recovery after photobleaching: application to nuclear proteins. Adv.Biochem Eng Biotechnol. 2005;95:177–199. doi: 10.1007/b102214. [DOI] [PubMed] [Google Scholar]
- 2.Sprague BL, McNally JG. FRAP analysis of binding: proper and fitting. Trends Cell Biol. 2005;15:84–91. doi: 10.1016/j.tcb.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 3.Hager GL, McNally JG, Misteli T. Transcription dynamics. Mol.Cell. 2009;35:741–753. doi: 10.1016/j.molcel.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phair RD, Misteli T. Kinetic modelling approaches to in vivo imaging. Nat Rev Mol Cell Biol. 2001;2:898–907. doi: 10.1038/35103000. [DOI] [PubMed] [Google Scholar]
- 5.Michelman-Ribeiro A, Mazza D, Rosales T, Stasevich TJ, Boukari H, Rishi V, Vinson C, Knutson JR, McNally JG. Direct measurement of association and dissociation rates of DNA binding in live cells by fluorescence correlation spectroscopy. Biophys J. 2009;97:337–346. doi: 10.1016/j.bpj.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sprague BL, Pego RL, Stavreva DA, McNally JG. Analysis of binding reactions by fluorescence recovery after photobleaching. Biophys J. 2004;86:3473–3495. doi: 10.1529/biophysj.103.026765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dundr M, Hoffmann-Rohrer U, Hu Q, Grummt I, Rothblum LI, Phair RD, Misteli T. A Kinetic Framework for a Mammalian RNA Polymerase in Vivo. Science. 2002;298:1623–1626. doi: 10.1126/science.1076164. [DOI] [PubMed] [Google Scholar]
- 8.Phair RD, Scaffidi P, Elbi C, Vecerova J, Dey A, Ozato K, Brown DT, Hager G, Bustin M, Misteli T. Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol Cell Biol. 2004;24:6393–6402. doi: 10.1128/MCB.24.14.6393-6402.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Darzacq X, Shav-Tal Y, de TV, Brody Y, Shenoy SM, Phair RD, Singer RH. In vivo dynamics of RNA polymerase II transcription. Nat Struct Mol Biol. 2007;14:796–806. doi: 10.1038/nsmb1280. A thorough investigation of polymerase II kinetics including drug inhibition analysis.
- 10.Sprague BL, Muller F, Pego RL, Bungay PM, Stavreva DA, McNally JG. Analysis of binding at a single spatially localized cluster of binding sites by fluorescence recovery after photobleaching. Biophys J. 2006;91:1169–1191. doi: 10.1529/biophysj.105.073676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carrero G, McDonald D, Crawford E, de Vries G, Hendzel MJ. Using FRAP and mathematical modeling to determine the in vivo kinetics of nuclear proteins. Methods. 2003;29:14–28. doi: 10.1016/s1046-2023(02)00288-8. [DOI] [PubMed] [Google Scholar]
- 12. Hinow P, Rogers CE, Barbieri CE, Pietenpol JA, Kenworthy AK, DiBenedetto E. The DNA binding activity of p53 displays reaction-diffusion kinetics. Biophys J. 2006;91:330–342. doi: 10.1529/biophysj.105.078303. A mathematical approach to solve the kinetic model equations including a procedure to evaluate when additional model parameters are superfluous.
- 13. Beaudouin J, Mora-Bermudez F, Klee T, Daigle N, Ellenberg J. Dissecting the contribution of diffusion and interactions to the mobility of nuclear proteins. Biophys J. 2006;90:1878–1894. doi: 10.1529/biophysj.105.071241. A numerical/computational approach to solve the kinetic model equations including a novel strategy to account for heterogeneous binding using the acquired image data.
- 14. Gorski SA, Snyder SK, John S, Grummt I, Misteli T. Modulation of RNA polymerase assembly dynamics in transcriptional regulation. Molecular Cell. 2008;30:486–497. doi: 10.1016/j.molcel.2008.04.021. Investigation of pol I kinetics including comparison of model predictions at different cell stages and in the presence of a dominant negative mutant.
- 15. Mueller F, Wach P, McNally JG. Evidence for a common mode of transcription factor interaction with chromatin as revealed by improved quantitative fluorescence recovery after photobleaching. Biophys J. 2008;94:3323–3339. doi: 10.1529/biophysj.107.123182. Resolves discrepant predictions among three different kinetic modeling procedures and identifies the errors responsible.
- 16.Minton AP. How can biochemical reactions within cells differ from those in test tubes? Journal of Cell Science. 2006;119:2863–2869. doi: 10.1242/jcs.03063. [DOI] [PubMed] [Google Scholar]
- 17. Müller KP, Erdel F, Caudron-Herger M, Marth C, Fodor BD, Richter M, Scaranaro M, Beaudouin J, Wachsmuth M, Rippe K. Multiscale analysis of dynamics and interactions of heterochromatin protein 1 by fluorescence fluctuation microscopy. Biophys J. 2009;97:2876–2885. doi: 10.1016/j.bpj.2009.08.057. Cross validation approach using an assortment of techniques to analyze kinetics of HP1.
- 18.Mikuni S, Tamura M, Kinjo M. Analysis of intranuclear binding process of glucocorticoid receptor using fluorescence correlation spectroscopy. FEBS Lett. 2007;581:389–393. doi: 10.1016/j.febslet.2006.12.038. [DOI] [PubMed] [Google Scholar]
- 19.Renz M, Langowski J. Dynamics of the CapG actin-binding protein in the cell nucleus studied by FRAP and FCS. Chromosome Res. 2008;16:427–437. doi: 10.1007/s10577-008-1234-6. [DOI] [PubMed] [Google Scholar]
- 20.Schmiedeberg L, Weisshart K, Diekmann S, Meyer Zu HG, Hemmerich P. High- and low-mobility populations of HP1 in heterochromatin of mammalian cells. Mol Biol Cell. 2004;15:2819–2833. doi: 10.1091/mbc.E03-11-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Elf J, Li GW, Xie XS. Probing transcription factor dynamics at the single-molecule level in a living cell. Science. 2007;316:1191–1194. doi: 10.1126/science.1141967. The first in-vivo measurement of transcription factor binding at the single molecule level.
- 22.Grünwald D, Martin RM, Buschmann V, Bazett-Jones DP, Leonhardt H, Kubitscheck U, Cardoso MC. Probing intranuclear environments at the single-molecule level. Biophys J. 2008;94:2847–2858. doi: 10.1529/biophysj.107.115014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nalley K, Johnston SA, Kodadek T. Proteolytic turnover of the Gal4 transcription factor is not required for function in vivo. Nature. 2006;442:1054–1057. doi: 10.1038/nature05067. Investigation of the in-vivo binding behavior of a transcription factor at a single promoter using competition ChIP.
- 24.van Werven FJ, van Teeffelen HA, Holstege FC, Timmers HT. Distinct promoter dynamics of the basal transcription factor TBP across the yeast genome. Nat Struct Mol Biol. 2009;16:1043–1048. doi: 10.1038/nsmb.1674. [DOI] [PubMed] [Google Scholar]
- 25.Weidemann T, Wachsmuth M, Knoch TA, Muller G, Waldeck W, Langowski J. Counting nucleosomes in living cells with a combination of fluorescence correlation spectroscopy and confocal imaging. J Mol Biol. 2003;334:229–240. doi: 10.1016/j.jmb.2003.08.063. [DOI] [PubMed] [Google Scholar]
- 26.Delon A, Usson Y, Derouard J, Biben T, Souchier C. Continuous photobleaching in vesicles and living cells: a measure of diffusion and compartmentation. Biophys J. 2006;90:2548–2562. doi: 10.1529/biophysj.105.069815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Royen ME, Cunha SM, Brink MC, Mattern KA, Nigg AL, Dubbink HJ, Verschure PJ, Trapman J, Houtsmuller AB. Compartmentalization of androgen receptor protein-protein interactions in living cells. J Cell Biol. 2007;177:63–72. doi: 10.1083/jcb.200609178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dobrucki JW, Feret D, Noatynska A. Scattering of exciting light by live cells in fluorescence confocal imaging: phototoxic effects and relevance for FRAP studies. Biophys J. 2007;93:1778–1786. doi: 10.1529/biophysj.106.096636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kimura H, Sugaya K, Cook PR. The transcription cycle of RNA polymerase II in living cells. J Cell Biol. 2002;159:777–782. doi: 10.1083/jcb.200206019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boireau S, Maiuri P, Basyuk E, de la MM, Knezevich A, Pradet-Balade B, Backer V, Kornblihtt A, Marcello A, Bertrand E. The transcriptional cycle of HIV-1 in real-time and live cells. J Cell Biol. 2007;179:291–304. doi: 10.1083/jcb.200706018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yao J, Ardehali MB, Fecko CJ, Webb WW, Lis JT. Intranuclear distribution and local dynamics of RNA polymerase II during transcription activation. Mol Cell. 2007;28:978–990. doi: 10.1016/j.molcel.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 32.Farla P, Hersmus R, Trapman J, Houtsmuller AB. Antiandrogens prevent stable DNA-binding of the androgen receptor. J Cell Sci. 2005;118:4187–4198. doi: 10.1242/jcs.02546. [DOI] [PubMed] [Google Scholar]
- 33.Raghuram N, Carrero G, Th'ng J, Hendzel MJ. Molecular dynamics of histone H1. Biochemistry and Cell Biology-Biochimie et Biologie Cellulaire. 2009;87:189–206. doi: 10.1139/O08-127. [DOI] [PubMed] [Google Scholar]
- 34.Lele T, Wagner SR, Nickerson JA, Ingber DE. Methods for measuring rates of protein binding to insoluble scaffolds in living cells: Histone H1-chromatin interactions. J Cell Biochem. 2006;99:1334–1342. doi: 10.1002/jcb.20997. [DOI] [PubMed] [Google Scholar]
- 35.Stasevich TJ, Mueller F, Brown DT, McNally JG. Dissecting the binding mechanism of the linker histone in live cells: an integrated FRAP analysis. EMBO J. 2010 doi: 10.1038/emboj.2010.24. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. van Royen ME, Farla P, Mattern KA, Geverts B, Trapman J, Houtsmuller AB. Fluorescence Recovery After Photobleaching (FRAP) to Study Nuclear Protein Dynamics in Living Cells. Methods Mol Biol. 2009;464:363–385. doi: 10.1007/978-1-60327-461-6_20. A detailed description of a computational method to include complex 3D geometries into the kinetic model.
- 37.Mazza D, Cella F, Vicidomini G, Krol S, Diaspro A. Role of three-dimensional bleach distribution in confocal and two-photon fluorescence recovery after photobleaching experiments. Appl Opt. 2007;46:7401–7411. doi: 10.1364/ao.46.007401. [DOI] [PubMed] [Google Scholar]
- 38.Hallen MA, Layton AT. Expanding the scope of quantitative FRAP analysis. J Theor Biol. 2010;262:295–305. doi: 10.1016/j.jtbi.2009.10.020. [DOI] [PubMed] [Google Scholar]
- 39.Kang M, Kenworthy AK. A closed-form analytic expression for FRAP formula for the binding diffusion model. Biophys J. 2008;95:L13–L15. doi: 10.1529/biophysj.108.135913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kang M, Day CA, Drake K, Kenworthy AK, DiBenedetto E. A generalization of theory for two-dimensional fluorescence recovery after photobleaching applicable to confocal laser scanning microscopes. Biophys.J. 2009;97:1501–1511. doi: 10.1016/j.bpj.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Braga J, Desterro JM, Carmo-Fonseca M. Intracellular macromolecular mobility measured by fluorescence recovery after photobleaching with confocal laser scanning microscopes. Mol.Biol.Cell. 2004;15:4749–4760. doi: 10.1091/mbc.E04-06-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sinnecker D, Voigt P, Hellwig N, Schaefer M. Reversible photobleaching of enhanced green fluorescent proteins. Biochemistry. 2005;44:7085–7094. doi: 10.1021/bi047881x. [DOI] [PubMed] [Google Scholar]
- 43.Saxton MJ. Anomalous subdiffusion in fluorescence photobleaching recovery: a Monte Carlo study. Biophys J. 2001;81:2226–2240. doi: 10.1016/S0006-3495(01)75870-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guigas G, Weiss M. Sampling the cell with anomalous diffusion - the discovery of slowness. Biophys J. 2008;94:90–94. doi: 10.1529/biophysj.107.117044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lubelski A, Klafter J. Fluorescence recovery after photobleaching: the case of anomalous diffusion. Biophys J. 2008;94:4646–4653. doi: 10.1529/biophysj.107.119081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bancaud A, Huet S, Daigle N, Mozziconacci J, Beaudouin J, Ellenberg J. Molecular crowding affects diffusion and binding of nuclear proteins in heterochromatin and reveals the fractal organization of chromatin. EMBO J. 2009 doi: 10.1038/emboj.2009.340. Introduces novel experiments and theory to address the roles of more complex diffusion and binding behaviors.
- 47.Phair RD, Gorski SA, Misteli T. Measurement of dynamic protein binding to chromatin in vivo, using photobleaching microscopy. Methods Enzymol. 2004;375:393–414. doi: 10.1016/s0076-6879(03)75025-3. [DOI] [PubMed] [Google Scholar]
- 48.Gerlich D, Koch B, Dupeux F, Peters JM, Ellenberg J. Live-cell imaging reveals a stable cohesin-chromatin interaction after but not before DNA replication. Current Biology. 2006;16:1571–1578. doi: 10.1016/j.cub.2006.06.068. [DOI] [PubMed] [Google Scholar]
- 49.Kopelman R. Fractal Reaction Kinetics. Science. 1988;241:1620–1626. doi: 10.1126/science.241.4873.1620. 1988 Sep 23;241(4873):16. [DOI] [PubMed] [Google Scholar]
- 50.Mazza D, Braeckmans K, Cella F, Testa I, Vercauteren D, Demeester J, De Smedt SS, Diaspro A. A new FRAP/FRAPa method for three-dimensional diffusion measurements based on multiphoton excitation microscopy. Biophys J. 2008;95:3457–3469. doi: 10.1529/biophysj.108.133637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matsuda T, Miyawaki A, Nagai T. Direct measurement of protein dynamics inside cells using a rationally designed photoconvertible protein. Nature Methods. 2008;5:339–345. doi: 10.1038/nmeth.1193. [DOI] [PubMed] [Google Scholar]
- 52.Dross N, Spriet C, Zwerger M, Muller G, Waldeck W, Langowski J. Mapping eGFP oligomer mobility in living cell nuclei. PLoS One. 2009;4:e5041. doi: 10.1371/journal.pone.0005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen Y, Muller JD, Ruan Q, Gratton E. Molecular brightness characterization of EGFP in vivo by fluorescence fluctuation spectroscopy. Biophys J. 2002;82:133–144. doi: 10.1016/S0006-3495(02)75380-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xouri G, Squire A, Dimaki M, Geverts B, Verveer PJ, Taraviras S, Nishitani H, Houtsmuller AB, Bastiaens PIH, Lygerou Z. Cdt1 associates dynamically with chromatin throughout G1 and recruits Geminin onto chromatin. Embo Journal. 2007;26:1303–1314. doi: 10.1038/sj.emboj.7601597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grunwald D, Spottke B, Buschmann V, Kubitscheck U. Intranuclear binding kinetics and mobility of single native U1 snRNP particles in living cells. Mol Biol Cell. 2006;17:5017–5027. doi: 10.1091/mbc.E06-06-0559. [DOI] [PMC free article] [PubMed] [Google Scholar]