Abstract
The mussel Mytilus trossulus can develop a neoplasia of the haemolymph, which occurs with high frequency (up to 40%) in nature. Associated with this disease are pro-apoptotic tumor suppressor protein p53 isoforms, which are highly conserved between molluscs and vertebrates. The vertebrate wildtype p53 protein is maintained at low levels by the MDM2 protein in non-stressed cells to prevent undesired apoptosis. Identification of a putative invertebrate MDM-like homolog suggests early evolution of this mechanism of p53 regulation. The M. trossulus MDM homolog consists of a conserved NH2-terminal p53 binding domain, an acidic domain with highly conserved phosphorylation sites, and a highly conserved C-terminal RING-finger Zn-binding domain. Although BLAST queries predict this homologue to be more similar to vertebrate MDM2 than to MDM4, phylogenetic analysis suggests that it may be an ancestral form to both vertebrate MDM genes. Using yeast-two-hybrid assays and pull-down assays, we show that this molluscan MDM is able to bind to its p53 counterpart. We also show that MDM expression levels are directly correlated with p53 expression levels in healthy and in neoplastic haemocytes, but not with other p53 isoforms or with the proto-oncogene RAS. The combination of expression levels of five gene transcripts (p53, mdm, ras, ΔNp63/73, TAp63/73) is significantly correlated with late-stage haemic neoplasia in M. trossulus.
Keywords: haemic neoplasia, bivalve, Mytilus, cancer, p53, mdm2, ras
1. Introduction
Haemic neoplasia is a fatal leukemia-like proliferative disorder of the invertebrate haemolymph, which occurs in a number of molluscan species (Barber 2004). Populations of the bay mussel, Mytilus trossulus, can exhibit a disease prevalence of over 40 % in some locations on the northwest coast of North America (Bower 2006) and thus provide us with a naturally occurring out-breeding model of a neoplastic disorder. The investigation of molecular mechanisms in molluscan haemic neoplasia illuminates ancestral cell growth regulation pathways, which may underlie regulatory pathways that are affected in some human cancers (Walker et al. 2008). One such regulatory pathway that has been implicated in molluscan neoplasia is p53-mediated cell apoptosis in response to DNA damage (Böttger 2008).
The tumor suppressor protein p53 inhibits tumor formation in mammals by initiating apoptosis or cell cycle arrest pathways in response to DNA damage (Murray-Zmijewski et al. 2006). Its sister proteins p63 and p73 have important functions in embryonic development (Yang et al. 2002). Homologous proteins have been isolated in a number of invertebrates (Schmale et al. 1997; Kelley et al. 2001; Cox et al. 2003; Muttray et al. 2005; Goodson et al. 2006; Muttray et al. 2007); functional domains exhibit a high degree of similarity to their vertebrate homologues. We have shown previously that p53 mRNA and an N-terminal truncated isoform, ΔNp63/73, were expressed during late-stage haemic neoplasia in M. trossulus at levels higher than those seen in normal cells (Muttray et al. 2008).
Mammalian p53, p63 and p73 induce expression of MDM2, a negative regulator of the p53 family proteins (reviewed in Levrero et al. 2000; Harms et al. 2004). MDM2 represses p53 and p73 activity by binding to their N-termini, obstructing the transactivation domain and hence the ability of p53/p73 to transactivate downstream target genes. In addition, MDM2 functions as a p53-specific ubiquitin ligase, targeting the C-terminus and destining p53 for degradation by the proteasome. Although p63 activates MDM2, MDM2 activity on p63 remains controversial (Calabro 2002, Marine 2005). While homologous p53 family members have been characterized in different invertebrates, no MDM2 has been isolated to date in classical invertebrate models, such as Drosophila melanogaster or Caenorhabditis elegans. Given that molluscan p53 contains many of the functional domains found in vertebrate p53, including the MDM2 binding region, we hypothesized that a molluscan MDM-homologue likely exists and is able to bind to p53.
The proto-oncogene RAS was isolated in M. trossulus (Ciocan et al. 2006) and also may play a role in haemic neoplasia in bivalves. RAS is a signal transduction protein and proto-oncogene, which is often mutated in human cancers (Friday et al. 2005). An alternative splice variant, p19ras, can stimulate p73-dependent transcriptional activation of target genes (such as p21) through a mechanism involving MDM2 (Jeong 2006). In mussels, RAS was found to have a higher number of silent polymorphic variations and potentially elevated expression levels (estimated by band intensity after PCR amplification) in neoplastic haemolymph samples than in normal samples (Ciocan et al. 2006). We therefore wanted to investigate if significant correlations existed at the mRNA level between the molluscan p53 family members, MDM and RAS.
In this work, a mdm-like sequence was cloned from the bivalve, M. trossulus, and was found to be phylogenetically related to other known mdm2 and mdm4 homologues. Results from yeast-two-hybrid assays, confirmed by pull-down assays, indicated that this molluscan MDM homologue interacts with molluscan p53. Furthermore, expression of mdm correlated with that of p53, both of which are over-expressed in the disease haemic neoplasia. This was in contrast to ras, which was under-expressed. This supports the hypothesis that MDM-mediated regulation of p53 evolved in invertebrates prior to the evolutionary divergence of protostomes and deuterostomes, which opens the door to further studies of p53 regulation in the molluscan cancer models.
2. Materials and Methods
2.1. Mussel collection
M. trossulus of size 40–60 mm were collected from the intertidal area at Hopkins Landing, Sunshine Coast, British Columbia, in February 2007. Mussels were transferred to cages submerged at three locations: Two on the north shore of Burrard Inlet, West Vancouver, and one near Hopkins Landing, BC. They were sampled in October of 2007, after seven months of continuous submersion. Mussels were transported to the laboratory and kept at ambient temperature (11 °C) in seawater drawn from the same locations and depths of the cages at their previous habitat, with frequent seawater changes. Processing of mussel samples began on the same day and continued, if necessary, the following day. All animals were processed in accordance with the Department of Fisheries and Oceans License Authorizing Fish Collection for Scientific Purposes 06.18 and the University of British Columbia Animal Care Protocol A05-0057-R001.
2.2 Cloning and sequence analysis of putative M. trossulus Mdm-like cDNA
cDNA was synthesized from total RNA extracted from M. trossulus mantle and gill tissue (Omega Bio-tek, Doraville, GA) using SuperScript™ III Reverse Transcriptase (Invitrogen, Burlington, ON) and an oligo-dT(12–18) primer following the manufacturer’s instructions. An initial 607 bp fragment was amplified by PCR from mantle cDNA with primers designed to a conserved region in a putative mdm2 EST sequence from M. californianus (GenBank accession number ES394701), kindly provided by Andrew Gracey (University of Southern California). The primers were as follows (all 5’ to 3’): mdm2-F2, ATGGACGTGCACAGAATGT, and mdm2-R3, ATGTCCAGTGCAGCCATG. Cloning was performed using the TOPO TA Ligation kit (Invitrogen). Sequencing was performed by the Nucleic Acid and Protein Service Center at the University of British Columbia.
Nested 5’ and 3’ RACE primers were designed based on the initial sequence and used to amplify the 5’ and 3’ ends of RACE-ready cDNA prepared previously from M. trossulus gill RNA (BD Biosciences Clontech, Mississauga, ON). RACE-primer sequences were as follows: mdm-5’RACE-2, TGTAGTCTCTGGTAACCAGTTTGGACGTACT; mdm-5’RACE-1(nested), ACAATGTCCGCAGTGACCATGTAAAGGAG; mdm-3’RACE-1, CTCGGTCAGTGCTGTATTTGTTTCAGCCGGCC; and mdm-3’RACE-2(nested), CTGCCAGCATTATTCATGGCTGCACTGGAC. Nested 5’RACE yielded one band which was cloned and sequenced. Nested 3’RACE yielded two bands (1.2 and 1.4 kb), of which only the 1.4 kb could be cloned successfully. The nested RACE reactions were repeated and four more clones were obtained and sequenced. All sequences were assembled using the EMBOSS Merger program (Rice et al. 2000).
The complete mdm-like sequence was amplified from mantle cDNA, cloned and sequenced using primers at the 5’ start codon, mdm2-F4, ATGTGCAATAATCCAAATCTAATTGT, and just upstream of the polyA-tail, mdm2-R6, ATTGATGAGTGTACATATATCTGTGC. A total of five clones were sequenced and the consensus was submitted to GenBank, accession number HM004082. The putative protein sequence was submitted to Discontiguous Mega BLAST searches as well as to Conserved Domain searches (Marchler-Bauer et al. 2007). Pair-wise amino acid alignment of the deduced M. trossulus MDM-like protein sequence was done with selected vertebrate and invertebrate species using ClustalX 1.83 (residue substitution matrix Gonnet; Benner et al. 1994) to illustrate conserved domains (penalties: gap opening 10, gap extension 0.1). The alignment was edited to match highly conserved regions of the protein. A second pair-wise multiple alignment with a wider selection of vertebrate MDM2 and MDM4 sequences was performed to identify the MDM gene that we isolated from M. trossulus (Table 1). The unrooted phylogenetic tree was produced in ClustalX, bootstrapped 1000 times and displayed using TreeView32 (Page 1996). Percent protein sequence identities were obtained in ClustalX.
Table 1.
List of species, phylogenetic taxa, MDM protein type, and accession numbers used for phylogenetic analysis of MDM proteins in Figures 1 and 2.
| Lottia gigantea | Owl limpet | Invertebrate, Mollusc |
predicted protein similar to MDM |
http://genome.jgi-psf.org/ Lottia gigantean v1.0 database jgi|Lotgi1|161929|fgenesh2_pg.C_sca_31000171 |
| Mytilus trossulus | Bay mussel | Invertebrate, Mollusc |
MDM | HM004082 |
|
Strongylocentrotus purpuratus |
Purple sea urchin |
Invertebrate, Echinodermata |
predicted, similar to MDM2 |
XM_001188537 |
|
Nematostella vectensis |
Sea anemone | Invertebrate, Cnidaria |
predicted protein |
XM_001637981 |
2.3 Construction of yeast expression plasmids and yeast-two-hybrid assay
Mating-compatible yeast strains, EGY 40 and EGY 188, and expression plasmids, pEG202 and pJG4-5, were used in a yeast-two-hybrid assay to identify protein/protein interactions. All yeast strains and vectors were obtained from Origene Technologies (Rockville, MD). The M. trossulus mdm coding sequence was cloned into the pEG202 bait plasmid; the M. trossulus p53 coding sequence was inserted into the target plasmid pJG4-5. The mdm and p53 sequences were prepared for insertion by attaching linker fragments using PAGE-purified primers: pGE202Mdm2-F, GAATTCCCGGGGATCCGTCGACCAATGTGCAATAATCCAAATCTAATTGTACAA; pGE202Mdm2-R, AGGTCGACTCGAGCGGCCGCCATTAAGCAAAATAGTTTTTAATGATCTTTTGAATAGG; pFG4-5p53-F, ATGATGTGCCAGATTATGCCTCTCCCATGTCACAAGCTTCAGTTTCAACTACAT; pFG4-5p53-R, AAAGCTTCTCGAGTCGGCCGAATTCTCAAGTTCTTGATTTGATTCCTGGTCGA. PCR cycling was performed using iProof™ High-Fidelity DNA polymerase reaction mix (Bio-Rad, Hercules, CA) with a gradient of annealing temperatures between 60 and 72°C. Products were gel purified, quantified and cloned into the respective plasmid using the QuikChange® Lightning Site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to manufacturer’s instructions. Cycling was performed as follows: 2 min 95°C, 18 cycles of: 20 sec 95°C, 10 sec 60°C, 6 min (mdm plasmid) or 4 min (p53 plasmid) 68°C, and 5 min 68°C final extension. Resulting plasmids were digested with DpnI and transformed into XL-10 Gold® ultracompetent cells (Stratagene). Colonies were grown on LB medium plus ampicillin and cultures were screened for presence of plasmids by agarose gel electrophoresis. Plasmids were purified and inserts confirmed by sequencing. Any errors were rectified by site-directed mutagenesis using the QuikChange® Lightning Site-directed mutagenesis kit (Stratagene). Plasmids were designated pJG4-5-p53 and pEG202-Mdm2.
To examine possible protein/protein interactions, pEG202-Mdm2 was transformed with the LacZ reporter plasmid, pRB1840 (URA3, 8LexAop-lacZ) into yeast strain EGY40 (Mat _, trp1, his3, ura3, leu2::0LexAop-LEU2), forming a constitutively expressing LexA DNA binding domain fusion protein (bait); pJG4-5-p53 was transformed into yeast strain EGY188 (Mat a trp1, his3, ura3, leu2::2LexAop-LEU2), forming a galactose-dependent B42 transactivation domain fusion protein (target). Transformations were based on the lithium acetate method (Golemis et al. 1998; Gietz et al. 1992; Schiestl, 1989). Bait and target strains were mated (Munger et al. 1989) to obtain yeast with pNLexA-/Mdm-2, pRB1840, and pJG4-5-p53. Plasmid expression was confirmed by western blotting.
Dilutions of mated cells were plated on nutrient selective plates to obtain clones containing all three plasmids (Golemis et al. 1998). Basic yeast plate media was composed of yeast nitrogen base (YNB; QBiogene), complete supplement mixture (CSM) lacking histidine, uracil, tryptophan, leucine (CSM-his-ura-trp-leu; QBiogene), a carbon source of either glucose or galactose/raffinose, and the addition of any of the aforementioned amino acids when required (further reference to YNB includes the addition of CSM, carbon source and absent amino acids). Five colonies were patched to YNB plates containing glucose, but lacking histidine, uracil, and tryptophan. Following sufficient growth, these were replica-plated to four plate types: X-gal-treated LacZ reporter plates consisting of YNB minus histidine, uracil, and tryptophan, with either (1) glucose or (2) galactose/raffinose as the carbon source, and LEU2 reporter plates consisting of YNB minus histidine, uracil, tryptophan and leucine, with either (3) glucose or (4) galactose/raffinose as the carbon source. LacZ reporter plates were observed for blue/white screening, and LEU2 reporter plates were observed for the presence or absence of growth. Positive controls were supplied by OriGene; interaction-compatible empty plasmids were employed as controls for background autoactivation.
Because of strong autoactivation of the mdm reporter plasmid on the LEU2 reporter plates, activation of the Leu2 reporter was measured by dilution plating. Mated yeast were plated on three types of selective media: (1) galactose-based media containing leucine (Gal + Leu); (2) galactose-based media minus leucine (Gal −Leu); and (3) glucose-based media minus leucine (Glu −Leu). Colony growth was monitored over 144 hours for activation of the Leu2 reporter, measured as growth in the absence of leucine.
2.4 Pull-down assays
M. trossulus p53 and Mdm cDNA sequences were cloned into pTnT expression vectors (Promega, Madison, WI, USA) modified to allow expression of target proteins with N-terminal His tags. Inserts were verified by sequencing (University of Maine DNA Sequencing Facility). Proteins were synthesized in vitro, following manufacturer’s protocol (TnT SP6 coupled Rabbit Reticulocyte Lysate Systems, Promega). All target proteins were synthesized with 35S-methionine (PerkinElmer, Waltham, MA, USA).
Interactions were performed in protein pairs: M. trossulus MDM as a bait protein, M. trossulus p53 as a target protein, and the reciprocal. Interacting proteins were captured using the MagZ Protein Purification System (Promega) as per manufacturer’s instructions: Briefly, in vitro transcribed/translated proteins were incubated together at room temperature for 1.5 hours. MagZ Binding Particles were prepared and combined with the sample. The mix was rotated for an additional 30 minutes at room temperature. Proteins were eluted off the beads using an elution buffer mix (4× NuPAGE LDS Sample Buffer, Invitrogen, 10× Sample Reducing Agent, Invitrogen, and MagZ elution buffer, Promega, 2.5:1:6.5) heated for four minutes at 80 °C. Sample elution was repeated once. Proteins were run on SDS-PAGE gradient gels (NuPAGE Novex 4–12% Bis-Tris, Invitrogen) with MOPS NuPAGE running buffer (Invitrogen) per manufacturer’s instructions. Following electrophoresis, gels were soaked in fixative (45% methanol, 10% acetic acid), followed by drying solution (10% methanol, 7% acetic acid, 1% glycerol) and dried under vacuum (1.5 hours at 80°C). Dried gels were placed on Hyperfilm (Amersham) for approximately 17 hours.
2.5 Sampling of haemolymph
Haemolymph was withdrawn from the posterior adductor muscle using a pre-chilled 3 mL, 21G syringe and screened by phase contrast microscopy for contamination by gametes, bacteria and particulates, and presence/absence of haemic neoplasia, as previously described (Barber 2004, Muttray et al. 2008). Absence of haemic neoplasia was defined by the occurrence of only normal haemocytes (approximately 106 cells per mL), which were predominately granular and adhesive with pseudopodia. Haemic neoplasia was defined by the presence of only round, non-adhesive cells with no or very few, short pseudopodia at high cell density (approximately 108 cells per mL). Early-stage neoplastic samples and samples contaminated with other particles were excluded from further analysis. Aliquots (0.2 mL) of haemocyte samples were spun down (425 g, 3 min, 4–6 °C), the supernatant removed, and cells stored at −80 °C until RNA extraction.
2.6 RNA extraction and cDNA synthesis
Total RNA was extracted from haemocytes and species identity was confirmed as previously described (Muttray et al. 2008). Briefly, total RNA was extracted using the E.Z.N.A. Total RNA Kit (Omega Bio-tek) and eluted with 40 µl of hot diethylpyrocarbonate (DEPC)- treated water. Species identity was confirmed by RFLP-PCR of the internal transcribed spacer (ITS) region of 18S rDNA using the HhAI enzyme (Invitrogen). RNA extracts were treated with DNaseI (Invitrogen, Burlington, ON, Canada) according to manufacturer’s instructions. RNA concentrations were measured in triplicate using a NanoDrop® ND-1000 UV-Vis Spectrophotometer (NanoDrop Technologies, Wilmington, DE). 1 µg of RNA was used for reverse transcription (SuperScript ™ II, Invitrogen) with random hexamers, according to manufacturer’s instructions. Resulting cDNAs were stored at −20 °C until analysis.
2.7 Real-time quantitative PCR
Real-time PCR was performed using the qPCR MasterMix Plus for SYBR® Green I (Eurogentec North America, Inc., San Diego, CA, USA) and the ABI Prism 7000 Real Time PCR System (Applied Biosystems, Foster City, CA, USA). Primers specific to M. trossulus p53, TAp63/73 and ΔNp63/73 isoforms were described previously (Muttray et al. 2008). Primers for the mdm and ras transcripts were designed with the assistance of the Primer ExpressTM software (Applied Biosystems) as follows (5’-3’ direction): mdm2-F, CCTTGGTAAGACCAAGACGTGA; mdm2-R, CCCATGATGCACCTACTTGTTT; ras18-F, TGTGGTTGTTGGAGCTGGTG; ras68-R, AGTTGGATGGTTAATGCACTTTTG. Real-time PCR reactions were performed in triplicate, as previously described (Muttray et al. 2008). A mix of randomly selected control samples (neoplastic and normal) were used to develop a standard curve for all primer sets thus ensuring that the amplification efficiencies were similar between standard and samples. All results were expressed relative to these standard curves. Data analysis was carried out using the ABI Prism 7000 SDS Software Version 1.0. Threshold and baseline were set to 0.20 and cycles 6 to 15, respectively, for all plates.
2.8 Statistical Analysis
All statistical analyses were conducted using JMP IN 5.1 (SAS Institute Inc., Cary, NC, USA). All data sets were tested for normality using the Shapiro-Wilk test. The non-parametric Wilcoxon rank sum test was used to compare the means of leukaemic samples with the means of normal samples for each isoform (with α = 0.01). Pearson’s correlation test was used to test for potential correlations between the transcription levels of the three isoforms in mussels belonging to either the normal or neoplastic population; p-values < 0.01 were declared significant. We performed a discriminant analysis in order to determine whether the mRNA levels were predictive of haemocyte health status.
3. Results
3.1 Analysis of the putative M. trossulus Mdm sequence
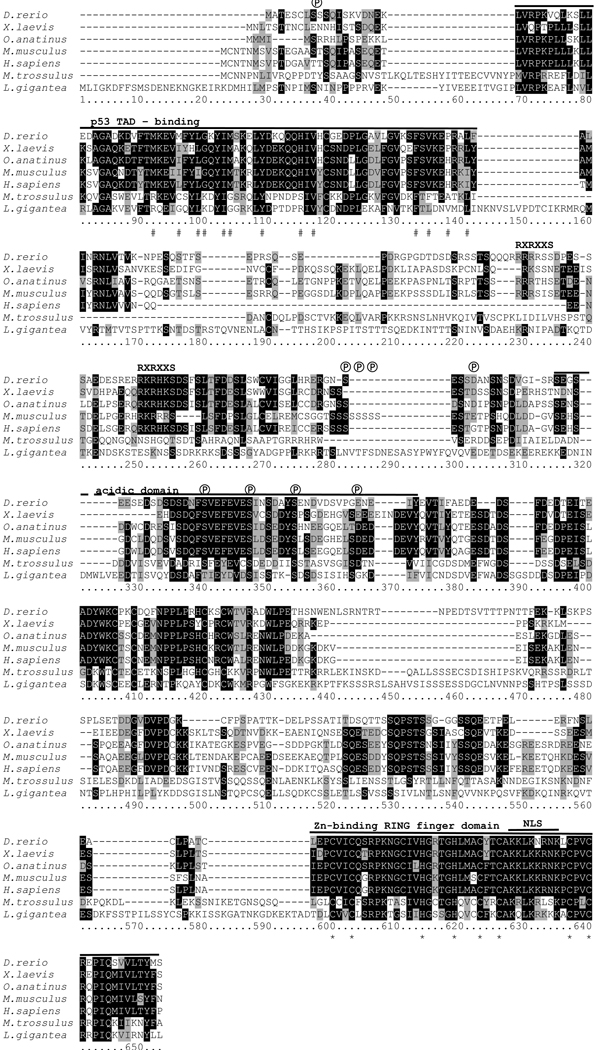
The consensus nucleic acid sequence for M. trossulus mdm was determined by RACE PCR and subsequently from five read-through clones, and was submitted to GenBank as HM004082. The total length of the cDNA is 2852 nt, with an open reading frame (ORF) of 1683 nt, predicting a protein of 561 amino acids with a molecular mass of 62.6 kDa, based on an average amino acid weight of 115 Da. This is longer than previously reported vertebrate MDM2 proteins, which are 12 to 56 kDa. The Mytilus MDM-like protein shows a second potential translational start site, 46 amino acids after the first methionine. This second start site aligns with N-termini of the p53-binding domain from vertebrate MDM2 (Figure 1).
Figure 1. Multiple pairwise alignment of predicted amino acid sequences of MDM-like homologues from M. trossulus, L. gigantea (predicted from genome sequence) and of predicted MDM2 sequences from representative vertebrate species.
Shading: white on black, identical residues; black on grey, similar residues; black on white, non-homologous residues. Abbreviations: #, hydrophobic amino acids identified as making contact with p53 in the p53 binding domain of MDM2 (Kussie et al. 1996); ℗, putative conserved phosphorylation sites. RXRXXS, consensus motif for association of protein kinases, such as AKT with vertebrate MDM2, not present in molluscan MDM; NLS, nuclear localization sequence; *, conserved amino acids required for E3 activity of MDM2 within the RING-finger domain. The solid black lines indicate conserved functional domains. GenBank accession numbers are provided in Table 1.
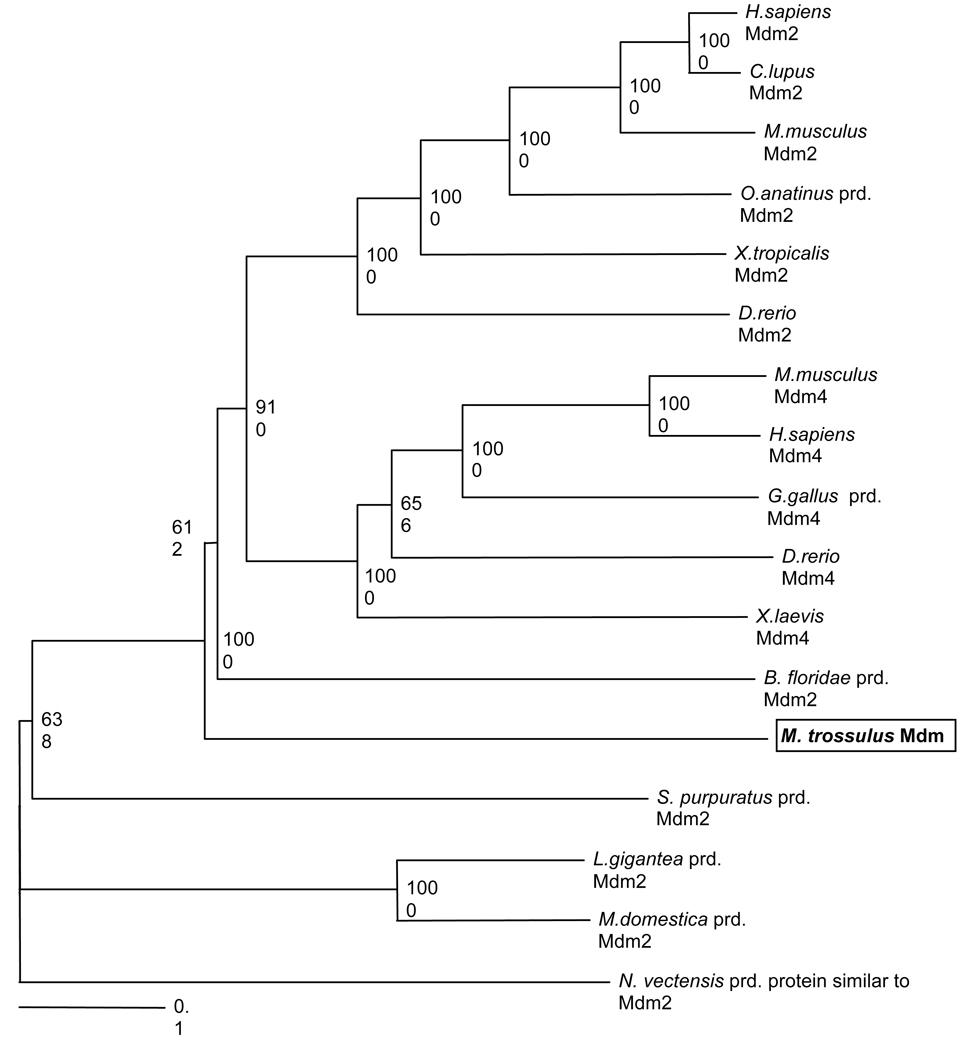
Comparative analyses among the putative Mytilus MDM, L. gigantea and representative vertebrate MDM2 are shown in Figures 1 and 2 and Table 1. Alignment with other MDM2 sequences showed strong similarities between the Mytilus MDM and vertebrate MDM2 within two conserved domains: the p53-binding domain near the N-terminus (E value of 5×10−8; NCBI Conserved domain database, Marchler-Bauer et al. 2007) and a Zn-binding RING-finger domain near the C-terminus (E value of 0.008). The p53-binding domain is characterized by 13 hydrophobic amino acids (Kussie et al. 1996; indicated by # in Figure 1), six of which are identical between Mytilus and the majority of vertebrate MDM2, while the remaining amino acids are similar in their hydrophobic nature. This domain forms a deep cleft, and interaction relies on van der Waales forces between the hydrophobic amino acids of MDM2 and p53. The RING-finger domain is characterized by a cross-brace motif consisting of eight highly characteristic amino acids (six cysteine, two histidine) responsible for zinc binding (Linke et al. 2008). All eight amino acids are present in M. trossulus MDM (indicated by * in Figure 1). The E3 ubiquitin ligase activity of MDM2 is dependent on the RING finger domain. The central acidic domain is less similar between vertebrate MDM2 and molluscan MDM, but contains four highly conserved phosphorylation sites (indicated by ℗ in Figure 1). Phosphorylation of the acidic domain contributes to interactions with various proteins, such as p300, ARF, and Rb in vertebrate MDM2 (Meek et al. 2003). The RXRXXS consensus motif, responsible for association between protein kinases, such as AKT, and vertebrate MDM2, is not present in molluscan MDM, suggesting that this signaling pathway is not used in these invertebrates. Overall percent sequence similarity between the Mytilus MDM and representative vertebrate MDM2 is 25–27 %, while similarities to representative MDM4 proteins is only 21–22% (ClustalX). However, the un-rooted neighborhood joining tree illustrates that the Mytilus MDM is phylogenetically more closely related to vertebrate MDM2 and MDM4 than to other predicted invertebrate MDM2 sequences (Figure 2). The Mytilus MDM may constitute an ancestral form of the vertebrate MDMs. Surprisingly, another (predicted) molluscan MDM2 protein from the snail L. gigantea clusters with a predicted MDM2 from M. domestica (short-tailed opossum, a marsupial) on a separate branch from other invertebrate MDM2. Neither protein is thus far supported by cDNA sequence data and it is likely that their protein sequence and hence location in the MDM2-MDM4 phylogenetic tree will be revised in future studies.
Figure 2. Phylogenetic relationship among MDM2 and MDM4 proteins of diverse species indicates that the Mytilus MDM protein may be an ancestral form of vertebrate MDM2 and MDM4.
The un-rooted neighbor-joined consensus tree was based on a pairwise ClustalX alignment, bootstrapped 1000 times. The bottom scale measures genetic distances in substitutions per nucleotide. Records labeled “prd” (predicted) were derived from published genomic sequences without supporting cDNA evidence and annotated using the gene prediction method GNOMON by NCBI. GenBank accession numbers are provided in Table 1.
3.2 p53-MDM protein/protein interactions
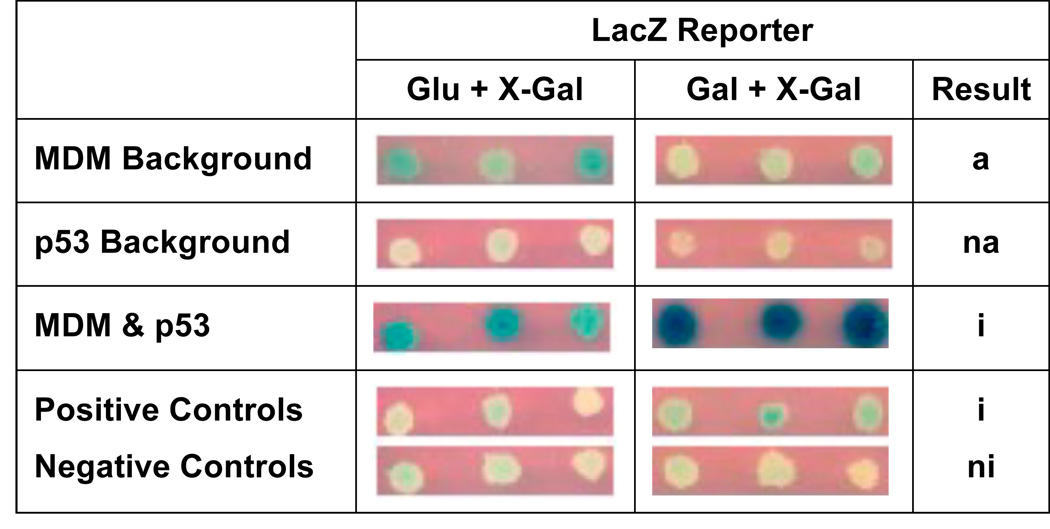
Interaction of Mytilus MDM with p53 was indicated by activation of the LacZ reporter in the yeast-two- hybrid assays on galactose-based media containing X-gal (Gal + X-Gal) (Figure 3). No β-galactosidase activity was expected on glucose-based media containing X-gal (Glu + X-Gal), since the pJG4-5 construct is under Gal1 promoter control. Positive controls (provided by Origene) acted as expected, with LacZ reporter being activated only slightly on Gal + X-gal, since a low-sensitivity reporter plasmid containing only one LexA operon (pRB1840) was used in this assay (Figure 3). Negative controls of mated yeast containing both the pJG4-5 and the pEG202 plasmids showed no LacZ reporter activation, as expected.
Figure 3. LacZ selection with replica plating in the yeast-two-hybrid assay indicates interaction between M. trossulus p53 and MDM.
Positive and negative controls acted as expected with only slight activation of the LacZ reporter due to the use of the most sensitive reporter plasmid (pBR1840). Abbreviations: na, no autoactivation; a, slight autoactivation; A, strong autoactivation; i, interaction; ni, no interaction.
There was no observable autoactivation of p53-B42. However, progeny yeast expressing the MDM-LexA fusion protein showed slight autoactivation on Glu + X-Gal (Figure 3). Yeast mated to test the interaction of p53 and MDM activated the LacZ reporter on Glu + X-Gal at the same intensity as the MDM background control, but strongly activated the LacZ reporter on Gal + X-Gal (Figure 3), indicating a positive interaction of M. trossulus MDM with M. trossulus p53. It is uncertain why the MDM-LexA fusion plasmid was able to initiate reporter gene transcription independently, especially since MDM2 is not known to contain a transcriptional activating domain.
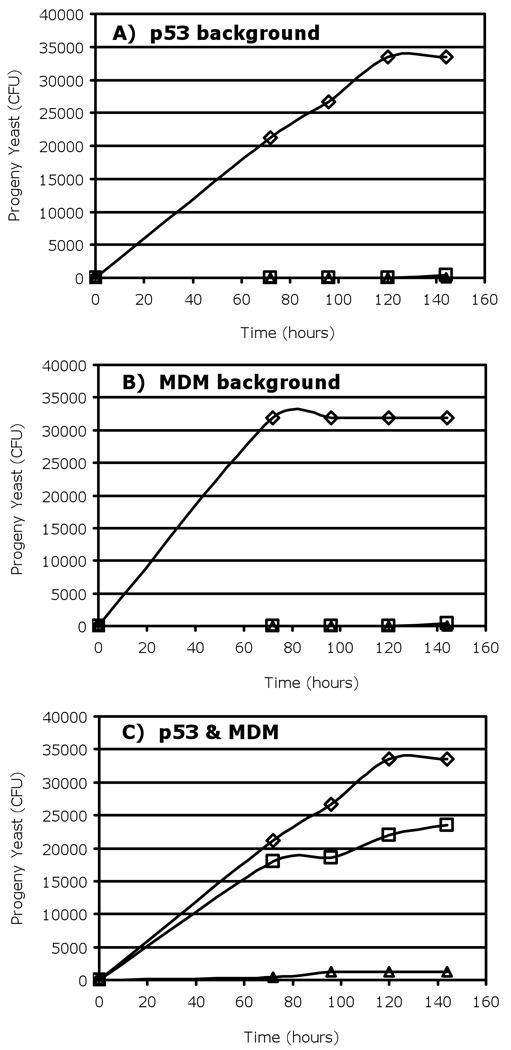
Activation of the Leu2 reporter was observed by dilution plating, with the growth of mated yeast (measured in colony forming units, CFU, over time) on Gal−Leu media, which minimized MDM autoactivation (Figure 4). The activation of the Leu2 reporter by progeny yeast containing both the M. trossulus MDM-LexA and the M. trossulus p53-B42 fusion plasmids was much greater than any background autoactivation produced by either fusion protein alone suggesting that these proteins do interact.
Figure 4. Leu2 selection measured by dilution plating in the yeast-two-hybrid assay indicates interaction of M. trossulus MDM with p53 in galactose-based, leu- medium.
A) Mdm background: Yeast cells transfected with LexA-Mdm fusion plasmid, B) P53 background: Yeast cells transfected with B42-p53 fusion plasmid, C) MDM and p53: Mated yeast containing both plasmids. Symbols: ◇, galactose-based media containing leucine (Gal + Leu); □, galactose-based media, no leucine (Gal −Leu); and △, glucose-based media, no leucine (Glu −Leu).
Interaction of M. trossulus p53 and MDM was confirmed by pull-down assays (Figure 5). The 35S-methionine labeled p53 protein was strongly expressed in the reticulocyte lysate (lane A1, Figure 5A) at the expected 53 kDa size. His-tagged MDM bait, attached to the MagZ™ beads, tightly bound p53. This was evident from repeated washes, in which no p53 was detected (lane A2 for the first wash, subsequent washes not shown), as well as from a strong 53 kDa band in the first and second eluates (lanes A3 and A4). A weak 53 kDa band in the supernatant of the initial mix of target, bait and beads, before loading the column (lane A5) indicates that p53 was slightly in excess of MDM due to its strong expression in the rabbit reticulocyte system. The reciprocal assay also indicated binding between the two proteins (Figure 5B). A variety of different-sized labeled products were produced in the rabbit reticulocytes transfected with the mdm expression plasmid (lane B1). However, only two products, one of the expected 64 kDa size and one unexpected approximately 50 kDa large product, were bound by His-tagged p53 and subsequently eluted (lanes B3 and B4). None of the unspecific labeled bands were detected in the washes (lane B2), probably due to their low concentration in the washing buffers. However, lane B5, which contains the supernatant from the initial mix of beads, bait and target, shows that all labeled targets were attached to the beads.
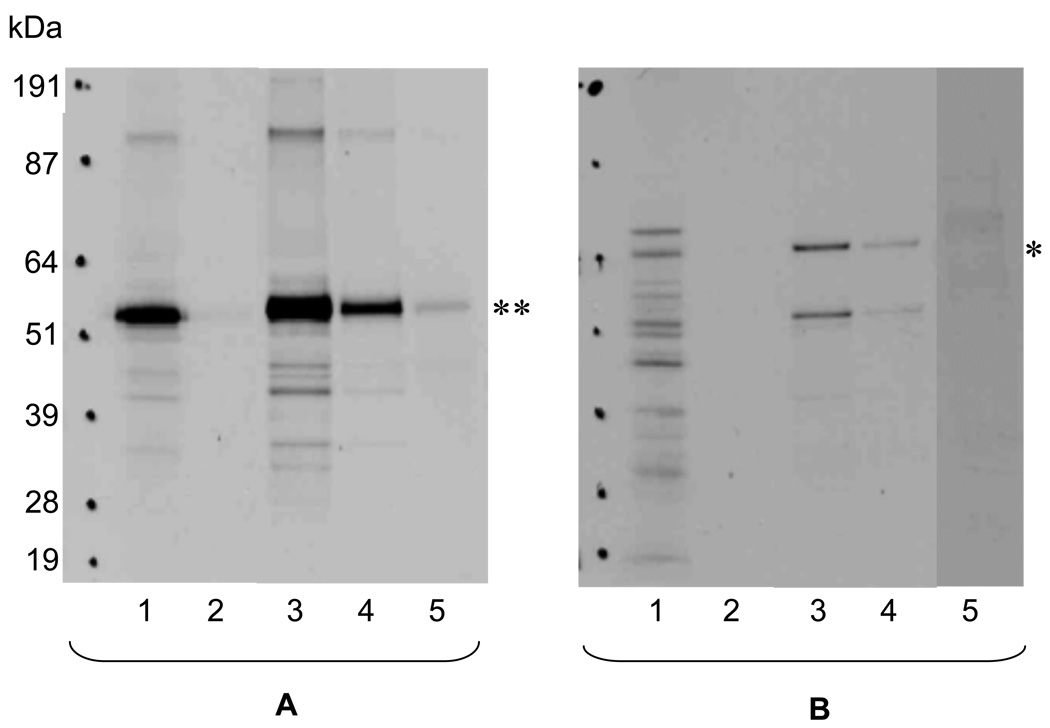
Figure 5. SDS-PAGE gradient gel for pull-down assays of p53 as target (A) and MDM as target (B) indicating interaction of the proteins.
Lanes 1, 35S-methionine labeled proteins synthesized in vitro in the TnT SP6 coupled reticulocyte lysate system; lanes 2, first column wash (three more washes were performed with similar results, data not shown); lanes 3, first elution; lanes 4, second elution; lanes 5, supernatant from initial binding step (bait plus target plus beads). The Hyperfilm (Amersham Biosciences) was exposed for approximately 17 hours. Labeled p53 target (**) has an expected size of approximately 53 kDa. Labeled MDM target (*) has an expected size of approximately 64 kDa, as well as a smaller 50 kDa band.
3.3 Quantification of mRNA expression
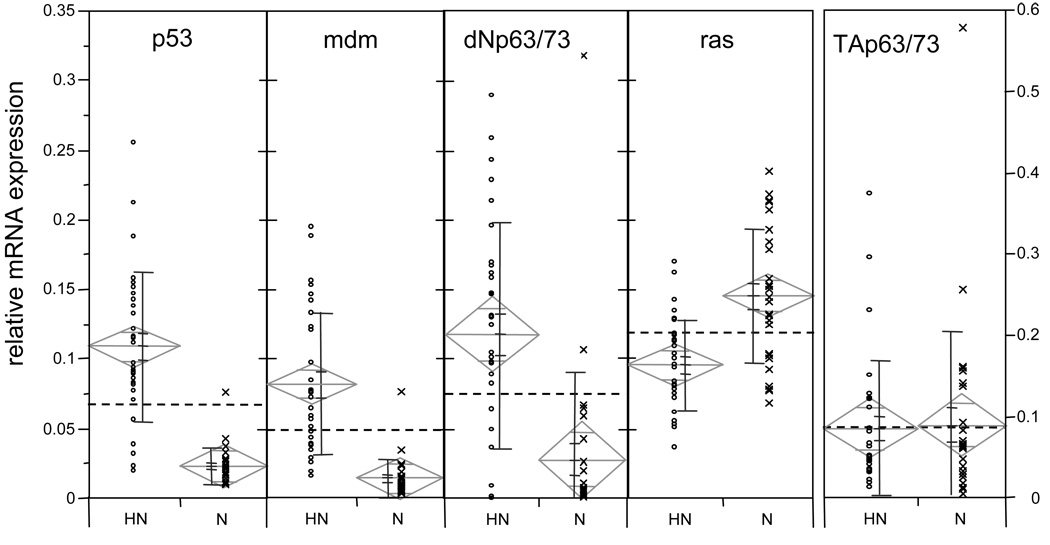
Using real time PCR, we investigated whether mRNA expression levels of three p53 family members as well as mdm and the proto-oncogene ras were different in normal and neoplastic mussel haemocytes. All mRNA species were ubiquitously expressed in both neoplastic and normal haemocytes (Figure 6). Six out of the ten data sets had a non-normal distribution (Shapiro Wilk test, α=0.01). We therefore used the non-parametric Wilcoxon rank sum test to compare the means of neoplastic cells with the means of normal sample groups for each mRNA (α = 0.01). The mRNA levels of p53, ΔNp63/73, and mdm were on average four to five times higher in neoplastic than in normal haemocytes (p<0.0001; Figure 6). In contrast, the ras mRNA was expressed on average at a 1.5 times higher level in normal than in neoplastic haemocytes. The TAp63/73 isoform was not differentially expressed between the two types of haemocytes. Overall, the lowest mRNA levels were found in the mdm mRNA species. We observed that the inter-individual variances in gene expression were greater for the neoplastic than for the normal animals. Six out of a total of ten data sets contained a small number of outliers (as defined by being outside the 1.5 × interquartile range), all of which were included in the statistical analyses. Classification of samples by morphology and cell density may not exclude the presence of a small number of varying cell types in an otherwise homogeneous sample.
Figure 6. Association between relative transcription levels for mdm, ras, p53-, ΔNp63/73- and TAp63/73-like mRNA isoforms and haemocyte status, neoplastic (L; n=30) and normal (N; n=27), in M. trossulus.
mRNA expression levels are relative to a standard dilution curve. Data points (◦, x) represent the average of triplicate wells. The bars next to the individual data points represent, from top to bottom, standard deviation, standard error of the mean, and mean. The means diamonds graphic illustrates the group mean (center horizontal line), the 95 % confidence interval (vertical points of diamond), overlap marks (top and bottom horizontal lines), and the width of the diamond indicates sample size. The dotted line shows the grand mean. If the 95 % confidence intervals do not overlap, the group means are significantly different (but the reverse is not necessarily true).
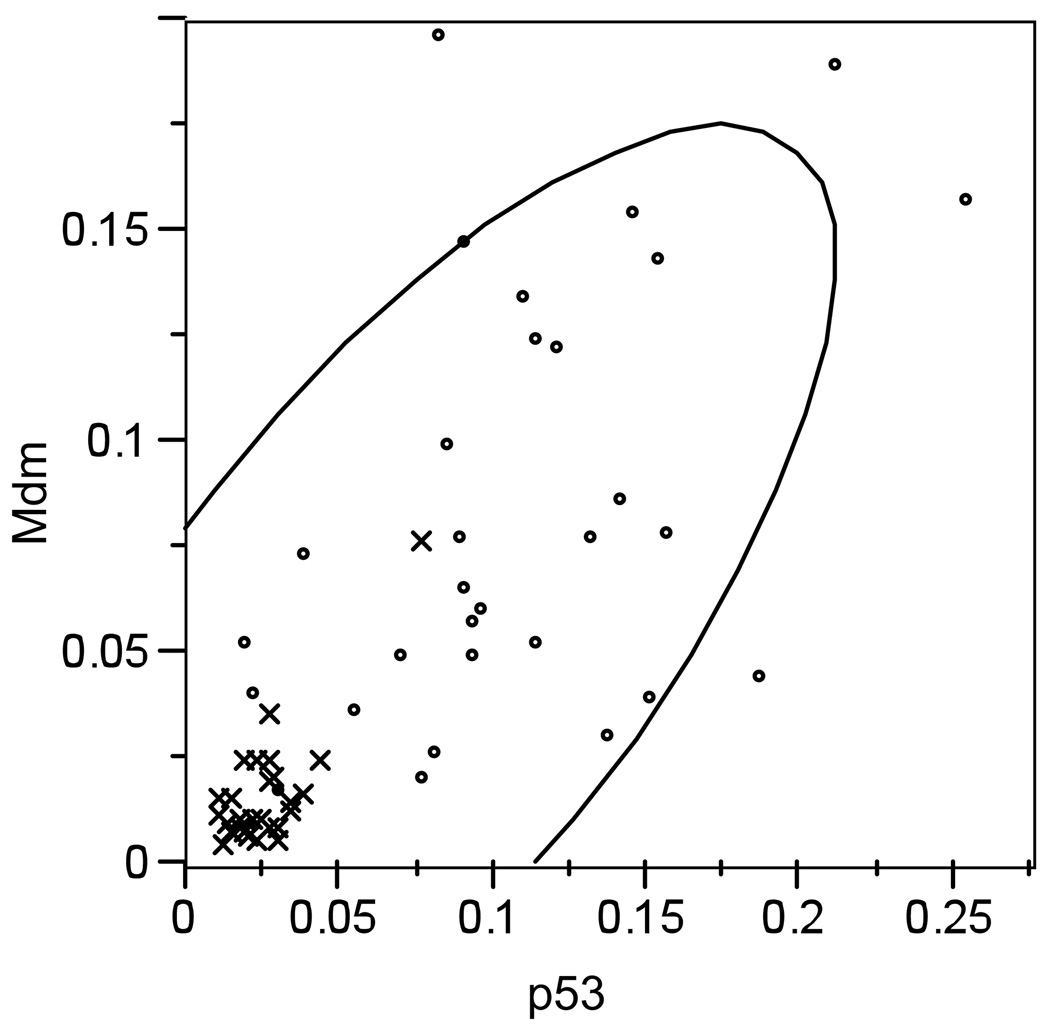
We performed the Pearson’s correlation test to investigate whether correlations exist in expression levels between the different p53 isoforms or between the p53 isoforms and the mdm regulatory protein or the ras proto-oncogene. Pearson’s correlation is considered more sensitive than other, non-parametric correlations. We found a strong positive linear correlation between expression of p53 and mdm in the combined (leukemic plus normal) dataset (R2=0.742, p<0.0001). Although weaker, this correlation persisted when data were sorted into the normal and neoplastic groups (normal group: R2=0.771, p<0.0001; leukemic group: R2=0.476, p=0.0079) (Figure 7). mdm was also correlated with TAp63/73 (R2=0.615, p=0.0003), but only in the neoplastic dataset.
Figure 7.
Positive Pearson’s correlation between mdm and p53 in the combined dataset for normal (x) and neoplastic (◦) haemocytes (R2=0.742, p<0.0001).
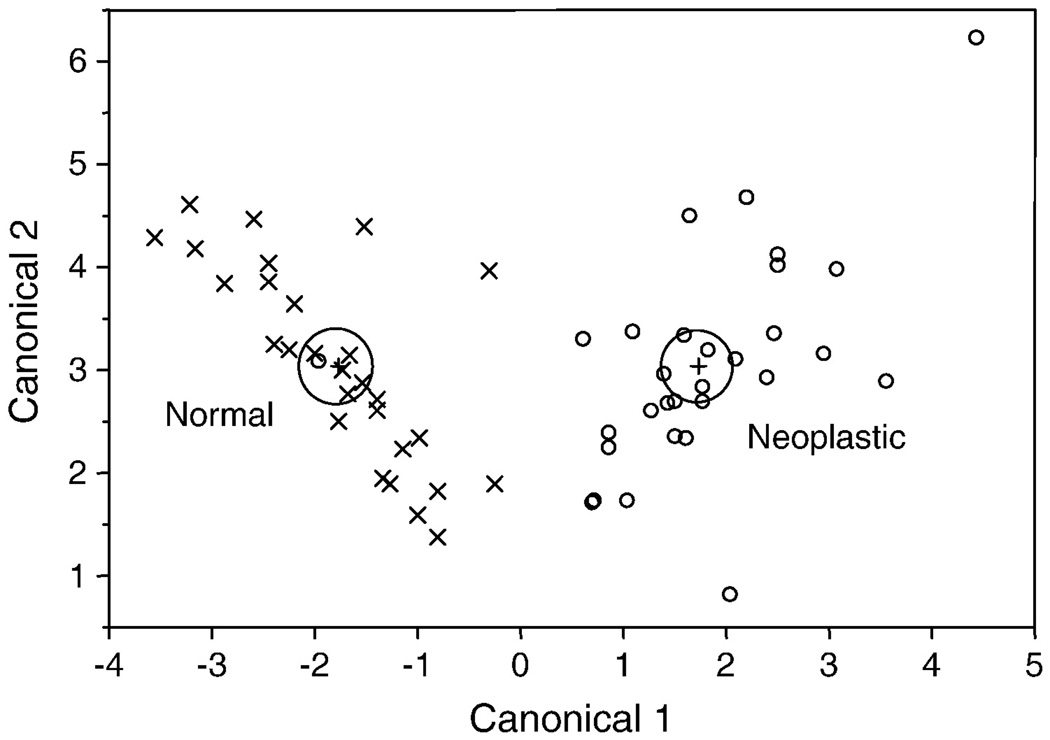
We performed a linear discriminant function analysis in order to determine whether the levels of these five mRNAs were predictive of health status, and if so, which mRNA species contributed most to a correct prediction. Only one data point out of 58, originating from a neoplastic sample, was misclassified as a normal. All other data points were predicted correctly with high probability (0.92 to 1.00), when all five mRNA species were included in the analysis (Figure 8). Stepwise discriminant analysis, based on F values, showed that the best prediction was provided by p53, followed by ras > ΔNp63/73 > mdm > TAp63/73.
Figure 8. Canonical plot illustrating the results of the linear discriminant analysis predictingthe health status of normal (x) and neoplastic (◦) haemocyte samples.
Except for one sample, all samples are classified correctly using five gene transcript levels (p53, mdm, ras, ΔNp63/73 and TAp63/73). Large circles are the 95% confidence limits for the means.
4. Discussion
Using sequence analysis, yeast-two-hybrid assay, and pull-down assays, we showed that the Mytilus p53 and MDM are able to interact and bind to each other. To our knowledge, this is the first detailed report of a cloned MDM-like mRNA in invertebrates. Its primary structure closely resembles vertebrate MDM2 and MDM4 in highly conserved regions that are characteristic for these proteins: the N-terminal p53-binding binding domain and C-terminal zinc-binding RING domain (Figure 1). In addition, five phosphorylation sites are conserved in the central acidic domain. It is known from studies in vertebrate models that MDM2 is responsible for p53 binding and subsequent ubiquitination (Honda et al. 1997). Since the molluscan p53 and MDM homologues are phylogenetically closely related to their vertebrate counterparts, we hypothesized that the two proteins can interact. This interaction was also suggested by interacting soft-shell clam p53 (Mya arenaria) with human MDM2 in a human cell background (Holbrook et al. 2009) and by recent interaction modelling for deer tick (Ixodes scapularis) p53 and Mdm2-like sequences (Lane et al. 2010).
The structure of the minimal MDM2-binding site within the transactivation domain on the human p53 was shown to consist of three critical amino acids (F19, L22, W23) (Lin et al. 1994; Picksley et al. 1994). These critical residues are highly conserved in the molluscan (M. arenaria and M. trossulus) p53 and p63/73. Holbrook et al. (2009) suggested that protein/protein interactions appear to be maintained, however, other substitutions in this site in p53 (E20/Y21 instead of S20/D21 in mammals) suggest that the regulation of the p53/MDM2 feedback loop may be different in molluscs. Phosphorylation of S20 modulates activity of human p53, presumably by inhibiting p53/MDM2 binding and stabilization of the p53 protein (Unger et al. 1999). However, glutamic acid (E20) found in molluscs is also conserved in some of the vertebrate sequences (such as Xenopus) suggesting that the substitution of S20 by another polar amino acid may not have been random. Unfortunately, functional data on the molluscan p53 is limited (Fernandes et al. 2008) and the effect of the substitution is unknown.
MDM2 contains a 12 kDa structural domain at its NH2-terminus (residues 17 to125 in X. laevis MDM2) that is highly conserved among vertebrate species (71 to 91 % identity, (Kussie et al. 1996)). This domain forms the cleft that is required for binding of the p53 protein. The M. trossulus binding domain is 39% identical to its vertebrate counterparts. Notably, six out of the14 conserved hydrophobic and aromatic amino acids that are thought to be important for p53 binding via van der Waals forces are identical, while the remaining residues share similarity in their hydrophobicity. These conserved structural homologies in both the p53 TAD and the MDM p53-binding domain in M. trossulus point to a strong likelihood that the two proteins are able to interact with each other and that Mdm may inhibit p53 activity at the transcriptional level.
Interactions between molluscan p53 and human MDM2 were investigated recently (Holbrook et al. 2009). Transfection of a p53-null, human non-small-cell-lung-carcinoma cell line with mollscan p53 from M. arenaria induced expression of endogenous HDM2, suggesting that the molluscan p53 might be able to participate in the MDM2 feedback loop. However, expression of the molluscan p53 protein in the human cell line was minimal, did not elicit expression of downstream genes (p21, Rb), and cell growth was not inhibited, suggesting that either the activities of the molluscan and the human p53 are not similar, or that the molluscan p53 could not function normally in the human cell. Elucidation of molluscan MDM sequences and establishment of molluscan cell lines (Walker et al. 2009) now allow for a more direct approach to the study of p53-MDM interactions in the native invertebrate cell environment. The results of the yeast-two-hybrid assay and pull-downs presented here are strong indicators that Mytilus p53 and MDM are able to bind to each other.
Fruit fly (D. melanogaster) and nematode (C. elegans) have been shown to have p53 homologues, but these homologues are phylogenetically distant and are not known to be regulated by an MDM2 homologue (Lu et al. 2006). It was thus thought that MDM2-mediated regulation of p53 appeared only in the vertebrate lineage after the divergence of protostomes and deuterostomes (Lu et al. 2006). However, our data and others (Holbrook et al. 2009, Lane et al. 2010) suggest that the evolutionary development of p53 was closely correlated with the evolution of a core p53 regulator, an MDM-like homologue, and is a much more ancient development than was previously thought.
It has been suggested that the molluscan p53 is more likely to be an ancestral p63 without the C-terminal SAM domain (Yang et al. 2002; Goodson et al. 2006; Muttray et al. 2008). Interestingly, the human p63, and especially the TAp63-γ isoform, which also lacks the SAM domain (Murray-Zmijewski et al. 2006) similar to the molluscan p53, has been shown to interact with MDM2 at the transactivation domain, but the degradation of human TAp63-γ did not require MDM2 and proceeded in an unknown MDM2-independent manner (Ying et al. 2005). This is surprising because the three hydrophobic amino acids, F19 W23 L26, which are critical for the MDM2-mediated degradation of the human p53 protein (Kussie et al. 1996) are conserved in human TAp63-γ (and in bivalve p53 and TAp63/73). It will therefore be important to establish whether or not molluscan MDM is able to ubiquitinate p53 and induce its degradation, or whether molluscan MDM/p53 binding has an affect on p53 transcriptional activity, as has been described for vertebrate MDM4 (Marine et al. 2006). Although BLAST queries indicate that the Mytilus MDM protein is more closely related to vertebrate MDM2 than MDM4, its placement in the phylogenetic tree indicates that it may be considered an ancestral form of both MDM2 and MDM4 with a functional spectrum different from the vertebrate MDM proteins. It is also not a foregone conclusion that other MDM variants may exist in some invertebrates.
We have shown previously that p53 and ΔNp63/73 isoforms were significantly up-regulated in neoplastic mussel haemocytes, while TAp63/73 expression levels did not change (Muttray et al. 2008). This new independent dataset confirms our previous findings. We now also present the mRNA expression levels for Mdm and ras. Based on accounts from human cancers (Friday et al. 2005) and from M. trossulus haemocytes (Ciocan et al. 2006) we expected to find an increased level of expression of the proto-oncogene ras in neoplastic mussel haemocytes. However, this was clearly not the case as expression levels of ras were 1.5× lower in neoplastic haemocytes. Previously, using a semi-quantitative PCR technique, Ciocan and co-workers (2006) found that ras expression may have been induced to a larger extent in neoplastic than in normal M. trossulus haemocytes. Ciocan also indicated that ras from neoplastic samples displayed silent gene polymorphic variations not found in normal samples, one of which coincided with a mutational hotspot (codon 13) which, in mammals leads to diminished GTPase activity and uncontrolled cell division (Bos 1989). Thus, the role of ras expression and silent mutations remain an interesting but unsolved component of the processes that may lead to haemic neoplasia in mussels.
In contrast to ras, Mdm is up-regulated by an average factor of 5.8 in neoplastic haemocytes compared to normal cells. Since it is known that vertebrate p53 and p63/p73 induce Mdm transcription, we investigated whether there was a correlation between p53 and TAp63/73 mRNA levels on one hand and Mdm mRNA levels on the other hand, assuming that p53 and TAp63/73 mRNA measured by us were translated and active at the protein level. We found that a strong pair-wise correlation existed between Mdm and p53 in both normal and neoplastic datasets indicating that the balance that exists between the expression levels in normal haemocytes is not disturbed in end-stage haemic neoplasia. MDM2 is the central part of several negative autoregulatory feedback loops in the p53 circuitry identified in mammals (Harris et al. 2005). An increase in p53 expression or activity results in an increased transcription of Mdm2, which will then modulate down p53 activity, thereby keeping p53 activity under tight control. We also found a strong correlation between TAp63/73 and Mdm in leukemic cells, but not in normal cells.
In mammalian systems, an alternatively spliced form of c-H-RAS, p19ras, can activate p73 by interacting with MDM2 and abrogating the MDM2-p73 inhibitory interaction (Harms et al. 2006). It was observed that an increase in p19ras expression was correlated with a decrease in MDM2 protein abundance. While it is too premature to say whether similar RAS splice forms and regulatory pathways exist in mollusks, it is worth pointing out that we observed opposing trends in mean levels of ras and Mdm mRNA in our data as well. We do not know whether this had an affect on TAp63/73 protein activity. RAS also has a regulatory role in the MDM2-p53 feedback loop via the mitogen-activated protein kinase pathway (Michael et al. 2002). RAS can activate mRNA transcription and translation of Mdm2 and p53, as well as induce ARF, a tumor-suppressor protein that binds MDM2 and blocks MDM2-mediated p53 degradation in a cell-type dependent manner. ARF may be missing in the molluscan genomes, but HES1 and HEY1, which behave in an analogous manner by blocking MDM2 (Huang et al. 2004), are highly conserved at least in the L. gigantean genome (L. gigantean v1.0 database, http://genome.jgi-psf.org/, C. Walker, personal communication). Interestingly, the conserved phosphorylation sites, Ser 166 and Ser 186, both within RXRXXS motifs that inhibit interaction of ARF with MDM2 when phosphorylated (Zhou et al. 2000), are missing in Mytilus MDM, further pointing to the possibility that ARF regulation is absent. We found that over-expression of ras in normal haemocytes did not lead to a higher expression of p53 or Mdm as found in vertebrates. We hypothesize that RAS may function by blocking MDM via HES1 or HEY1 and thus stabilize p53 at the protein level.
The measurement of gene expression has the potential to provide an assessment of the health status of an organism before adverse health effects become manifest and to test for sub-lethal endpoints in environmental monitoring. Mussels are widely used in environmental monitoring (Goldberg 1975). We hypothesized that correlating the expression of several genes in a pathway, such as those involved in cell proliferation or DNA damage response, has a higher predictive power than monitoring of single genes. The discriminant function analysis of our gene expression data shows that the combined data for all five mRNA species are highly predictive of the health status of haemocytes, with p53 and ras as the strongest predictors, followed by ΔNp63/73 and Mdm. This shows that mRNA levels can be used as statistical predictors of disease, but that a combination of at least four (p53, ras, ΔNp63/73 and Mdm) has to be applied for the prediction to be accurate. Combining expression of these four genes in a discriminant analysis gives a higher correlation coefficient than any of the single gene correlations. This also supports the use of these mRNA expressions as biomarkers for this disease. It remains to be investigated whether these mRNA species can also be used to predict earlier transitional stages of the disease in sub-tidally submerged animals.
5. Conclusions
The data presented here show that the highly conserved molluscan p53 is able to interact with and bind to a molluscan MDM-like protein. This opens the possibility that p53 activity can be modulated by an MDM-like protein in the molluscan species, similar to regulation of p53 activity in vertebrate species. Much functional analysis, especially at the protein level, needs to be completed before regulatory feedback loops can be established for certain in these invertebrate models. Gene expression analysis indicates that a combination of a limited number of key gene transcripts rather than single transcripts can be a better predictor of the health status of an organism and thus could be developed into biomarkers of haemic neoplasia in mussels. As mussels are widely used in environmental monitoring, this naturally occurring neoplastic disease provides the cancer research community with an accessible model that shares some of the exposure pathways to potential disease triggers and molecular mechanisms with similar human diseases.
Acknowledgements
Financial support was provided by NSERC grant CRDPJ 349918 and by Metro Vancouver to SAB, by NIH/NIEHS grant ES012066 and MAFES (Maine Agricultural and Forestry Experiment Station) ME08509 to RJVB, and INBRE Undergraduate Summer Research Fellowship (NCRR 1 P20 RR16463-01) to T.F.O. We would like to extend our gratitude to Farida Bishay and Ekaterina Vassilenko for assistance during field work, to Charles Walker for insightful discussions, and to the anonymous reviewers for their comments. This is MAFES publication number 3089.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barber BJ. Neoplastic diseases of commercially important marine bivalves. Aquat. Living Resour. 2004;17:449–466. [Google Scholar]
- Benner SA, Cohen MA, Gonnet GH. Amino acid substitution during functionally constrained divergent evolution of protein sequences. Protein Eng. 1994;7:1323–1332. doi: 10.1093/protein/7.11.1323. [DOI] [PubMed] [Google Scholar]
- Böttger S, Jerszyk E, Low B, Walker C. Genotoxic stress–induced expression of p53 and apoptosis in leukemic clam hemocytes with cytoplasmically sequestered p53. Cancer Res. 2008;68:777–782. doi: 10.1158/0008-5472.CAN-06-0968. [DOI] [PubMed] [Google Scholar]
- Bos JL. ras Oncogenes in Human Cancer: A Review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- Bower SM. [Last accessed: October 20, 2009];Synopsis of infectious diseases and parasites of commercially exploited shellfish: Haemocytic neoplasia of mussels. 2006 http://www.pac.dfo-mpo.gc.ca/science/species-especes/shellfish-coquillages/diseases-maladies/pages/hcnmu-eng.htm.
- Calabro V, Mansueto G, Parisi T, Vivo M, Calogero RA, La Mantia G. The human MDM2oncoprotein increases the transcriptional activity and the protein level of the p53 homolog p63. J Biol. Chem. 2002;277:2674–2681. doi: 10.1074/jbc.M107173200. [DOI] [PubMed] [Google Scholar]
- Ciocan CM, Moore JD, Rotchell JM. The role of ras gene in the development of haemic neoplasia in Mytilus trossulus. Mar. Environ. Res. 2006;62:S147–S150. doi: 10.1016/j.marenvres.2006.04.020. [DOI] [PubMed] [Google Scholar]
- Cox RL, Stephens RE, Reinisch CL. p63/73 homologues in surf clam: novel signaling motifs and implications for control of expression. Gene. 2003;320:49–58. doi: 10.1016/j.gene.2003.07.001. [DOI] [PubMed] [Google Scholar]
- Fernandes AD, Atchley WR. Biochemical and functional evidence of p53 homology is inconsistent with molecular phylogenetics for distant sequences. J. Mol. Evol. 2008;67:51–67. doi: 10.1007/s00239-008-9124-2. [DOI] [PubMed] [Google Scholar]
- Friday BB, Adjei AA. K-ras as a target for cancer therapy. Biochimica et Biophysica Acta (BBA) - Reviews on Cancer: The KRAS Oncogene. 2005;1756:127–144. doi: 10.1016/j.bbcan.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Gietz D, St. Jean A, Woods RA, Schiestl RH. Improved method for high-efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg ED. The mussel watch - a first step in global marine monitoring. Mar. Pollut. Bull. 1975;6:111. [Google Scholar]
- Golemis EA, Serebriiskii I, Finely RL, Kolonin MG, Gyuris J, Brent R. Interaction trap/two-hybrid system to identify interacting proteins. In: Coligan JE, Dunn BM, Speicher DW, Wingfield PT, editors. Analysis of Protein–Protein Interactions, Current Protocols in Protein Science. USA: Wiley; 1998. pp. 19.2.1–19.2.40. 1998. [Google Scholar]
- Goodson MS, Crookes-Goodson WJ, Kimbell JR, McFall-Ngai MJ. Characterization and role of p53 family members in the symbiont-induced morphogenesis of the Euprymna scolopes light organ. Biol. Bull. 2006;211:7–17. doi: 10.2307/4134573. [DOI] [PubMed] [Google Scholar]
- Harms K, Nozell S, Chen X. The common and distinct target genes of the p53 family transcription factors. Cell. Mol. Life Sci. 2004;61:822–842. doi: 10.1007/s00018-003-3304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms KL, Chen X. p19ras brings a new twist to the regulation of p73 by Mdm2. Sci STKE 2006. 2006;337:pe24. doi: 10.1126/stke.3372006pe24. [DOI] [PubMed] [Google Scholar]
- Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. 2005;24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- Holbrook LAC, Butler RA, Cashon RE, Van Beneden RJ. Soft-shell clam (Mya arenaria) p53: A structural and functional comparison to human p53. Gene. 2009;433:81–87. doi: 10.1016/j.gene.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- Huang Q, Raya A, DeJesus P, Chao S-H, Quon KC, Caldwell JS, Chanda SK, Izpisua-Belmonte JC, Schultz PG. Identification of p53 regulators by genome-wide functional analysis. Proc Natl Acad Sci USA. 2004;101:3456–3461. doi: 10.1073/pnas.0308562100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong MH, Bae J, Kim WH, Yoo SM, Kim JW, Song PI, Choi KH. p19ras interacts with and activates p73 by involving the MDM2 protein. J Biol Chem. 2006;281:8707–8715. doi: 10.1074/jbc.M513853200. [DOI] [PubMed] [Google Scholar]
- Kelley ML, Winge P, Heaney JD, Stephens RE, Farell JH, Van Beneden RJ, Reinisch CL, Lesser MP, Walker CW. Expression of homologues for p53 and p73 in the softshell clam (Mya arenaria), a naturally-occurring model for human cancer. Oncogene. 2001;20:748–758. doi: 10.1038/sj.onc.1204144. [DOI] [PubMed] [Google Scholar]
- Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, Pavletich NP. Structure of the Mdm2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- Lane DP, Cheok CF, Brown CJ, Madhumalar A, Ghadessy FJ, Verma C. The Mdm2 and p53 genes are conserved in the Arachnids. Cell Cycle. 2010;9:748–754. doi: 10.4161/cc.9.4.10616. [DOI] [PubMed] [Google Scholar]
- Levrero M, De Laurenzi V, Costanzo A, Gong J, Wang J, Melino G. The p53/p63/p73 family of transcription factors: overlapping and distinct functions. J. Cell Sci. 2000;113:1661–1670. doi: 10.1242/jcs.113.10.1661. [DOI] [PubMed] [Google Scholar]
- Lin J, Chen J, Elenbaas B, Levine AJ. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes Dev. 1994;8:1235–1246. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- Linke K, Mace PD, Smith CA, Vaux DL, Silke J, Day CL. Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death Differ. 2008;15:841–848. doi: 10.1038/sj.cdd.4402309. [DOI] [PubMed] [Google Scholar]
- Lu WJ, Abrams JM. Lessons from p53 in non-mammalian models. Cell Death Differ. 2006;13:909–912. doi: 10.1038/sj.cdd.4401922. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Derbyshire MK, DeWeese-Scott C, Gonzales NR, Gwadz M, Hao L, He S, Hurwitz DI, Jackson JD, Ke Z, Krylov D, Lanczycki CJ, Liebert CA, Liu C, Lu F, Lu S, Marchler GH, Mullokandov M, Song JS, Thanki N, Yamashita RA, Yin JJ, Zhang D, Bryant SH. CDD: a conserved domain database for interactive domain family analysis. Nucl. Acids Res. 2007;35:D237–D240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marine J-C, Francoz S, Maetens M, Wahl G, Toledo F, Lozano G. Keeping p53 in check: essential and synergistic functions of Mdm2 and Mdm4. Cell Death Differ. 2006;13:927–934. doi: 10.1038/sj.cdd.4401912. [DOI] [PubMed] [Google Scholar]
- Marine J-C, Jochemsen AG. Mdmx as an essential regulator of p53 activity. Biochem. Biophys. Res. Com. 2005;331:750–760. doi: 10.1016/j.bbrc.2005.03.151. [DOI] [PubMed] [Google Scholar]
- Meek DW, Knippschild U. Posttranslational Modification of MDM2. Mol. Cancer Res. 2003;1:1017–1026. [PubMed] [Google Scholar]
- Michael D, Oren M. The p53 and Mdm2 families in cancer. Curr. Opin. Genet. Dev. 2002;12:53–59. doi: 10.1016/s0959-437x(01)00264-7. [DOI] [PubMed] [Google Scholar]
- Munger K, Phelps WC, Bubb V, Howley PM, Schlegel R. The E6 and E7 genes of the human papillomavirus type 16 together are necessary and sufficient for transformation of primary human keratinocytes. J. Virol. 1989;63:4417–4421. doi: 10.1128/jvi.63.10.4417-4421.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray-Zmijewski F, Lane DP, Bourdon J-C. p53/63/73 isoforms: an orchestra of isoforms to harmonize cell differentiation and response to stress. Cell Death Differ. 2006;13:962–972. doi: 10.1038/sj.cdd.4401914. [DOI] [PubMed] [Google Scholar]
- Muttray AF, Cox RL, Reinisch CL, Baldwin SA. Identification of DeltaN isoform and polyadenylation site choice variants in molluscan p63/p73-like homologues. Mar. Biotechnol. 2007;9:217–230. doi: 10.1007/s10126-006-6045-1. [DOI] [PubMed] [Google Scholar]
- Muttray AF, Cox RL, St-Jean SD, van Poppelen P, Reinisch CL. Identification and phylogenetic comparison of p53 in two distinct mussel species (Mytilus) Comp. Biochem. Physiol. C. Pharmacol. Toxicol. Endocrinol. 2005;140:237–250. doi: 10.1016/j.cca.2005.02.011. [DOI] [PubMed] [Google Scholar]
- Muttray AF, Schulte PM, Baldwin SA. Invertebrate p53-like mRNA isoforms are differentially expressed in mussel haemic neoplasia. Mar. Environ. Res. 2008;66:412–421. doi: 10.1016/j.marenvres.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Page RDM. TREEVIEW: An application to display phylogenetic trees on personal computers. Comp. Appl. Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Picksley SM, Vojtesek B, Sparks AK, Lane D. Immunochemical analysis of the interaction of p53 with MDM2;-fine mapping of the MDM2 binding site on p53 using synthetic peptides. Oncogene. 1994;9:2523–2529. [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Schiestl RH, Gietz D. High-efficiency transformation of intact yeast cells using nucleic acids as a carrier. Curr. Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Schmale H, Bamberger C. A novel protein with strong homology to the tumor suppressor p53. Oncogene. 1997;15:1363–1367. doi: 10.1038/sj.onc.1201500. [DOI] [PubMed] [Google Scholar]
- Unger T, Juven-Gershon T, Moallem E, Berger M, Sionov RV, Lozano G, Oren M, Haupt Y. Critical role for Ser20 of human p53 in the negative regulation of p53 by Mdm2. The EMBO Journal. 1999;18:1805–1814. doi: 10.1093/emboj/18.7.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker C, Bottger SA, Mulkern J, Jerszyk E, Litvaitis M, Lesser M. Mass Culture and Characterization of Tumor Cells From a Naturally Occurring Invertebrate Cancer Model: Applications for Human and Animal Disease and Environmental Health. Biol. Bull. 2009;216:23–39. doi: 10.1086/BBLv216n1p23. [DOI] [PubMed] [Google Scholar]
- Walker CW, Böttger S. A naturally occurring cancer with molecular connectivity to human diseases. Cell Cycle. 2008;7:1–3. doi: 10.4161/cc.6366. [DOI] [PubMed] [Google Scholar]
- Yang A, Kaghad M, Caput D, McKeon F. On the shoulders of giants: p63, p73 and the rise of p53. Trends Genet. 2002;18:90–95. doi: 10.1016/s0168-9525(02)02595-7. [DOI] [PubMed] [Google Scholar]
- Ying H, Chang DLF, Zheng H, McKeon F, Xiao Z-XJ. DNA-Binding and Transactivation Activities Are Essential for TAp63 Protein Degradation. Mol. Cell. Biol. 2005;25:6154–6164. doi: 10.1128/MCB.25.14.6154-6164.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]