Abstract
Long non-coding RNAs (lncRNAs) are mRNA-like, non-protein coding RNAs that are pervasively transcribed throughout eukaryotic genomes. Rather than silently accumulating in the nucleus, many of these are now known or suspected to play important roles in nuclear architecture or in the regulation of gene expression. In this review, we highlight some recent progress in how lncRNAs regulate these important nuclear processes at the molecular level.
Introduction
In comparison to constitutively expressed ‘housekeeping’ ncRNAs and small regulatory RNAs, lncRNAs (larger than 200 nucleotides) are mRNA-like, non-protein coding RNAs that are pervasively transcribed throughout eukaryotic genomes [1-3]. Despite a few well-characterized lncRNAs, such as Xist [4,5] and H19 [6], lncRNAs were only appreciated as a significant new transcript class following the large-scale sequencing of a full-length cDNA library in mouse [1]. Most recently, the increased sensitivity of genome tiling arrays [3,7], along with several other techniques [8-10] have demonstrated widespread antisense and lncRNA transcription in mammalian genomes. These studies reveal that the eukaryotic transcriptome is surprisingly complex, with lncRNAs often overlapping with, or interspersed between multiple protein-coding and non-coding transcripts.
LncRNAs have been implicated in a number of important nuclear events, such as chromatin remodeling [11-16], transcriptional regulation [17-21], and the integrity of subcellular compartments [22-25]. In this review, we highlight some recent advances in our understanding of how lncRNAs are involved in the regulation of nuclear processes.
LncRNA-mediated chromatin remodeling
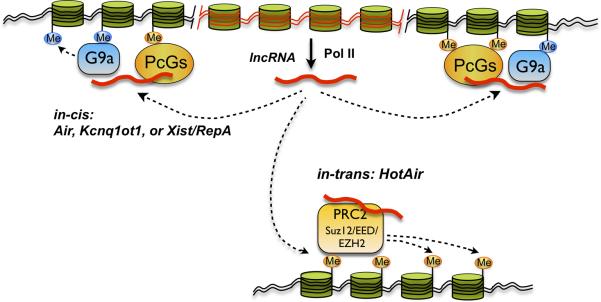
Many lncRNAs are associated with chromatin-modifying complexes and affect gene expression. A recent study found about 20% of 3,300 human long intergenic ncRNAs were bound by Polycomb Repressive Complex 2 (PRC2) or other chromatin-modifying complexes [26]. Although mechanisms are not yet completely clear, there are several cases that illustrate how lncRNAs can recruit transcriptional repressive complexes to silence specific genomic regions (Figure 1).
Figure 1.
Long non-coding RNA-mediated chromatin remodeling. Some lncRNAs that are transcribed by RNA polymerase II recruit transcriptional repressive complexes including PcGs and G9a to silence specific genomic regions, both in cis (top) and in trans (bottom). See text for details.
During X-chromosome inactivation (XCI), one of the two X chromosomes in female mammals must be inactivated in order to achieve an equal level expression of X-chromosome genes. A number of lncRNAs including Xist, Tsix, and Xite participate in this process [27,28]. Since the transcription of Xist on the Xi is required for the maintenance of XCI [29], it has been hypothesized that Xist recruits chromatin modeling complexes to silence Xi. Recent findings, however, suggested a more complex mechanism. Zhao et al [11] discovered a 1.6 kb ncRNA, RepA, which comprises sequences also contained in the 5’ region of Xist and which directly binds PRC2. In pre-XCI cells, RepA initially recruits PRC2 to the future Xi, although the lncRNA Tsix, which is antisense to Xist and has an established role as an Xist antagonist (reviewed in [27,30]), inhibits this interaction by binding PRC2, thus competing with RepA for this factor. During Xi initiation, Tsix is downregulated on the future Xi, hence RepA can now productively engage PRC2 and activate full-length Xist transcription. The upregulated Xist in turn preferentially binds to PRC2 through its RepA sequence, resulting in the spread of Xist along Xi and the distribution of PRC2 and trimethylated histone H3 lysine27 (H3K27) throughout the Xi [11]. Supporting this model, RepA depletion abolishes full-length Xist induction and trimethylation on H3K27 of the Xi. Likewise, PRC2 deficiency compromises Xist upregulation [11]. Therefore, RepA and Xist are capable of recruiting PRC2 to establish the local chromatin modification, which is required for the initiation and spread of XCI (Figure 1).
Similar mechanisms have been observed during genomic imprinting [12-14] and tumorigenesis [15]. Air, 108 kb in length, is required for allele-specific silencing of the cis-linked Slc22a3, Slc22a2, and Igf2r genes [31]. Air uniquely interacts with the Slc22a3 promoter chromatin and the H3K9-specific histone methyltransferase G9a in placenta [14]. Depletion of G9a fails to silence Slc22a3 and results in non-imprinted transcription. Truncation of Air fails to accumulate at the Slc22a3 promoter and results in reduced G9a recruitment and biallelictranscription (Fig. 1) [14]. Similarly, the 90.5 kb long Kcnq1ot1 [12] has been linked to the bidirectional silencing of about 10 paternally imprinted genes in the Kcnq1 domain [32]. Here the mechanism involves the interaction between Kcnq1ot1 and G9a and PRC2 in a lineage-specific manner (Figure 1) [12,13]. In addition, the p15 antisense (p15AS) ncRNA of the tumor suppressor gene p15 has been implicated in leukemia, and their transcription is inversely correlated. By introducing p15AS expression constructs into mammalian cells, p15 silencing is induced through heterochromatin formation [15]. Although the protein mediators of this silencing process remain to be determined, p15AS and other natural antisense ncRNA might serve as triggers for heterochromatin formation in tumor suppressor gene silencing.
Finally, lncRNA-mediated chromatin remodeling also occurs in trans. Hundreds of HOX ncRNAs were identified along the human HOX loci [16], among which, the 2.2 kb long HOTAIR (HOX antisense intergenic RNA) resides in a regulatory boundary in the HOXC locus. Surprisingly, knockdown of HOTAIR does not lead to any changes in the HOXC locus, but instead represses transcription across 40 kb of the HOXD locus, as shown by the loss of the PRC2 occupancy and H3K27 trimethylation, in the HOXD cluster which is located on a different chromosome from HOXC (Figure 1). Pulldown assays show a specific interaction between HOTAIR and PRC2 key components, Suz12 and Ezh2 [16].
LncRNA-mediated transcription regulation
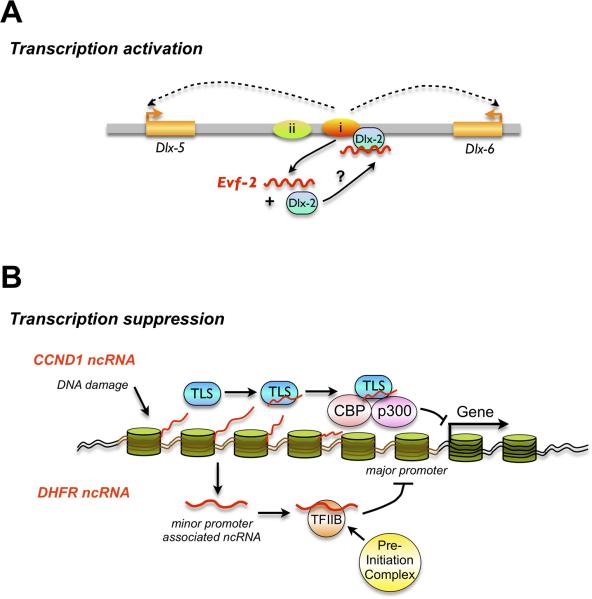
Some lncRNAs can directly influence transcription. Recent advances in DNA sequencing have revealed widespread transcription of promoter-associated transcripts from yeast to mammals [3,7,33,34]. Although the functional mechanisms remain largely unknown, emerging evidence suggests that at least some of these ncRNAs may regulate transcription by serving as “ligands” for transcription factors. First, some lncRNAs act as transcription co-activators. Vertebrate Dlx genes play critical roles in neuronal development and patterning [17]. The Dlx genes are regulated by two ultraconserved intergenic enhancers, which are located in the Dlx-5/6 locus. One of the ultraconserved enhancers is transcribed to a 3.8kb ncRNA, Evf-2, which forms a stable complex with the homeodomain protein Dlx-2 and thus activates Dlx-2 acting a transcriptional enhancer of Dlx-5/6 (Fig. 2A) [17]. Second, some lncRNAs suppress transcription. Wang et al [18] reported that DNA damage signals could induce a set of single-stranded, low-copy-number ncRNAs transcribed from the 5' regulatory region of the cyclin D1 (CCND1) gene. These ncRNAs could allosterically modulate the activity of an RNA-binding protein, TLS (translocated in liposarcoma). The modified TLS in turn inhibited CREB-binding protein (CBP) and p300 histone acetyltransferase activities, which subsequently inhibited CCND1 transcription (Figure 2B). Third, some lncRNAs might compete with transcription factors to inhibit gene transcription. The gene encoding dihydrofolate reductase (DHFR) contains a minor and a major promoter, with the latter being transcriptionally suppressed in quiescent cells. Transcriptional repression of the major promoter depends on a ncRNA initiated from the upstream minor promoter and which is involved in the formation of a stable complex between the ncRNA, the major promoter, and the general transcription factor IIB [19] (Figure 2B). Taken together, these studies suggest that ncRNAs transcribed from regulatory regions of transcription units can affect RNA-binding co-regulators.
Figure 2.
Long non-coding RNA-mediated transcription regulation. A. Transcription activation by lncRNA. In this example, Evf-2 is transcribed from an ultraconserved enhancer and forms a stable complex with Dlx-2, which in turn activates Dlx-2 as a transcriptional enhancer. B. Transcription suppression by lncRNA. Top: in response to DNA damage, lncRNAs are transcribed from the 5’-upstream region of the CCND1 gene and recruit the RNA-binding protein TLS to modulate CBP and p300 to inhibit CCND1 transcription. Bottom: lncRNA transcribed from the upstream of the minor promoter of DHFR gene competes with transcription factors to inhibit the major promoter transcription in quiescent cells.
In addition, a number of RNA polymerase III-transcribed human Alu and mouse B2 RNAs are induced during heat shock. Some of these transcripts can act to block RNA polymerase II (RNAP II) transcription in trans by binding directly and tightly to RNAP II and co-occupying the promoters of repressed genes [20]. Further studies showed that they prevent interactions between RNAP II and the promoter during closed complex formation, resulting in complexes with an altered conformation that are transcriptionally inert [21]. Given the abundance of B2 and Alu RNAs in the genome and their possible evolutionary roles, this transcription repression effect could be profound.
LncRNAs in nuclear architecture and subnuclear compartments
The eukaryotic nucleus is a highly compartmentalized organelle and contains a variety of membraneless subnuclear bodies (reviewed in [35]). Relatively little is known about how these nuclear domains assemble and function, but some are associated with distinct lncRNAs. One interesting example is that of paraspeckles. These are cell-cycle-regulated nuclear foci that depend on RNA for their structural integrity [36,37]. While the function of paraspeckles is not yet completely clear, some studies have suggested that they could be sites of nuclear retention of at least a subset of mRNAs that have undergone adenosine (A)-to-inosine (I) editing [25,38,39].
Paraspeckles contain three DBHS (Drosophila Behavior and Human Splicing) family proteins: PSP1α, p54nrb, and PSF. PSP1α serves as a paraspeckle marker [36], yet overexpression of PSP1α does not induce paraspeckle numbers [23], and PSP1α depletion has little effect on their integrity [24], suggesting it is not essential in paraspeckle assembly [23]. p54nrb and PSF are multifunctional proteins that have been implicated in a variety of nuclear processes (reviewed in [40]). Most recently, Sasaki et al [24] reported that p54nrb and PSF may be important for paraspeckle integrity since individual knockdowns resulted in paraspeckle disintegration.
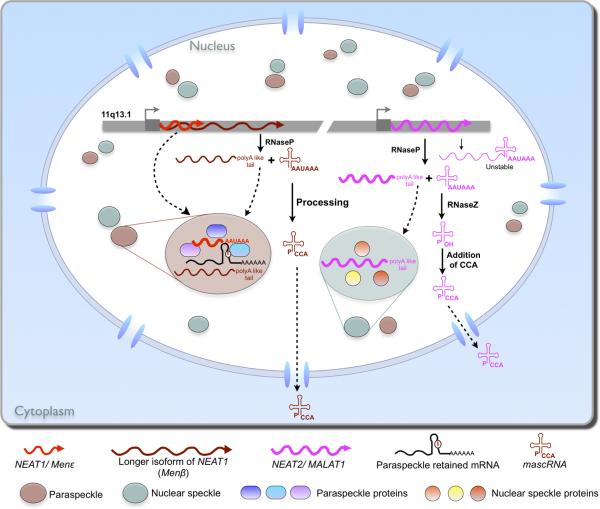
In addition to protein components, the lncRNA, NEAT1 (Men ε/β in mouse), plays a crucial role in paraspeckle structural integrity [22-25]. The polyadenylated NEAT1 and Men ε are 3.7 kb and 3.2 kb respectively and are enriched in paraspeckles in the nucleus [22-25]. A longer transcript (>17 kb in mouse, and >19 kb in human) [22,24,41] was also detected. This longer isoform is not polyadenylated [22], also localizes to paraspeckles and likely plays a similar role in the de novo paraspeckle formation [22,24] (Figure 3). Depletion of NEAT1 [23,25] or Men ε/β [22,24] disrupts paraspeckles, and overexpression of Men ε increases paraspeckle number in both NIH3T3 and HeLa cells [23], suggesting that NEAT1 and Men ε/β are essential paraspeckle components and may be functionally conserved. Furthermore, Men ε/β transcripts fail to remain colocalized with the paraspeckle marker protein PSP1α after drug-induced transcriptional arrest [22,24]. In addition, depletion of Men ε/β transcripts results in suppression of paraspeckle reformation after release from transcriptional arrest [22,24]. Interestingly, Men ε/β transcripts are upregulated during mouse myoblast differentiation into myotubes [22], suggesting a role in developmental regulation. Consistent with this notion, the transcription of NEAT1 is strongly suppressed in human embryonic stem cells (ESCs) but is induced upon differentiation to trophoblasts, and, consistent with the role of NEAT1 in paraspeckle assembly, paraspeckles are absent in human ESCs and appear during differentiation [25].
Figure 3.
Long non-coding RNAs in nuclear subcompartments. Human NEAT1 (Menε in mouse) localizes to paraspeckles and is required for paraspeckle structural integrity. NEAT2 (MALAT1) localizes to splicing speckles but is not required for their structural integrity. See text for details. Nascent Menβ and MALAT1 transcripts can each be processed by the unusual mechanism of RNase P cleavage to generate the 5’ end of mascRNA (MALAT1-associated small cytoplasmic RNA) and the 3’ end of the mature Menβ and MALAT1 transcripts, which localize to paraspeckles and splicing speckles, respectively.
Biochemical analyses demonstrated that NEAT1 and Men ε/β interact with paraspeckle DBHS proteins [22-25], indicating that it might serve as a platform for the assembly of large macromolecular complexes. These large complexes that generate paraspeckles may function directly in the regulation of some gene expression. Human ESCs express all key DBHS proteins but lack NEAT1, and the nuclear retention pathway for A-to-I edited RNAs is also not functional [25]. Furthermore, knockdown of NEAT1 in HeLa cells results both in loss of paraspeckles and in enhanced nucleocytoplasmic export of mRNAs containing inverted repeats of Alus [25], suggesting NEAT1 might be a key factor in the regulation of the nuclear retention of edited or structured mRNAs in paraspeckles. However, how this subset of retained transcripts is regulated by NEAT1 remains unclear. It will be of interest to determine whether NEAT1 directly associates with the retained mRNAs and thereby controls their nuclear export or whether retained mRNAs bind to proteins that are assembled in paraspeckles by NEAT1 RNA.
In addition to NEAT1, there are a number of lncRNAs that localize to different subnuclear regions. MALAT1 (metastasis-associated lung adenocarcinoma transcript 1) (NEAT2 in human), is transcribed from the downstream region of Men ε/β (NEAT1) gene, and is specifically localized to splicing speckles” [41] (Figure 3). However, the structure of splicing speckles is largely unaffected in cells with reduced NEAT2/MALAT1 expression [23], suggesting that this lncRNA is not required for the assembly or integrity of speckles. In addition to NEAT1 and MALAT1, lncRNAs that are involved in chromatin silencing also localize to distinct regions in the nucleus. While Air specifically envelops paternal Slc22a3 [14], Xist and Kcnq1ot1 both localize to the perinucleolar region during the S phase of the cell cycle [12,42,43]. Finally, several repeat-associated lncRNAs have been localized to specific nuclear regions. A subclass of Sat III (Satellite III) is transcribed upon heat shock in human cells and these lncRNAs are associated with nuclear stress bodies, which are assembled on specific pericentromeric heterochromatic domains containing Sat III DNA, several transcription and splicing factors, but are devoid of heterochromatin markers [44,45].
Involvement of lncRNAs in a wide variety of other biological processes
Apart from the above-discussed roles in nuclear processes, lncRNAs have also been implicated in the regulation of a number of diverse other biological events. For example, some lncRNAs can act as precursors for small RNAs, either by an RNase III-like cleavage from the sense and antisense duplexes, such as Xist/Tsix [46] or by a tRNA-like 3’ end processing of MALAT1 [47] (Fig. 3). NRON (ncRNA repressor of the nuclear factor of activated T cells [NFAT]) ncRNA is involved in NFAT intracellular trafficking by interacting with multiple proteins including members of the importin β superfamily [48]. During heat shock response, HSR1 (heat shock RNA-1) cooperatively works with translation elongation factor eEF1A to activate the heat shock transcription factor 1 and induce the expression of heat-shock and other cytoprotective proteins [49]. Furthermore, a natural antisense transcript (Zeb2-AS) blocks alternative splicing by overlapping a splice site of its sense-coding transcript (Zeb2), therefore altering Zeb translation [50]. Also, lncRNAs have been implicated in stem cell pluripotency and differentiation, as shown by a recent study that over 900 lncRNAs are specifically associated with mouse ESC differentiation into embryoid bodies [51].
Finally, the expression of lncRNAs has been linked to a number of human diseases, including cancer and neurological diseases (reviewed in [52,53]). Most recently, lncRNAs have been reported as additional players in the regulation of some well-documented disorders, such as Alzheimer's disease [54] and Fragile X syndrome [55]. Overall, although our understanding of how these lncRNAs cause diseases lags far behind that of their protein partners, lncRNAs may serve as additional clinical targets in the treatment of diseases in the future.
Perspectives
While lncRNAs do not possess protein-coding capacity, they have been found to have previously unexpected impacts on the programming and regulation of mammalian genome. Despite recent rapid progress in the functional study of many of them, some important questions remain to be addressed.
Unlike proteins, where sequences can often be classified into motifs, which are usually indicative of function, the primary sequences of lncRNAs often contain insufficient information to predict their function. They frequently exhibit sequence divergence yet conserved function between species. For instance, mouse B2 and human Alu RNAs are not similar in sequence or overall secondary structure [20], but surprisingly share identical mechanisms of repression of RNAP II in both organisms. Furthermore, human NEAT1 and mouse Menε/β, both of which are transcribed from similar genomic contexts, share only very low sequence similarity [41], but are functionally conserved in mouse and human in the assembly of paraspeckle structure integrity [22-25]. Moreover, PRC2 proteins are associated with a number of lncRNAs whose lengths vary from 1.6 kb to over 90 kb [11,12,15,16] (Figure 1). It is almost certain that very long lncRNAs do not interact exclusively with these proteins. Therefore, it is of great interest to decipher the sequences and the structural motifs for their functional significance.
Another challenging unanswered question is how do protein partners interact with lncRNAs to allow the specialized functions? In one model, lncRNAs might recruit and then “guide” its protein partners to proper chromosomal destinations. Specific sequences within the lncRNAs could recognize specific chromatin regions via sequence complementarity, therefore bringing the associated proteins to the targeted region. For instance, RepA and Xist recruit PRC2 to establish local chromatin modifications on the inactive X [11]. In the case of lncRNAs recruiting proteins at a distance [14] or in trans [16], the tertiary structure of the higher-order of chromatin might help bring distant chromatin regions together. Alternatively, lncRNAs might induce allosteric structural modifications of their protein partners to either enhance [17] or suppress [18] their normal activities. Although these hypotheses remain to be experimentally confirmed, they might not be independent as one lncRNA might be able to both “guide” and “modify” its protein partner(s) during the same biological process. For instance, CCND1 upstream associated ncRNA not only allosterically “modifies” the activity of TLS but also “guides” the modified complex to the CCND1 upstream region to perform transcriptional repression [18] (Figure 2A).
Acknowledgments
This work was supported by grant CA04382 from the National Cancer Institute and awards from the State of Connecticut under the Connecticut Stem Cell Research Grants Program to L-L.C. and G.G.C. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the State of Connecticut, the Department of Public Health of the State of Connecticut, or Connecticut Innovations, Inc. Owing to length constraints, we have not been able to cite many articles that are of interest and importance in the areas discussed, and for this we apologize.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest have been highlighted:
* Of special interest
** Of outstanding interest
- 1.Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 2.Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- 3.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 4.Borsani G, Tonlorenzi R, Simmler MC, Dandolo L, Arnaud D, Capra V, Grompe M, Pizzuti A, Muzny D, Lawrence C, et al. Characterization of a murine gene expressed from the inactive X chromosome. Nature. 1991;351:325–329. doi: 10.1038/351325a0. [DOI] [PubMed] [Google Scholar]
- 5.Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence J, Willard HF. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 6.Bartolomei MS, Zemel S, Tilghman SM. Parental imprinting of the mouse H19 gene. Nature. 1991;351:153–155. doi: 10.1038/351153a0. [DOI] [PubMed] [Google Scholar]
- 7.Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- 8.He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. The antisense transcriptomes of human cells. Science. 2008;322:1855–1857. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10*.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [Authors used ChIP-Seq to map K4–K36 trimethylation domains genome widely. Further, they identified K4–K36 structures that reside outside known protein-coding gene loci to systematically discover new functional long non-coding RNAs.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11*.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [This study described a 1.6 kb ncRNA RepA from the Xist A-repeat region. RepA, together with PRC2, is required for the initiation and spread of XCI.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 13.Terranova R, Yokobayashi S, Stadler MB, Otte AP, van Lohuizen M, Orkin SH, Peters AH. Polycomb group proteins Ezh2 and Rnf2 direct genomic contraction and imprinted repression in early mouse embryos. Dev Cell. 2008;15:668–679. doi: 10.1016/j.devcel.2008.08.015. [DOI] [PubMed] [Google Scholar]
- 14**.Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–1720. doi: 10.1126/science.1163802. [By genetic knockout of G9a and truncation of Air, this study demonstrated that Air specifically interacts with the promoter chromatin of one of its imprinted gene, Slc22a3 to epigenetically silence imprinted genes.] [DOI] [PubMed] [Google Scholar]
- 15.Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16*.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [This paper studied the transcriptional activity of the human HOX loci using tiling arrays and identified 231 HOX ncRNAs. Among these ncRNAs, HOTAIR was shown to interact with PRC2 and repress transcription of the HOXD locus in trans.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 20.Mariner PD, Walters RD, Espinoza CA, Drullinger LF, Wagner SD, Kugel JF, Goodrich JA. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell. 2008;29:499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 21.Yakovchuk P, Goodrich JA, Kugel JF. B2 RNA and Alu RNA repress transcription by disrupting contacts between RNA polymerase II and promoter DNA within assembled complexes. Proc Natl Acad Sci U S A. 2009;106:5569–5574. doi: 10.1073/pnas.0810738106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. MEN {epsilon}/{beta} nuclear retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2008 doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci U S A. 2009;106:2525–2530. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25**.Chen L-L, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: Functional role of a nuclear noncoding RNA. Mol. Cell. 2009;35:467–478. doi: 10.1016/j.molcel.2009.06.027. [By working with human embryonic stem cells before and during differentiaiton, this work showed that NEAT1 is not only essential for paraspeckle formation, but also might be a key factor in the regulation of the nuclear retention of edited or structured mRNAs in nuclear paraspeckles.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26**.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [By chromatin immunoprecipitation assays, this paper showed that about 20% of 3,300 human long intergenic ncRNAs were bound by PRC2 or other chromatin-modifying complexes. In addition, depletion of certain lncRNAs associated with PRC2 resulted in the upregulated expression of genes normally silenced by PRC2.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chow J, Heard E. X inactivation and the complexities of silencing a sex chromosome. Curr Opin Cell Biol. 2009;21:359–366. doi: 10.1016/j.ceb.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 28.Leeb M, Steffen PA, Wutz A. X chromosome inactivation sparked by non-coding RNAs. RNA Biol. 2009;6 doi: 10.4161/rna.6.2.7716. [DOI] [PubMed] [Google Scholar]
- 29.Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 30.Senner CE, Brockdorff N. Xist gene regulation at the onset of X inactivation. Curr Opin Genet Dev. 2009;19:122–126. doi: 10.1016/j.gde.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 31.Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- 32.Mancini-Dinardo D, Steele SJ, Levorse JM, Ingram RS, Tilghman SM. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev. 2006;20:1268–1282. doi: 10.1101/gad.1416906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neil H, Malabat C, d'Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–1042. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- 35.Spector DL. Nuclear domains. J Cell Sci. 2001;114:2891–2893. doi: 10.1242/jcs.114.16.2891. [DOI] [PubMed] [Google Scholar]
- 36.Fox AH, Lam YW, Leung AK, Lyon CE, Andersen J, Mann M, Lamond AI. Paraspeckles. A novel nuclear domain. Curr. Biol. 2002;12:13–25. doi: 10.1016/s0960-9822(01)00632-7. [DOI] [PubMed] [Google Scholar]
- 37.Fox AH, Bond CS, Lamond AI. P54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in an RNA-dependent manner. Mol Biol Cell. 2005;16:5304–5315. doi: 10.1091/mbc.E05-06-0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, Zhang MQ, Spector DL. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 39.Chen LL, DeCerbo JN, Carmichael GG. Alu element-mediated gene silencing. EMBO J. 2008;27:1694–1705. doi: 10.1038/emboj.2008.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen LL, Carmichael GG. Gene regulation by SINES and inosines: Biological consequences of A-to-I editing of Alu element inverted repeats. Cell Cycle. 2008;7:3294–3301. doi: 10.4161/cc.7.21.6927. [DOI] [PubMed] [Google Scholar]
- 41.Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang LF, Huynh KD, Lee JT. Perinucleolar targeting of the inactive X during S phase: evidence for a role in the maintenance of silencing. Cell. 2007;129:693–706. doi: 10.1016/j.cell.2007.03.036. [DOI] [PubMed] [Google Scholar]
- 43.Mohammad F, Pandey RR, Nagano T, Chakalova L, Mondal T, Fraser P, Kanduri C. Kcnq1ot1/Lit1 noncoding RNA mediates transcriptional silencing by targeting to the perinucleolar region. Mol Cell Biol. 2008;28:3713–3728. doi: 10.1128/MCB.02263-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rizzi N, Denegri M, Chiodi I, Corioni M, Valgardsdottir R, Cobianchi F, Riva S, Biamonti G. Transcriptional activation of a constitutive heterochromatic domain of the human genome in response to heat shock. Mol Biol Cell. 2004;15:543–551. doi: 10.1091/mbc.E03-07-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valgardsdottir R, Chiodi I, Giordano M, Cobianchi F, Riva S, Biamonti G. Structural and functional characterization of noncoding repetitive RNAs transcribed in stressed human cells. Mol Biol Cell. 2005;16:2597–2604. doi: 10.1091/mbc.E04-12-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46*.Ogawa Y, Sun BK, Lee JT. Intersection of the RNA interference and X-inactivation pathways. Science. 2008;320:1336–1341. doi: 10.1126/science.1157676. [This study described that small RNAs can be processed from the duplex that is formed betwwen Xist/Tsix. These small RNAs, between 25 nt to 42 nt in length, are most likely generated in a Dicer-dependent manner as Dicer deletion inhibits small RNA production and gives rise to a series of X-inactivation defects, such as the derepression of Xist and the loss of Xist accumulation and H3K27 on Xi.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47*.Wilusz JE, Freier SM, Spector DL. 3' end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–932. doi: 10.1016/j.cell.2008.10.012. [This work identified a highly conserved 61 nt-tRNA like mascRNA that was processed from the 3’ region of MALAT1 by two sequential RNase cleavages in mouse cells. This cleavage is important for MALAT1 maturation but its functional significance is not known.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, Hogenesch JB, Schultz PG. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309:1570–1573. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- 49.Shamovsky I, Ivannikov M, Kandel ES, Gershon D, Nudler E. RNA-mediated response to heat shock in mammalian cells. Nature. 2006;440:556–560. doi: 10.1038/nature04518. [DOI] [PubMed] [Google Scholar]
- 50.Beltran M, Puig I, Pena C, Garcia JM, Alvarez AB, Pena R, Bonilla F, de Herreros AG. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008;22:756–769. doi: 10.1101/gad.455708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Solda G, Simons C, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Costa FF. Non-coding RNAs: new players in eukaryotic biology. Gene. 2005;357:83–94. doi: 10.1016/j.gene.2005.06.019. [DOI] [PubMed] [Google Scholar]
- 53.Prasanth KV, Spector DL. Eukaryotic regulatory RNAs: an answer to the ‘genome complexity’ conundrum. Genes Dev. 2007;21:11–42. doi: 10.1101/gad.1484207. [DOI] [PubMed] [Google Scholar]
- 54.Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G, 3rd, Kenny PJ, Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Khalil AM, Faghihi MA, Modarresi F, Brothers SP, Wahlestedt C. A novel RNA transcript with antiapoptotic function is silenced in fragile X syndrome. PLoS One. 2008;3:e1486. doi: 10.1371/journal.pone.0001486. [DOI] [PMC free article] [PubMed] [Google Scholar]