Abstract
RNA is arguably the most versatile biological macromolecule due to its ability both to encode and to manipulate genetic information. The diverse roles of RNA depend on its ability to fold back on itself to form biologically functional structures that bind small molecules and large protein ligands, to change conformation, and to affect the cellular regulatory state. These features of RNA biology can be structurally interrogated using chemical mapping experiments. The usefulness and applications of RNA chemical probing technologies have expanded dramatically over the past five years due to several critical advances. These innovations include new sequence-independent RNA chemistries, algorithmic tools for high-throughput analysis of complex data sets composed of thousands of measurements, new approaches for interpreting chemical probing data for both secondary and tertiary structure prediction, facile methods for following time-dependent processes, and the willingness of individual research groups to tackle increasingly bold problems in RNA structural biology.
Introduction
In most cases, nascent RNA molecules exist in an unstructured or fully single-stranded form for only an instant. Within milliseconds, significant elements of an RNA fold to form higher-order structures. In many regions, the structure may initially consist primarily of simple base paired elements. Over the second to minute timeframe, some elements will continue to fold into compact elements stabilized by higher-order tertiary interactions. For regions that do not form either base pairing or tertiary interactions, this lack of structure may facilitate interactions with proteins, small regulatory RNAs, or the translation initiation or splice site selection machineries. RNA structure is also dynamic, meaning that a structure that plays an important functional role in one biological state may not persist in other regulatory regimes. A full understanding of RNA-mediated biology, regulation, and disease will require a deep understanding of the varied structures accessible to RNA. A powerful way to understand these structures, especially for large RNAs in solution, is by evaluating their conformations using chemical probing technologies.
Background
Chemical probing of RNA is a venerable field whose basic methods were worked out 25–30 years ago [1–3]. An RNA of interest is treated with a chemical reagent that “modifies” the RNA in some way. The reagent can be a small organic molecule, a metal ion, or an RNase enzyme. Modification can be performed using purified components in vitro or in a complex biological environment in situ or in vivo. The experiment is performed such that reaction with the RNA is relatively limited and any two modification events are uncorrelated. Modification can result either in cleavage of the RNA or in formation of a covalent chemical adduct between the RNA and probe molecule. Chemical cleavage is usually detected by resolving end-labeled RNA fragments by size. Both classes of modification, cleavage and adduct formation, can be detected as a stop to primer extension mediated by a reverse transcriptase enzyme with sites of modification inferred from the length of the resulting cDNA fragments. There exist many excellent reviews describing conventional chemical probing experiments.
Structural Information Obtainable by Chemical Mapping
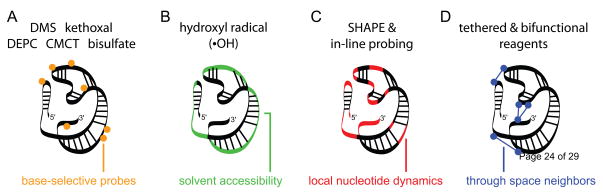
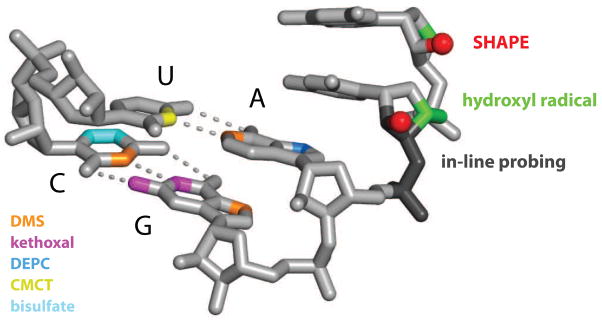
In general, four distinct types of information about RNA structure can be obtained from chemical probing experiments. The first group includes traditional chemical mapping reagents that form stable covalent adducts with one or more of the four RNA base groups (Fig. 1A). These base-specific reagents, which include dimethyl sulfate (DMS), kethoxal, diethyl pyrocarbonate (DEPC), CMCT and bisulfate yield structural information about the base stacking, hydrogen bonding, and electrostatic environment immediately adjacent to the base (sites of modification are shown in Fig. 2) [2,4]. Second, hydroxyl radicals, an OH group with an unpaired electron (•OH), can be generated in solution from hydrogen peroxide or molecular oxygen using an Fe(II)-EDTA catalyst. The hydroxyl radical cleaves the RNA backbone, likely by abstracting a proton from either the ribose C4′ or C5′, or both, positions [5] (in green, Figs. 1B & 2) providing information on the solvent accessibility of the backbone [5]. Third, two recently introduced experiments, in-line probing [6] and selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) [7,8], involve reaction with functional groups in the RNA backbone and ultimately measure local nucleotide flexibility or dynamics (Figs. 1C & 2). Finally, through use of bifunctional reagents or a group that is tethered to one part of an RNA and able to react with a second region, it is possible to use chemical mapping methods to obtain through-space information regarding RNA structure (Fig. 1D). Classic experiments that probe long-range structure include UV and chemically-induced cross-linking and site-directed cleavage and cross-linking.
Figure 1.
Classes of RNA structure information obtained by chemical probing include (A) base selective data, (B) solvent accessibility information, (C) measurements of nucleotide dynamics, and (D) constraints on long-range interactions.
Figure 2.
Sites of RNA modification for base-selective (left) and sequence-independent (right) modification chemistries. DEPC and CMCT also react with guanosine (not shown).
Advance 1 -- Sequence-Independent Chemistries for RNA Structure Analysis
The traditional, base-selective chemical reagents have yielded many important insights regarding RNA structure-function relationships. However, in order to interrogate every nucleotide in an RNA, multiple reagents need to be used together. Some of the more useful reagents, like DMS, react at different functional groups depending on the nucleotide (in orange, Fig. 2). Methods that interrogate all nucleotides in an RNA simultaneously are especially useful and yield concise and highly informative experiments.
In-line probing has proven useful for developing secondary structure models for small RNAs and for quantifying ligand-induced conformational changes in RNA [6]. In-line probing is typically performed at pH 9 for 40 hours and measures local nucleotide flexibility by virtue of the ability of a nucleotide to undergo “spontaneous” cleavage that reflects base-catalyzed attack of the 2′-hydroxyl group on the adjacent phosphate diester to form a 2′,3′-cyclic phosphate product (Fig. 2) [6].
SHAPE chemistry interrogates local nucleotide dynamics in RNAs of virtually any size in a single experiment. A SHAPE experiment employs hydroxyl-selective electrophiles that react with the 2′-hydroxyl group to form a 2′-O-ester adduct over periods of a few minutes [7,9,10], but that can be as short as 1 sec [11]. SHAPE measures local nucleotide dynamics because flexible nucleotides are better able to adopt conformations conducive to electrophilic attack by the 2′-hydroxyl group. Mechanistic studies show that SHAPE reactivities are largely independent of nucleotide type [12] and correlate strongly with the model-free order parameter calculated from NMR relaxation measurements on RNA [13], suggesting that this chemistry provides a quantitative measure of local nucleotide flexibility.
In addition to chemical probes, several RNases have been shown to react with single-stranded or unstructured regions in RNA, independent of the underlying sequence. E. coli RNase I was one of the first RNases to be characterized and reacts with RNA, independent of sequence. RNase I has proven highly useful in developing structural models for RNA regulatory elements [14]. RNase J1 from B. subtilis also cleaves single-stranded RNA regions with low sequence specificity and has been used to probe secondary structure and RNA-ribosome interactions [15].
These sequence-independent chemistries and RNase reagents make it possible to interrogate the local nucleotide environment at almost every position in RNA simultaneously. Used in conjunction with the hydroxyl radical reagent, which also reacts broadly with RNA (Fig. 2), these experiments afford an impressively comprehensive view of RNA secondary and tertiary interactions (Figs. 1B, C).
Advance 2 -- Software for High-Throughput RNA Structure Analysis
In principle, the experimental component of a chemical mapping experiment is straightforward. An RNA is incubated in an instructive cellular, ex vivo, or in vitro environment, allowed to react with a chemical or RNase probe, and the structural interrogation phase is complete. The results of a given experiment are then obtained by resolving and quantifying the fragments resulting from either RNA cleavage or primer extension at nucleotide resolution.
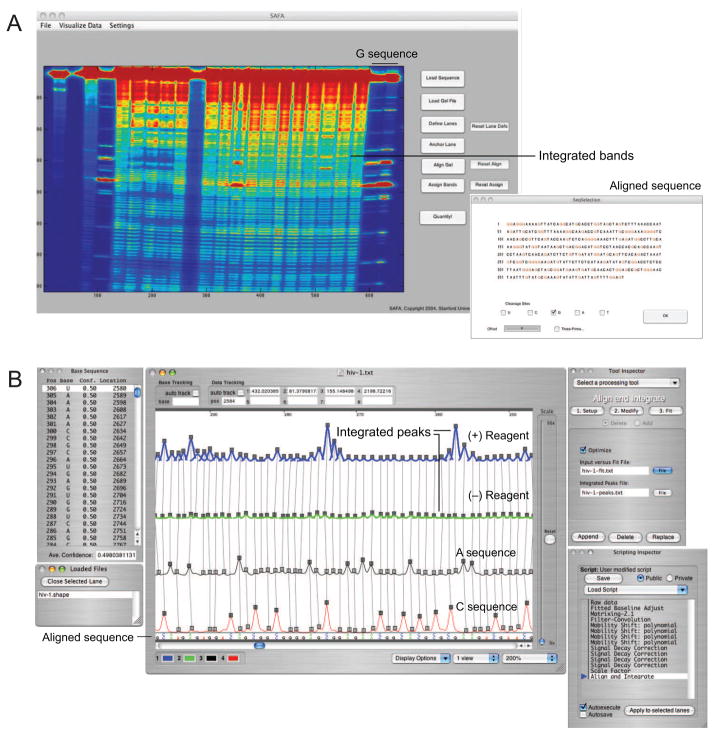
These steps have been traditionally accomplished by resolving nucleotide fragments in sequencing gels and estimating or individually quantifying band intensities. The steps required to read out the chemical probing information are time-consuming and labor-intensive. As a practical matter, individuals focusing on chemical probing analysis of RNA have spent more time and energy running sequencing gels and quantifying bands than actually designing experiments or interpreting their result. This bottleneck was addressed in a major way with the development of the Semi-Automated Footprinting Analysis (SAFA) program, which streamlines quantification of chemical mapping data as resolved in sequencing gels (Fig. 3A) [16]. A complete gel image, often containing thousands of reactivity measurements, can be quantified in 20–60 minutes. This represents roughly an order of magnitude improvement over manual methods.
Figure 3.
Automated analysis of chemical probing experiments using the (A) SAFA and (B) ShapeFinder programs. Figures adapted from [18,56].
In principle, gel electrophoresis could be replaced by any of the technologies now used in DNA sequencing. Commercial capillary electrophoresis instruments, widely used for routine DNA sequencing, yield single-nucleotide resolution read lengths of up to 1000 nts. Two software systems have been developed that make possible high-throughput analysis of RNA chemical mapping experiments, resolved by capillary electrophoresis. These are the Capillary Automated Footprinting Analysis (CAFA) [17] and ShapeFinder [18] programs. CAFA and ShapeFinder have different strengths. CAFA can be easier to set up and requires the use of only a single user-created fluorescently-labeled DNA primer. Good alignments between different experiments can be achieved for up to 250 nts in a single capillary electrophoresis read. CAFA has been optimized for repeated analysis of the same RNA, which is important in biophysical studies of RNA folding [17,19]. ShapeFinder requires at least two fluorescently labeled DNA primers and involves greater effort in the initial set up and alignment phase, but is generally a good choice when analyzing long and novel RNAs (Fig. 3B). With ShapeFinder, read lengths of 400–650 nts are routinely achievable [20–22]. As data analysis software continues to improve, it will become possible to tackle biological problems involving RNA structure analysis in increasingly adventurous and comprehensive ways.
Advance 3 -- Secondary Structure Determination
An absolute prerequisite for understanding structure-function relationships involving RNA is an accurate understanding of the pattern of base pairing, or secondary structure. The most reliable way to establish an RNA secondary structure is by comparative sequence analysis. Unfortunately, this approach is applicable to only the small fraction of RNAs that have a large number of diverse, and alignable, homologs. RNA secondary structure can also be predicted from sequence alone based on free energy minimization, a field which continues to make strong advances [23,24].
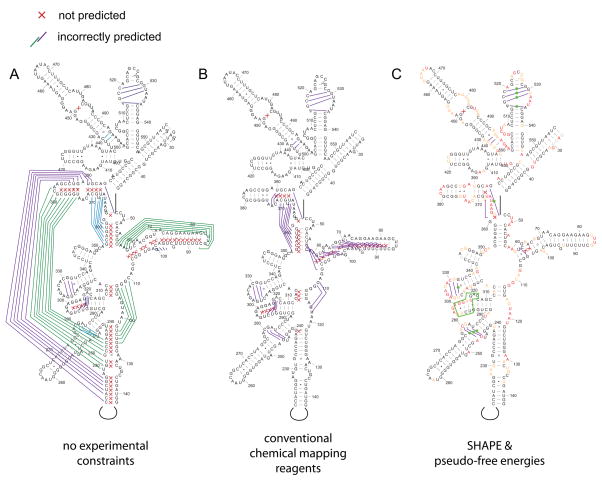
Many features of RNA are difficult to account for in a secondary structure folding algorithm. Such features include folding kinetics, the role of higher-order interactions, and the requirement to develop accurate energy rules for all possible RNA structure motifs. For example, using one of the most accurate currently available dynamic programming algorithms, the prediction sensitivity for E. coli 16S ribosomal RNA is less than 50% (errors within the 5′ domain of the 16S RNA are shown in Fig. 4A).
Figure 4.
Comparison of secondary structure prediction accuracy in the 5′ domain of E. coli 16S rRNA using free energy minimization (A) alone, (B) with conventional chemical modification reagents, and (C) with SHAPE-derived pseudo-free energy change terms. Missed base pairs are indicated by red x’s; incorrectly predicted base pairs are represented by colored lines. Regions where experimental information supports local RNA refolding are indicated with green boxes and spheres in panel C. Figure adapted from [27].
A possible solution to the inability to predict RNA secondary structure accurately, especially for large RNAs, lies in incorporating experimental chemical mapping data into the prediction algorithm. A major advance in this field was the demonstration that, when chemical mapping data are rigorously incorporated into the dynamic programming algorithms widely used to predict RNA secondary structure, accuracies improve significantly [25]. In one example, if nucleotides that are reactive towards chemical reagents are prohibited from forming internal base pairs in a helix, the prediction accuracy for 16S RNA improved from 50 to 72% (Fig. 4B). At this level of accuracy, many structures are predicted correctly but significant errors remain. These residual errors likely reflect a number of features. Two critical issues are that most base-selective chemical agents appear to be sensitive to both solvent accessibility and electrostatic features [4,26] and it is difficult to place reactivity measurements obtained from multiple reagents on a uniform scale (see Fig. 2, left).
The SHAPE experiment is helpful here because reactivity measurements are obtained for almost every nucleotide in an RNA in a single experiment and these measurements can be quantitatively compared. 2′-Hydroxyl reactivity is roughly inversely proportional to the probability that nucleotide forms a canonical base pair [7,13] and it is therefore possible to convert SHAPE reactivities into pseudo-free energy change terms. These pseudo-free energy terms provide an experimentally-informed adjustment to the nearest neighbor model for RNA secondary structure folding [27]. This blended experimental chemical mapping and thermodynamic approach yields very high prediction accuracies for many RNAs. For example, the SHAPE-directed prediction for 16S ribosomal RNA includes up to 97% of the base pairs in the accepted structure [27] (Fig. 4C).
Advance 4 -- Time-Resolved RNA Structure Analysis
A full understanding of the structures of RNA and RNA-protein complexes requires knowledge of the mechanisms by which these systems fold and assemble as a function of time. It is becoming increasingly clear that many RNAs fold by complex pathways and via energy landscapes that are commonly characterized as “rugged”, which means there exist many stable intermediate and non-functional states. A major set of advances have focused on developing efficient and information-rich approaches for performing chemical mapping as a function of time. The basic experiment involves initiating an RNA folding reaction, assessing structure at specific times by addition of a chemical probe, quenching the reagent (if needed), and quantifying the folding behavior as a function of time.
A method for time-resolved hydroxyl radical footprinting with very short (ms) time resolution was initially developed by exposing RNA to synchrotron radiation and controlling exposure time with a rapid mixing device [28]. More recently, the requirement for a synchrotron source has been eliminated by the development of “fast Fenton” methods [19,29]. Fast Fenton hydroxyl radical probing makes use of a rapid mixing and quench device and Fe(II)-EDTA-catalyzed reduction of oxygen to perform time-resolved footprinting in a conventional laboratory setting. Time-resolved hydroxyl radical footprinting provides a rich view of RNA folding on time scales spanning sub-second to many minutes [19,29] and for folding and assembly events involving small molecule-riboswitch interactions [30], large RNAs [31,32], and the ribosome [33,34].
The reaction of hydroxyl-selective electrophiles used in a SHAPE experiment can be tuned by varying the substituents near the reactive center [9,35]. Hydroxyl reactive electrophiles also undergo inactivation via the competing hydrolysis reaction with water. Benzoyl cyanide (BzCN) has a half-life in water of ~0.2 sec at 37 °C [11,36]. Thus, time-resolved analysis of local nucleotide flexibility can be followed in a simple way by initiating an RNA folding reaction and then obtaining ~1 sec snapshots of the RNA structure by mixing aliquots of the evolving reaction with pre-aliquoted BzCN reagent. Since the reagent undergoes auto-inactivation, a quench step is unnecessary if 1 sec time intervals are sufficient to monitor the dynamic processes of interest. This approach was used to develop a nucleotide-resolution model for folding of an RNase P RNA [11] and to show that a slow conformational change involving a single C2′-endo nucleotide represents the rate determining step for structural biogenesis of a 150-nt RNA domain [37].
Advance 5 -- Tertiary Structure Constraints
Many RNAs form both base-paired secondary structures and also complex higher-order, or tertiary, structure interactions. The chemical mapping reagents discussed thus far all react with RNA in ways that are sensitive to precise, very local, features of RNA structure (Fig. 1A–C). Tertiary interactions can be inferred from these local reactivity measurements in several ways. For example, most RNA nucleotides that form simple A-form helices or loop structures have roughly uniform and high solvent accessibility at the ribose backbone. Therefore, observation of nucleotides that are protected from hydroxyl radical-mediated cleavage can be taken as evidence of higher-order contacts [5].
Alternatively, if the tertiary structure of an RNA is known from high-resolution NMR or crystallography analysis, then comparison of the known structure and the observed reactivity pattern using a chemical mapping reagent can be used to infer whether a given tertiary interaction is formed in distinct RNA states or upon introducing mutations to the sequence. A number of recent studies have elegantly melded knowledge of higher-order structure with chemical mapping to follow folding pathways [38], monitor ligand binding [39,40], measure the thermodynamics and thermal stability of individual RNA structural elements [8,41], assess the effect of RNA mutations [42,43], evaluate the structure of a small RNA in complex with its biological partners [44], and explore RNA contacts within a crystal lattice [45].
A current challenge is to use chemical mapping information for de novo refinement of three-dimensional structures of RNA, especially for structures containing extensive and nontrivial long-range tertiary interactions. This kind of analysis requires knowledge of through-space connectivity information (Fig. 1D). Like single-site chemical modification, the use of chemical approaches to obtain through-space information for RNA is well-established. Classic methods include UV and chemical cross-linking and analysis of cleavage originating from a site-specific cleavage group. UV and chemical cross-linking experiments can, in principle, give a large amount of through-space connectivity information but tend to be very difficult to analyze; in contrast, RNAs containing site-directed cleavage groups tend to be laborious to create but analysis of the resulting through-space information is easier.
The ideal chemical mapping strategy for generating useful through-space connectivity information should (i) involve a simple approach for performing the initial chemical analysis, (ii) produce a large number of high-quality, short distance, through-space constraints, (iii) be straightforward to interpret, and (iv) be applicable to RNAs of arbitrary size and complexity. Although no current method fully meets all of these criteria, substantial progress has been made in developing broadly useful approaches for obtaining through-space information for RNA structure.
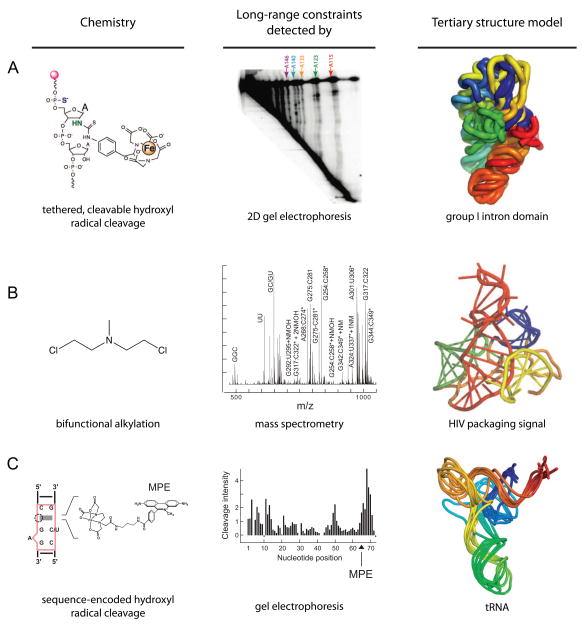
In multiplexed hydroxyl radical cleavage analysis (MOHCA) [46], RNAs are created in which nucleotides are randomly incorporated such that the probe nucleotide is ultimately linked to two reporter chemistries. An Fe(II)-EDTA moiety is tethered at the 2′-ribose position and is used to induce through-space cleavage of nearby (within ~25 Å) residues in the RNA. The site from which cleavage was initiated is identified by site-specific cleavage of the probe nucleotide using iodine-mediate cleavage of a 5′ phosphorothioate label (Fig. 5A). Sites of Fe(II)-EDTA mediated cleavage and the location of the probe nucleotide are identified by information-rich two-dimensional gel electrophoresis. This approach is straightforward to set up and yields a large number of high-quality, medium-distance constraints. The two-dimensional gel electrophoresis approach can be challenging to implement, especially as RNA size increases. MOHCA was used both to refine a model for the structure of the Tetrahymena group I intron P546 domain and to develop a model for a folding intermediate of this domain (Fig. 5A) [46].
Figure 5.
Recent developments using chemical probing to obtain through-space constraints on RNA tertiary structure. (A) MOHCA, (B) MS3D, and (C) sequence-encoded cleavage. Figures are adapted from images in [46,48,49].
The information-rich, but complex, cross-linking patterns obtained using conventional bifunctional cross-linking reagents can be read out by mass spectrometry in an approach termed MS3D [47,48]. Nitrogen mustard [bis(2-chloroethyl)methylamine] reacts preferentially at the N7 position of guanosine and N3 of adenosine and contains a relatively short spacer group (~9 Å, Fig. 5B). The RNA is digested with ribonuclease and the nucleotides that formed cross-links are detected by mass analysis and tandem sequencing by mass spectrometry. The initial cross-linking experiment is very easy to implement and MS3D yields a large number of short distance contacts. The final analysis of pairs of cross-linked nucleotides requires sophisticated instrumentation and considerable expertise. MS3D has been used to refine models for a mouse mammary tumor virus pseudoknot [47] and for an RNA involved in HIV-1 packaging [48] (Fig. 5B).
Site-directed cleavage can be accomplished by linking an Fe(II)-EDTA group to a molecule that binds selectively to a user-defined site in an RNA [49]. For example, MPE (methidiumpropyl-EDTA) binds selectively to a simple bulged RNA element [49,50]. An MPE binding site, and thus a tethered Fe(II)-EDTA group, can be placed at many sites by simply mutating the sequence of an RNA helix (Fig. 5C). Sequence-encoded, site-directed cleavage yields a large number of medium distance (25–35 Å) constraints that can be analyzed in a simple one-dimensional gel electrophoresis experiment. Site-directed cleavage requires that a new mutant RNA be created for each experiment and that careful controls be performed to ensure that cleavage agent binding and sequence changes do not disrupt the native RNA structure. Sequence-encoded cleavage was used to refine the structure of tRNAAsp to roughly nucleotide resolution (4–6 Å RMSD) [49] (Fig. 5C).
To create useful three-dimensional models of an RNA, through-space information must be interpreted via a constraint-satisfaction algorithm. This “algorithm” can be computer-assisted manual modeling or can make use of computationally based automated methods. Recent approaches for refining three-dimensional RNA structures include adapting all-atom algorithms commonly employed for crystallographic and NMR refinements [48,51] and use of simplified approaches in which each RNA nucleotide is represented as a single sphere [52,53] or as three beads (corresponding to the phosphate, ribose and-based groups) [49,54].
Advance 6 – Recent Ambitious Experiments
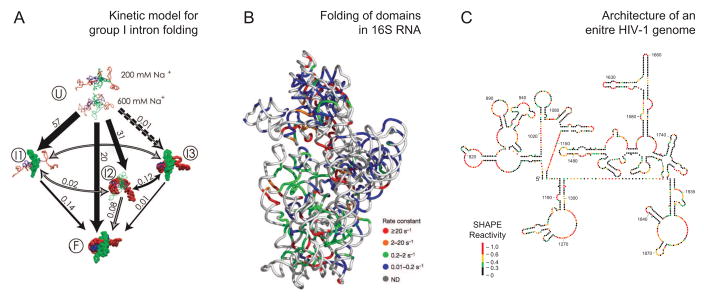
The recent and ongoing confluence of advances in mapping chemistries, data analysis, and interest in understanding complex RNAs and RNPs have made it possible to address structure-function relationships in remarkably large systems. In some ways, new trends in RNA structure analysis contain an element of going back to the future. Among the first applications of chemical probing were analyses of the structure of E. coli 16S RNA in a free state and in intact 30S subunits [3]. Representative recent ambitious studies have returned to analysis of 16S RNA [34] and have tackled the group I intron [31] and an entire HIV-1 genome [20,55]. A current theme is that chemical mapping approaches may be entering a new era in which novel and biologically important inferences can be obtained, especially when large, nucleotide resolution, and complete data sets are obtained.
Time-resolved fast Fenton hydroxyl radical cleavage was used to obtain a global analysis of the folding of a group I ribozyme along a time trajectory from very early to late steps and to characterize nearly all transitions between the initial, intermediate, and final conformational states [31]. The results of this analysis emphasize the importance of early events in determining the flux through distinct folding pathways (Fig. 6A).
Figure 6.
Representative examples of the use of chemical probing technologies to address ambitious problems in biology. Adapted from [31,34,55].
Synchrotron-mediated, time-resolved hydroxyl radical footprinting was used to follow assembly of the E. coli 16S ribosomal RNA (1542 nts) with all 30 small subunit proteins [34]. Observed kinetic profiles were used to infer RNA-RNA and RNA-protein interactions over time domains spanning four orders of magnitude. Some RNA nucleotides showed protection from hydroxyl radical cleavage in less than 20 ms whereas others were not protected until 1–3 minutes later. These data were interpreted in terms of a “chaotic” model for small subunit assembly involving concurrent nucleation events at many positions along the RNA, local refolding during assembly, and rapid appearance of native interactions throughout the RNA (Fig. 6B).
High-throughput SHAPE chemistry was used to analyze the overall architecture and develop a secondary structure model for an entire HIV-1 RNA genome (~9200 nts) using authentic viral RNA [55]. The SHAPE data support a model in which the genome contains more than 20 highly structured domains. Evolutionary comparisons and direct functional analysis indicate that many of these domains play important, and previously unrecognized, functions in viral replication (Fig. 6C).
Perspective
Chemical probing approaches for analyzing RNA structure are undergoing a renaissance in usefulness and impact. For many RNAs, including large structurally dynamic RNAs and conformational and functional intermediates, chemical mapping represents the best approach for obtaining structure information. In designing chemical probing experiments, researchers should consider using a reagent that reacts broadly with all four nucleotides as a first choice. The mechanical and experimental elements of chemical probing experiments are becoming easier: it is now possible to analyze RNAs that are sufficiently long to fully represent most systems of interest. One should try to obtain sufficient equilibrium or kinetic data such that a particular system can be understood as completely as possible. At the present, de novo structure prediction of RNA secondary and tertiary structure is fraught with challenges. However, melding experimental information with computational approaches for structure analysis will likely make possible sophisticated understanding of the numerous roles of RNA structure in all areas of biology.
Acknowledgments
Many important contributions and insightful reviews, especially those published prior to 2005, could not be included due to space constraints. I am indebted to the colleagues who allowed images from their primary research work to be included in this review. The US National Institutes of Health and National Science Foundation support work in my laboratory focused on creating probing technologies for understanding the structural and functional biology of RNA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Peattie DA, Gilbert W. Chemical Probes for Higher-Order Structure in RNA. Proc Natl Acad Sci USA. 1980;77:4679–4682. doi: 10.1073/pnas.77.8.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ehresmann C, Baudin F, Mougel M, Romby P, Ebel J-P, Ehresmann B. Probing the Structure of RNAs in Solution. Nucleic Acids Res. 1987;15:9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stern S, Moazed D, Noller HF. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- 4.Lavery R, Pullman A. A New Theoretical Index of Biochemical Reactivity Combining Steric and Electrostatic Factors. Biophys Chem. 1984;19:171–181. doi: 10.1016/0301-4622(84)85017-6. [DOI] [PubMed] [Google Scholar]
- 5.Tullius TD, Greenbaum JA. Mapping nucleic acid structure by hydroxyl radical cleavage. Curr Opin Struct Biol. 2005;9:127–134. doi: 10.1016/j.cbpa.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 6.Regulski EE, Breaker RR. In-line probing analysis of riboswitches. Methods Mol Biol. 2008;419:53–67. doi: 10.1007/978-1-59745-033-1_4. [DOI] [PubMed] [Google Scholar]
- 7.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by Selective 2′-Hydroxyl Acylation and Primer Extension (SHAPE) J Am Chem Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 8.Wilkinson KA, Merino EJ, Weeks KM. RNA SHAPE chemistry reveals nonhierarchical interactions dominate equilibrium structural transition in tRNAAsp transcripts. J Am ChemSoc. 2005;127:4659–4667. doi: 10.1021/ja0436749. [DOI] [PubMed] [Google Scholar]
- 9.Mortimer SA, Weeks KM. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J Am Chem Soc. 2007;129:4144–4145. doi: 10.1021/ja0704028. [DOI] [PubMed] [Google Scholar]
- 10.Wilkinson KA, Merino EJ, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nature Protocols. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 11.Mortimer SA, Weeks KM. Time-resolved RNA SHAPE chemistry. J Am Chem Soc. 2008;130:16178–16180. doi: 10.1021/ja8061216. [DOI] [PubMed] [Google Scholar]
- 12.Wilkinson KA, Vasa SM, Deigan KE, Mortimer SA, Giddings MC, Weeks KM. Influence of nucleotide identity on ribose 2′-hydroxyl reactivity in RNA. RNA. 2009;15:1314–1321. doi: 10.1261/rna.1536209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gherghe CM, Shajani Z, Wilkinson KA, Varani G, Weeks KM. Strong correlation between SHAPE chemistry and the generalized NMR order parameter (S2) in RNA. 2008;130:12244–12245. doi: 10.1021/ja804541s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z, Treder K, Miller WA. Structure of a viral cap-independent translation element that functions via high affinity binding to the eIF4E subunit of eIF4F. J Biol Chem. 2009;284:14189–14202. doi: 10.1074/jbc.M808841200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daou-Chabo R, Condon C. RNase J1 endonuclease activity as a probe of RNA secondary structure. RNA. 2009;15:1417–1425. doi: 10.1261/rna.1574309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16 •.Das R, Laederach A, Pearlman SM, Herschlag D, Altman RB. SAFA: Semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA. 2005;11:344–354. doi: 10.1261/rna.7214405. Chemical probing technologies become much more facile and attractive with the introduction of computationally facilitated data analysis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17 •.Mitra S, Shcherbakova IV, Altman RB, Brenowitz M, Laederach A. High-throughput single-nucleotide structural mapping by capillary automated footprinting analysis. Nucleic Acids Res. 2008;36:e63. doi: 10.1093/nar/gkn267. See annotation for ref. 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18 •.Vasa SM, Guex N, Wilkinson KA, Weeks KM, Giddings MC. ShapeFinder: A software system for high-throughput quantitative analysis of nucleic acid reactivity information resolved by capillary electrophoresis. RNA. 2008;14:1979–1990. doi: 10.1261/rna.1166808. See annotation for ref. 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shcherbakova I, Brenowitz M. Monitoring structural changes in nucleic acids with single residue spatial and millisecond time resolution by quantitative hydroxyl radical footprinting. Nature Protocols. 2008;3:288–302. doi: 10.1038/nprot.2007.533. [DOI] [PubMed] [Google Scholar]
- 20.Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biology. 2008;6:e96. doi: 10.1371/journal.pbio.0060096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duncan CD, Weeks KM. SHAPE analysis of long-range interactions reveals extensive and thermodynamically preferred misfolding in a fragile group I intron RNA. Biochemistry. 2008;47:8504–8513. doi: 10.1021/bi800207b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duncan CDS, Weeks KM. The Mrs1 splicing factor binds the bI3 group I intron at each of two tetraloop-receptor motifs. PloS ONE. 2010;5:e8983. doi: 10.1371/journal.pone.0008983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mathews DH, Turner DH. Prediction of RNA secondary structure by free energy minimization. Curr Opin Struct Biol. 2006;16:270–278. doi: 10.1016/j.sbi.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 24.Shapiro BA, Yingling YG, Kasprzak W, Bindewald E. Bridging the gap in RNA structure prediction. Curr Opin Chem Biol. 2007;17:157–165. doi: 10.1016/j.sbi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 25 ••.Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc Natl Acad Sci USA. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. A major advance in RNA secondary structure prediction algorithms showing that prediction accuracies improve significantly with the rigorous inclusion of experimental information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mortimer SA, Johnson JS, Weeks KM. Quantitative analysis of RNA solvent accessibility by N-silylation of guanosine. Biochemistry. 2009;48:2109–2114. doi: 10.1021/bi801939g. [DOI] [PubMed] [Google Scholar]
- 27 ••.Deigan KE, Li TW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA structure determination. Proc Natl Acad Sci USA. 2009;106:97–102. doi: 10.1073/pnas.0806929106. Experimentally constrained RNA secondary structure prediction at up to 95–100% accuracy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sclavi B, Woodson S, Sullivan M, Chance M, Brenowitz M. Following the folding of RNA with time-resolved synchrotron X-ray footprinting. Methods Enzymol. 1998;295:379–402. doi: 10.1016/s0076-6879(98)95050-9. [DOI] [PubMed] [Google Scholar]
- 29.Shcherbakova I, Mitra S, Beer RH, Brenowitz M. Fast Fenton footprinting: a laboratory-based method for the time-resolved analysis of DNA, RNA and proteins. Nucleic Acids Res. 2006;34:e48. doi: 10.1093/nar/gkl055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brooks KM, Hampel KJ. A rate-limiting conformational step in the catalytic pathway of the glmS ribozyme. Biochemistry. 2009;48:5669–5678. doi: 10.1021/bi900183r. [DOI] [PubMed] [Google Scholar]
- 31 ••.Laederach A, Shcherbakova I, Jonikas MA, Altman RB, Brenowitz M. Distinct contribution of electrostatics, initial conformational ensemble, and macromolecular stability in RNA folding. Proc Natl Acad Sci USA. 2007;104:7045–7050. doi: 10.1073/pnas.0608765104. These three papers (see also refs. 34 & 55) likely represent an important beginning in application of chemical probing technologies to address important, large-scale, and multidimensional problems in RNA biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Russell R, Tijerina P, Chadee AB, Bhaskaran H. Deletion of the P5abc peripheral element accelerates early and late folding steps of the Tetrahymena group I ribozyme. Biochemistry. 2007;46:4951–4961. doi: 10.1021/bi0620149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nguyenle T, Laurberg M, Brenowitz M, Noller HF. Following the dynamics of changes in solvent accessibility of 16 S and 23 S rRNA during ribosomal subunit association using synchrotron-generated hydroxyl radicals. J Mol Biol. 2006;359:1235–1248. doi: 10.1016/j.jmb.2006.04.030. [DOI] [PubMed] [Google Scholar]
- 34 ••.Adilakshmi T, Bellur DL, Woodson SA. Concurrent nucleation of 16S folding and induced fit in 30S ribosome assembly. Nature. 2008;455:1268–1272. doi: 10.1038/nature07298. See annotation for ref. 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gherghe CM, Mortimer SA, Krahn JM, Thompson NL, Weeks KM. Slow conformational dynamics at C2′-endo nucleotides in RNA. J Am Chem Soc. 2008;130:8884–8885. doi: 10.1021/ja802691e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mortimer SA, Weeks KM. Time-resolved RNA SHAPE chemistry: quantitative RNA structure analysis in one-second snapshots and at single-nucleotide resolution. Nature Protoc. 2009;4:1413–1421. doi: 10.1038/nprot.2009.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mortimer SA, Weeks KM. C2′-endo nucleotides as molecular timers suggested by the folding of an RNA domain. Proc Natl Acad Sci USA. 2009;106:15622–15627. doi: 10.1073/pnas.0901319106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dann C, Wakeman C, Sieling C, Baker S, Irnov I, Winkler W. Structure and Mechanism of a Metal-Sensing Regulatory RNA. Cell. 2007:878–892. doi: 10.1016/j.cell.2007.06.051. [DOI] [PubMed] [Google Scholar]
- 39.Gilbert SD, Rambo RP, Tyne DV, Batey RT. Structure of the SAM-II riboswitch bound to S-adenosylmethionine. Nature Struct Mol Biol. 2008;15:177–182. doi: 10.1038/nsmb.1371. [DOI] [PubMed] [Google Scholar]
- 40.Wang B, Wilkinson KA, Weeks KM. Complex ligand-induced conformational changes in tRNAAsp revealed by single nucleotide resolution SHAPE chemistry. Biochemistry. 2008;47:3454–3461. doi: 10.1021/bi702372x. [DOI] [PubMed] [Google Scholar]
- 41.Stoddard CD, Gilbert SD, Batey RT. Ligand-dependent folding of the three-way junction in the purine riboswitch. RNA. 2008;14:675–684. doi: 10.1261/rna.736908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Y, Fender J, Legassie JD, Jarstfer MB, Bryan TM, Varani G. Structure of stem-loop IV of Tetrahymena telomerase RNA. EMBO J. 2006;25:3156–3166. doi: 10.1038/sj.emboj.7601195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Legiewicz M, Badorrek CS, Turner KB, Fabris D, Hamm TE, Rekosh D, Hammarskjöld ML, Le Grice SF. Resistance to RevM10 inhibition reflects a conformational switch in the HIV-1 Rev response element. Proc Natl Acad Sci USA. 2008;105:14365–14370. doi: 10.1073/pnas.0804461105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Costantino DA, Pfingsten JS, Rambo RP, Kieft JS. tRNA-mRNA mimicry drives translation initiation from a viral IRES. Nature Struct Mol Biol. 2008;15:57–64. doi: 10.1038/nsmb1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vicens Q, Gooding AR, Laederach A, Cech TR. Local RNA structural changes induced by crystallization are revealed by SHAPE. RNA. 2007;13:536–548. doi: 10.1261/rna.400207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46 •.Das R, Kudaravalli M, Jonikas MA, Laederach A, Fong R, Schwans JP, Baker D, Piccirilli JA, Altman RB, Herschlag D. Structural inference of native and partially folded RNA by high-throughput contact mapping. Proc Natl Acad Sci USA. 2008;105:4144–4149. doi: 10.1073/pnas.0709032105. Three examples (see also refs. 48 & 49) of advances using chemical probing technologies to obtain through-space information for RNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu ET, Zhang Q, Fabris D. Untying the FIV frameshifting pseudoknot structure by MS3D. J Mol Biol. 2005;345:69–80. doi: 10.1016/j.jmb.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 48 •.Yu ET, Hawkins A, Eaton J, Fabris D. MS3D structural elucidation of the HIV-1 packaging signal. Proc Natl Acad Sci USA. 2008;105:12248–12253. doi: 10.1073/pnas.0800509105. See annotation for ref. 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49 •.Gherghe CM, Leonard CW, Ding F, Dokholyan NV, Weeks KM. Native-like RNA tertiary structures using a sequence-encoded cleavage agent and refinement by discrete molecular dynamics. J Am Chem Soc. 2009;131:2541–2546. doi: 10.1021/ja805460e. See annotation for ref. 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gooch BD, Krishnamurthy M, Shadid M, Beal PA. Binding of helix-threading peptides to E. coli 16S ribosomal RNA and inhibition of the S15-16S complex. ChemBioChem. 2005;6:2247–2254. doi: 10.1002/cbic.200500285. [DOI] [PubMed] [Google Scholar]
- 51.Badorrek CS, Gherghe CM, Weeks KM. Structure of an RNA switch that enforces stringent retroviral genomic RNA dimerization. Proc Natl Acad Sci USA. 2006;103:13640–13645. doi: 10.1073/pnas.0606156103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tan RKZ, Petrov AS, Harvey SC. YUP: A molecular simulation program for coarse-grained and multiscaled models. J Chem Theory Comput. 2006;2:529–540. doi: 10.1021/ct050323r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jonikas MA, Radmer RJ, Laederach A, Das R, Pearlman S, Herschlag D, Altman RB. Coarse-grained modeling of large RNA molecules with knowledge-based potentials and structural filters. RNA. 2009;15:189–199. doi: 10.1261/rna.1270809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ding F, Sharma S, Chalasani V, Demidov V, Broude NE, Dokholyan NV. Ab initio RNA folding by discrete molecular dynamics: from structure prediction to folding mechanisms. RNA. 2008;14:1164–1173. doi: 10.1261/rna.894608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55 ••.Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Swanstrom R, Burch CL, Weeks KM. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–716. doi: 10.1038/nature08237. See annotation for ref. 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Laederach A, Das R, Vicens Q, Pearlman SM, Brenowitz M, Herschlag D, Altman RB. Semiautomated and rapid quantification of nucleic acid footprinting and structure mapping experiments. Nature Protoc. 2008;3:1395–1401. doi: 10.1038/nprot.2008.134. [DOI] [PMC free article] [PubMed] [Google Scholar]