Abstract
RNA-binding proteins are critical effectors of gene expression. They guide mRNA localization, translation, and stability, and potentially play a role in regulating mRNA synthesis. The structural basis for RNA recognition by RNA-binding proteins is the key to understanding how they target specific transcripts for regulation. Compared to other metazoans, nematode genomes contain a significant expansion in several RNA-binding protein families, including Pumilio-FBF (PUF), TTP-like zinc finger (TZF), and argonaute-like (AGO) proteins. Genetic data suggest that individual members of each family have distinct functions, presumably due to sequence variations that alter RNA binding specificity or protein interaction partners. In this review, we highlight example structures and identify the variable regions that likely contribute to functional divergence in nematodes.
Introduction
RNA regulation is pervasive and impacts nearly every aspect of gene expression. RNA molecules function both as regulators and targets in diverse pathways to ensure appropriate decoding of the genome. RNA-binding proteins are central to this form of regulation. They act as effectors of RNA stability and translation efficiency, they guide transcripts to defined locations within a cell, they control the fidelity of gene decoding, and they function as cofactors to promote the activity of functional and structural RNA molecules.
The facile genetics, defined cellular lineage, and ease of observation has made the nematode Caenorhabditis elegans a popular model to study RNA regulatory mechanisms. A scan of the C. elegans and other nematode genomes reveals a surprising expansion of putative RNA-binding proteins relatives to other metazoans. For example, the RNA-binding protein Pumilio—discovered in flies—has two homologs in humans but eleven homologs in C. elegans [1, 2]. The CCCH-type tandem zinc finger family (TZF), typified by the mammalian protein tristetraprolin (TTP), has sixteen members in worms [3–6]. Finally, there are twenty-seven Argonaute homologs in C. elegans, including a clade of worm-specific argonautes (WAGOs) [7, 8].
It is not clear why RNA-binding protein families have expanded in nematodes. Forward and reverse genetic experiments indicate that many play distinct roles in germline development, gametogenesis, and early embryogenesis, where regulation of maternal RNAs plays a primary role. In this review, we outline representative structures from the PUF, TZF, and AGO families, and highlight data that identifies the basis for specialized function in the expanded set of nematode homologs.
The PUF family
PUF proteins in nematode germline development
The fem-3 binding factor (FBF) was the first Pumilio homolog identified in C. elegans [2]. Pumilio and FBF together comprise the founding members of the PUF family of RNA-binding proteins. FBF is encoded by two nearly identical genes, fbf-1 and fbf-2. Together, they act to maintain the population of progenitor cells in the distal region of the germline and promote the switch from spermatogenesis to oogenesis at the onset of adulthood [2, 9] (Fig. 1). FBF binds in a sequence specific fashion to the 3′ untranslated region (UTR) of several messenger RNAs, including fem-3 and gld-1 [10]. GLD-1 and FEM-3 promote spermatocyte differentiation, and GLD-1 promotes entry into meiosis [11–13]. FBF represses translation of gld-1 mRNA in the distal end of the germline, and it represses translation of gld-1 and fem-3 mRNA in developing oocytes [2, 9].
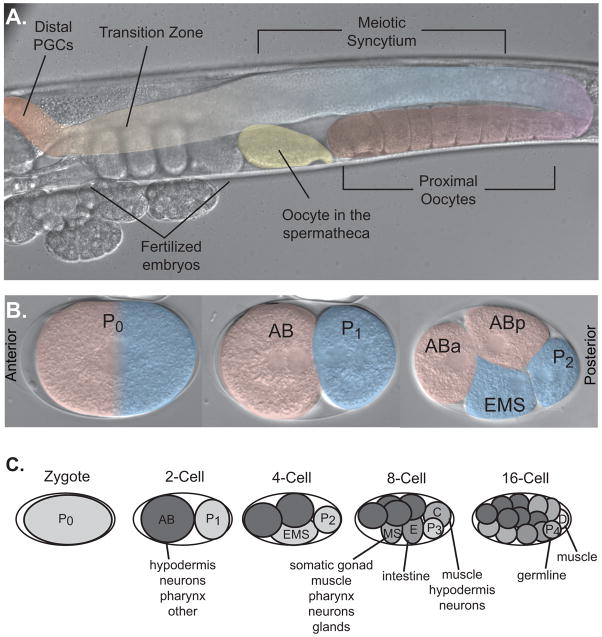
Figure 1.
Anatomy of C. elegans hermaphrodite reproduction. A. A single gonad arm from a hermaphrodite worm is shown. The gonad is highlighted in false color. The distal arm of the germline contains mitotically dividing progenitor cells (red). There is a transition (orange) from mitosis to meiosis concurrent with a transition from a single-celled state to a syncitial region (blue). Meoitic nuclei recellularize, first to form spermatocytes in the L4 larval stage that are stored in the spermatheca (yellow), and then switch to form oocytes (purple) at the onset of adulthood. B. Pattern of the first two cellular divisions after fertilization. The anterior and posterior poles are marked. C. Pattern of division and early lineage of embryogenesis. Several founder cells are established early in embryogenesis that go on to form different tissues in the adult. Adapted with permission from [50].
Nine additional puf genes, termed puf-3 to puf-12, are present in the C. elegans genome. Most have distinct biological functions defined by phenotypic differences, mRNA target specificity, or expression pattern. Three of these genes—puf-5, puf-6, and puf-7—are redundantly required for embryonic viability and oocyte maturation [14]. They prevent premature translation of glp-1 mRNA in oocytes. PUF-8 promotes mitosis in germline progenitor cells, similar to FBF, but binds to RNA with different sequence specificity and as such likely regulates a distinct set of target mRNAs. PUF-9 regulates hunchback-like (hbl-1) mRNA in the hypodermis and ventral nerve cord [15]. RNAi screens reveal important roles for PUF-3, PUF-4, PUF-11, and PUF-12 in oogenesis and early embryonic development, but their critical mRNA targets have not been identified [16, 17]. In the following sections, we review a recently published crystal structure of FBF and highlight biochemical experiments that define differences in RNA recognition in this family [18].
Biochemical insights into PUF binding specificity
Wickens and co-workers have dissected the RNA binding properties of several PUF proteins [10, 19–21]. The consensus sequence recognized by FBF, termed the FBF binding element (FBE), is 5′-UGURNNAUA-3′ [10]. The FBE is nine nucleotides in length and is partially degenerate at three positions. FBEs are present in the 3′-UTR of fem-3, gld-1, and numerous other mRNAs regulated by FBF in the germline. Mutation of the FBE in the 3′-UTR of fem-3 leads to derepression of FEM-3 and failure to switch from spermatogenesis to oogenesis [11].
PUF-8 and PUF-9, on the other hand, recognize an eight nucleotide consensus identical to that bound by human Pum1 (5′-UGUANAUA-3′) termed the Nanos Response Element (NRE) [21, 22]. The NRE is similar to the FBE but is a single nucleotide shorter. This difference is critical, as FBF discriminates between these two elements by more than 30-fold. Intriguingly, the specificity of PUF-8 can be converted to that of FBF by swapping a 64-amino acid fragment in the middle of the PUF domain, demonstrating that this region is critical for specificity.
PUF-5 and PUF-6/7 recognize a longer, partially degenerate consensus motif termed the PUF-5 Binding Element (5BE: 5′-CyCUGUAyyyUGU-3′, where y is a pyrimidine) [20]. PUF-11 binds three sets of RNA targets, 5′-CUGUGAAUA-3′, 5′-CUGUANAAUA-3′ and 5′-NUGUNAAAUA-3′, suggesting multiple modes of RNA recognition through a mechanism that is not immediately apparent [19]. Clearly, these experiments show that the nematode PUF family has diverged to expand the repertoire of sequences recognized by the PUF domain. Recent crystal structures begin to address the molecular basis for this variance.
Crystal structures of PUF proteins
The first structures of a PUF domain, including Drosophila Pumilio and human Pum1, were determined independently in 2001 [23, 24]. The structures revealed an architecture of eight repeat motifs comprised of three alpha helices. The repeats pack against each other to form an extended curved structure that vaguely resembles a banana. A subsequent structure of human Pum1 bound to RNA demonstrates that the concave surface comprises the RNA binding interface, where each repeat recognizes a single nucleotide (Fig. 2A) [22]. The amino acids that face the concave surface define the nucleotide specificity at each repeat, which has been reviewed previously [25]. This architecture immediately suggests a model where PUF proteins bind to RNA with modular specificity, such that changing the order of the repeats could modify RNA-binding specificity. Several experiments with chimeric PUF proteins support this model and suggest this domain is particularly amenable to protein engineering [19–21].
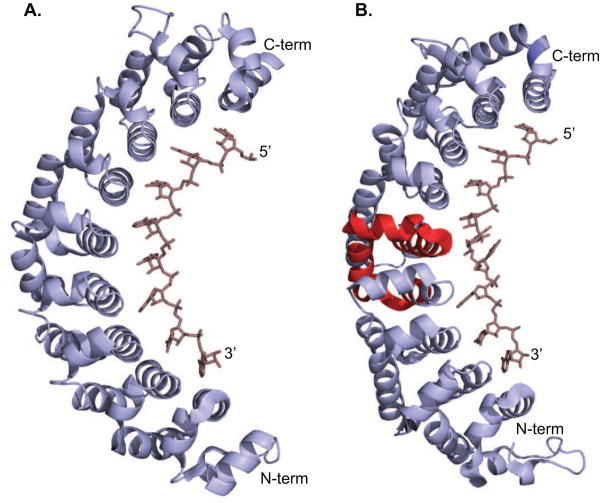
Figure 2.
Crystal structures of PUF domain proteins bound to RNA. A. The structure of human Pum1 bound to the NRE. Each PUF repeat specifies a single nucleotide [22]. B. Structure of C. elegans FBF bound to the FBE, similar to the NRE but containing an additional nucleotide [18]. Here, the reduced curvature of the protein enables recognition of a nine nucleotide consensus with an eight repeat PUF domain. One nucleotide flips away from the binding surface of the protein, and eight form interactions with each repeat. The region of the protein that confers FBF-like specificity when swapped into PUF-8 is shown in red.
All of the nematode PUF proteins are comprised of eight repeats, but many bind to a consensus element that contains more than eight nucleotides. To gain insight into the structural basis for recognition of longer elements by this domain, Hall and coworkers crystallized FBF-2 in complex with six different RNA sequences, including four naturally occuring sites [18]. This study reveals that FBF has an elongated structure with less curvature relative to other PUF domain proteins (Fig 2B). This elongated structure enables a single base to flip out and point away from the protein without affecting interactions with the other eight nucleotides. Thus, a slight variance of the curvature of the overall structure, governed by repeats 4–6, has a profound impact on the RNA-binding specificity.
It is possible that curvature driven base flipping accounts for the multiple modes of RNA recognition by PUF-11 [19]. Each mode contains two conserved regions, including a UGU trinucleotide and an element comprised of AAAUA. These may represent eight —core interactions with this protein. Each mode contains an insertion of one or more nucleotides into a distinct position within this core. If the inserted nucleotides flip out, a model similar to FBF would resolve the multiple modes of binding. The incomplete degeneracy of the inserted nucleotides may be partially explained by differential stacking free energy with neighboring nucleotides. A similar model could be proposed for PUF-5/6/7, where eight nucleotides are specified unambigously, and five more nucleotides are partially degenerate [20]. More structural work is needed to assess this hypothesis and define the basis for the variance in PUF specificity. It is also important to assess whether conformational flexibility contributes to binding specificity.
TTP-like CCCH tandem zinc finger proteins
TZF proteins in C. elegans early embryogenesis
TTP is a mammalian RNA-binding protein that regulates the immune response by promoting the turnover of the mRNA encoding the pro-inflammatory cytokine TNF-alpha [26, 27]. TTP is an AU-rich element (ARE) binding protein, which coordinate the stability of mRNAs containing extended repeats of UAUU in their 3′ UTRs. TTP has two CX8CX5CX3H zinc finger motifs. Each motif binds to a single UAUU repeat [28].
There are several TTP homologs in the C. elegans genome, many of which are required for worm fertility. A cascade of TZF proteins, including OMA-1/2, MOE-3, MEX-5/6, MEX-1, POS-1, and PIE-1, guide the progression from the oocyte to embryo. OMA-1/2 and MOE-3 are partially redundant factors that promote oocyte maturation, and inhibit embryonic gene expression prior to fertilization [4–6, 29]. MEX-5/6 are required for anterior patterning in the early embryo [5]. They are translated from maternally supplied mRNA shortly after fertilization, and migrate to the anterior of the embryo prior to the first cellular division. POS-1, PIE-1, and MEX-1 are also translated after fertilization, but accumulate in the posterior of the embryo in a pathway that depends upon MEX-5/6 anterior localization [30, 31]. All three proteins are required for posterior patterning and segregation of germline and somatic lineages, but have non-redundant functions [4, 29]. In addition to these well studied examples, there are eight additional TZF genes in the C. elegans genome. DCT-13 and possibly Y116A8C.20 promote germline tumor formation in a sensitized genetic background, while CCCH-1, CCCH-2, CCCH-5, F38C2.7, Y116A8C.19, and C35D6.4 have no known function [32].
NMR structure of a TZF family protein
Only one structure of a TZF protein has been determined to date. Wright and coworkers determined the solution structure of the Tis11D bound to the RNA sequence 5′-UUAUUUAUU-3′ (Fig. 3) [33]. Tis11D is a mammalian paralog of TTP that regulates mRNA stability in response to growth factors [3]. It binds to RNA with identical specificity to TTP. The structure reveals that each CX8CX5CX3H finger motif independently recognizes the four nucleotide sequence UAUU. A conserved motif with the sequence (R/K)YKTEL lies upstream of the first cysteine of each finger. This region makes numerous contacts with the RNA. These are primarily comprised of hydrogen bonds between the protein backbone and the Watson-Crick edges of the bases, and van der Waals interactions that specify the shape of the base at each position. In addition, the side chains of two conserved aromatic amino acids form stacking interactions between adjacent RNA bases at two positions within each finger. These amino acids are essential for high affinity binding, and may contribute to specificity through differential stacking propensity. This structure has thus far provided our only glimpse into RNA recognition by this class of RNA-binding proteins, and as such serves as the primary frame of reference for the interpretation of experiments for related factors.
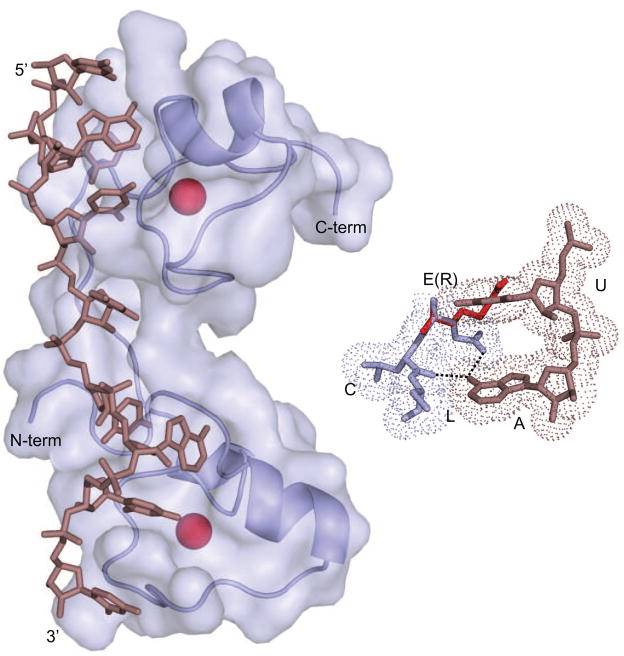
Figure 3.
NMR structure of Human Tis11D bound to RNA [33]. Each zinc finger domain independently recognizes the sequence UAUU through a combination of base-specific hydrogen bonding interactions and stacking interactions driven by aromatic side chains. The inset shows recognition of adenosine in the N-terminal finger. Three amino acids (blue), glutamate, leucine, and cysteine, come together to form an adenosine (violet) recognition pocket. The exocyclic amine hydrogen bonds with the glutamate side chain and the backbone carbonyl of the leucing. In MEX-5, the glutamate is replaced with an arginine (red), which is proposed to flip away from the adenosine and form non-specific interactions with the backbone of adjacent nucleotides [35].
Biochemical insights into nematode TZF binding specificity
In most cases, the RNA-binding activity of nematode TZF proteins has not been investigated in detail. The two exceptions are MEX-5 and POS-1, which bind to RNA but with different specificity compared to TTP, Tis11D, and each other [34, 35]. MEX-5 binds with high affinity but relaxed specificity to any uridine rich sequence, including polyuridine. This contrasts with TTP which binds >80-fold more tightly to AREs than polyuridine. POS-1 binds with high affinity to a consensus termed the POS-1 recognition element (PRE: 5′-UA(U2–3)RD(N1–3)G-3′, where R is any purine, D is A, G, or U, and N is any base). Compared to TTP binding sequence, the PRE is more degenerate and specifies three purines instead of two.
In Tis11D, three contiguous amino acids in each finger form an adenosine recognition pocket: glutamate, leucine, and the first cysteine of the CCCH motif (Fig. 3) [33]. The glutamate side chain accepts a hydrogen bond from the exocyclic amine of the adenosine. The leucine and the cysteine are conserved in both MEX-5 and POS-1, but the glutamate is not. In MEX-5, the analogous amino acids are arginine in the first finger and a lysine in the second. Mutating both to glutamate confers TTP-like specificity to MEX-5, suggesting they are critical specificity determinants [35]. In POS-1, an alanine and a valine occupy the analogous positions. It is not clear how these amino acids contribute to the differences in POS-1 specificity, or how this protein specifies three purines compared to two. Structural data are needed to resolve this problem. One nematode TZF protein, CCCH-1, has two glutamate residues in the analogous position similar to TTP. The rest have basic residues, small hydrophobic residues, or some combination thereof. It is expected that CCCH-1 will bind to RNA with TTP-like specificity, and that the others will bind to RNA with hybrid specificity, but this has not been experimentally demonstrated.
The worm Argonaute proteins
Biological functions of nematode Argonaute proteins
Argonautes are the primary effectors of small RNA silencing pathways, which have been the subject of intense investigation [36]. Twenty-seven Argonaute genes are annotated in the C. elegans genome. These fall into three paralogous groupings: 1) similar to Arabidopsis thaliana AGO1, 2) similar to Drosophila melanogaster PIWI, and 3) worm-specific argonautes (WAGOs) [37]. Some Argonaute proteins catalyze the cleavage of target RNAs recognized by a small RNA guide. Others function in regulation of mRNA translation in the microRNA pathway. And some are implicated in transcriptional gene silencing through modification of chromatin state.
The large number and apparent diversity of nematode Argonautes suggests a high degree of specialization. Canonical RNA interference triggered by exogenous double strand RNA (dsRNA) is mediated by RDE-1 [7]. Endogenously encoded siRNAs, which are proposed to control cellular homeostasis, are loaded into ERGO-1 [8]. SAGO-1 and SAGO-2 (members of the WAGO clade) are proposed to function in a systemic spreading mechanism, absent in flies and mammals, that leads to silencing of sequences upstream from loci targeted by primary small RNAs [8, 38–40]. ALG-1 and ALG-2 load microRNAs required for the temporal regulation of pattern formation during development [41]. The PIWI clade member PRG-1 loads 21U-RNAs and is required for germline maintenance and fertility [42]. More recently, WAGO-1 was shown to repress specific genes, transposons, pseudogenes, and cryptic loci in conjunction with a class of guide sequences termed the 22G-RNAs [43], and CSR-1 was shown to target euchromatic domains of the genome to enforce appropriate assembly of kinetochores and to facilitate segregation of the holocentric chromosomes [44, 45]. Several additional argonautes do not have clearly delineated function.
Argonaute structure
Eukaryotic Argonautes consist of four domains: the N-terminal, PAZ, MID, and PIWI domains [37]. A series of recent crystal structures of Argonaute-like RNA endonucleases from hyperthermophilic bacteria begin to define the basis for guide and target recognition as well as the mechanism of site-specific cleavage [46, 47]. As predicted, the structure shows that the 5′ monophosphate of the single stranded guide is lodged between the interface of the MID and PIWI domain while the 3′ end is held by the PAZ domain (Fig. 4) [46]. Target association is proposed to occur in two steps. First, the seed region of the guide pairs to the target [48]. Pairing is limited to the seed due to the —doubly anchored conformation of the guide. Once seed pairing is achieved, the helix propogates leading to dissociation of the 3′ end of the guide from the PAZ domain. This remodeling step positions the target adjacent to the metal-coordinated catalytic residues in the PIWI domain required for target cleavage. A minimum of fifteen contiguous base pairs is necessary to mediate the remodeling event, which explains why most microRNAs—which typically recognize their targets through incomplete pairing—do not guide cleavage of their mRNA targets.
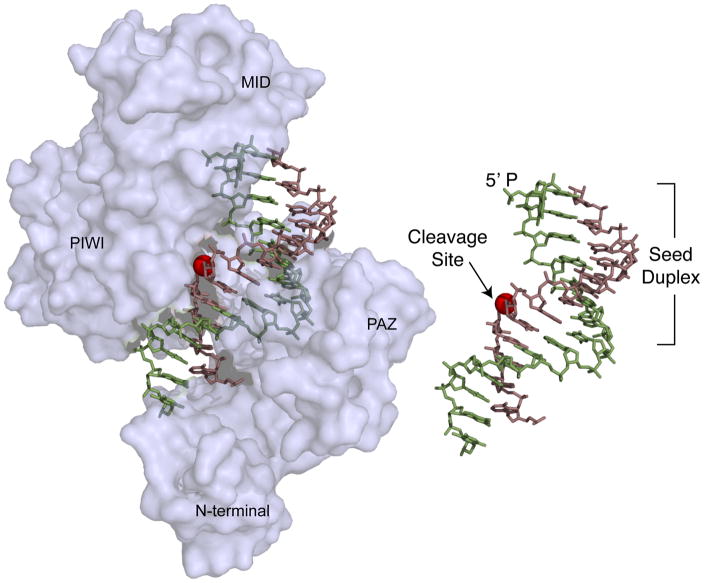
Figure 4.
Crystal structure of a Thermus thermophilus DNA-dependent RNA endonuclease related to eukaryotic Argonaute proteins [46]. The structure shows the protein bound to a DNA guide (green) and a target RNA (violet). The 5′ monophosphate end of the guide is anchored in a cleft between the MID and the PAZ domains. Nucleotides 2–8, which comprise the seed, are exposed on the surface of the protein complex and as such are positioned for substrate recognition. Pairing of the 3′-end of the guide with the target RNA aligns the scissile phosphate (red sphere) with the catalytic residues in the protein. The inset shows the guide target interaction in the absence of protein for clarity.
Implications for nematode-specific Argonautes
The features of the bacterial Argonaute structures shed light on the function of worm-specific Argonautes. WAGOs involved in systemic spreading lack the residues that coordinate the divalent metal ion required for target cleavage. This suggests that they do no regulate their RNA targets by guide directed cleavage [8]. However, they do load guide sequences that are at least in principle capable of completely base pairing with their target RNAs. It is not known if these proteins direct a two-step recognition process to bind to their RNA targets. If not, then complementarity between the 3′-end of the guide and the target RNA may be dispensable for function, increasing the number of potential targets as well as increasing the opportunity for off target effects. If so, then cleavage-independent RNA silencing must be possible in a conformation that includes significant pairing between the guide and the target.
In contrast, CSR-1 is capable of guide-directed RNA cleavage in worm extracts, implying but not proving that cleavage activity plays a role in their biological function [44]. The guide RNAs recognized by CSR-1 harbor a 5′-triphosphate moiety, unique among all Argonaute proteins studied. It is not clear how CSR-1 preferentially accommodates a triphosphate moiety in place of the canonical 5′-monophosphate. The 5′-triphosphate group enhances cleavage activity in extracts relative to the identical sequence with a 5′-monophosphate, suggesting this group functions in some aspect of target cleavage [49]. More work is needed to delineate the basis for 5′-end discrimination by CSR-1.
Concluding remarks
The function of RNA-binding proteins is dictated by their structure. For RNA-binding protein families where a common domain has evolved new binding specificity, it is important to understand how structural changes define the basis for novel function. While genetics and biochemical experiments can identify the critical sequence elements, they cannot in most cases address how these elements contribute to novel function in a mechanistic sense. Thus, it is important to continue to put effort into structural studies beyond the first structure in an RNA-binding protein family. Structural experiments can provide key insights needed to understand biological function.
Acknowledgments
The authors would like to thank Brian Farley, John Pagano, and members of the Zamore lab for helpful comments concerning this manuscript, and Dr. Philip Zamore for allowing W.L.M. the opportunity to contribute to this article. S.P.R. is supported by NIH R01 GM081422 and a Basil O’Connor Starter Scholar Award from the March of Dimes.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lehmann R, Nusslein-Volhard C. The maternal gene nanos has a central role in posterior pattern formation of the Drosophila embryo. Development. 1991;112:679–691. doi: 10.1242/dev.112.3.679. [DOI] [PubMed] [Google Scholar]
- 2.Zhang B, Gallegos M, Puoti A, Durkin E, Fields S, Kimble J, Wickens MP. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature. 1997;390:477–484. doi: 10.1038/37297. [DOI] [PubMed] [Google Scholar]
- 3.Varnum BC, Ma QF, Chi TH, Fletcher B, Herschman HR. The TIS11 primary response gene is a member of a gene family that encodes proteins with a highly conserved sequence containing an unusual Cys-His repeat. Mol Cell Biol. 1991;11:1754–1758. doi: 10.1128/mcb.11.3.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tabara H, Hill RJ, Mello CC, Priess JR, Kohara Y. pos-1 encodes a cytoplasmic zinc-finger protein essential for germline specification in C. elegans. Development. 1999;126:1–11. doi: 10.1242/dev.126.1.1. [DOI] [PubMed] [Google Scholar]
- 5.Schubert CM, Lin R, de Vries CJ, Plasterk RH, Priess JR. MEX-5 and MEX-6 function to establish soma/germline asymmetry in early C. elegans embryos. Mol Cell. 2000;5:671–682. doi: 10.1016/s1097-2765(00)80246-4. [DOI] [PubMed] [Google Scholar]
- 6.Detwiler MR, Reuben M, Li X, Rogers E, Lin R. Two zinc finger proteins, OMA-1 and OMA-2, are redundantly required for oocyte maturation in C. elegans. Dev Cell. 2001;1:187–199. doi: 10.1016/s1534-5807(01)00026-0. [DOI] [PubMed] [Google Scholar]
- 7.Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- 8.Yigit E, Batista PJ, Bei Y, Pang KM, Chen CC, Tolia NH, Joshua-Tor L, Mitani S, Simard MJ, Mello CC. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127:747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 9.Crittenden SL, Bernstein DS, Bachorik JL, Thompson BE, Gallegos M, Petcherski AG, Moulder G, Barstead R, Wickens M, Kimble J. A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature. 2002;417:660–63. doi: 10.1038/nature754. [DOI] [PubMed] [Google Scholar]
- 10.Bernstein D, Hook B, Hajarnavis A, Opperman L, Wickens M. Binding specificity and mRNA targets of a C. elegans PUF protein, FBF-1. RNA. 2005;11:447–458. doi: 10.1261/rna.7255805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahringer J, Kimble J. Control of the sperm-oocyte switch in Caenorhabditis elegans hermaphrodites by the fem-3 3′ untranslated region. Nature. 1991;349:346–38. doi: 10.1038/349346a0. [DOI] [PubMed] [Google Scholar]
- 12.Francis R, Barton MK, Kimble J, Schedl T. gld-1, a tumor suppressor gene required for oocyte development in Caenorhabditis elegans. Genetics. 1995;139:579–606. doi: 10.1093/genetics/139.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Francis R, Maine E, Schedl T. Analysis of the multiple roles of gld-1 in germline development: interactions with the sex determination cascade and the glp-1 signaling pathway. Genetics. 1995;139:607–30. doi: 10.1093/genetics/139.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lublin AL, Evans TC. The RNA-binding proteins PUF-5, PUF-6, and PUF-7 reveal multiple systems for maternal mRNA regulation during C. elegans oogenesis. Dev Biol. 2007;303:635–649. doi: 10.1016/j.ydbio.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 15.Nolde MJ, Saka N, Reinert KL, Slack FJ. The Caenorhabditis elegans pumilio homolog, puf-9, is required for the 3′UTR-mediated repression of the let-7 microRNA target gene, hbl-1. Dev Biol. 2007;305:551–563. doi: 10.1016/j.ydbio.2007.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- 17.Sonnichsen B, Koski LB, Walsh A, Marschall P, Neumann B, Brehm M, Alleaume AM, Artelt J, Bettencourt P, Cassin E, et al. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 2005;434:462–469. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- ••18.Wang Y, Opperman L, Wickens M, Hall TM. Structural basis for specific recognition of multiple mRNA targets by a PUF regulatory protein. Proc Natl Acad Sci USA. 2009;106:20186–20191. doi: 10.1073/pnas.0812076106. This article demonstrates the structural basis for differential binding site consensus sequence length between PUF proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••19.Koh YY, Opperman L, Stumpf C, Mandan A, Keles S, Wickens M. A single C. elegans PUF protein binds RNA in multiple modes. RNA. 2009;15:1090–1099. doi: 10.1261/rna.1545309. The diversity of sequence recognized by several nematode PUF domain proteins is explored in this research paper. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stumpf CR, Kimble J, Wickens M. A Caenorhabditis elegans PUF protein family with distinct RNA binding specificity. RNA. 2008;14:1550–1557. doi: 10.1261/rna.1095908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Opperman L, Hook B, DeFino M, Bernstein DS, Wickens M. A single spacer nucleotide determines the specificities of two mRNA regulatory proteins. Nat Struct Mol Biol. 2005;12:945–951. doi: 10.1038/nsmb1010. [DOI] [PubMed] [Google Scholar]
- 22.Wang X, McLachlan J, Zamore PD, Hall TM. Modular recognition of RNA by a human pumilio-homology domain. Cell. 2002;110:501–12. doi: 10.1016/s0092-8674(02)00873-5. [DOI] [PubMed] [Google Scholar]
- 23.Wang X, Zamore PD, Hall TM. Crystal structure of a Pumilio homology domain. Mol Cell. 2001;7:855–65. doi: 10.1016/s1097-2765(01)00229-5. [DOI] [PubMed] [Google Scholar]
- 24.Edwards TA, Pyle SE, Wharton RP, Aggarwal AK. Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell. 2001;105:281–29. doi: 10.1016/s0092-8674(01)00318-x. [DOI] [PubMed] [Google Scholar]
- 25.Lu G, Dolgner SJ, Hall TM. Understanding and engineering RNA sequence specificity of PUF proteins. Curr Opin Struct Biol. 2009;19:110–115. doi: 10.1016/j.sbi.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blackshear PJ. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem Soc Trans. 2002;30:945–952. doi: 10.1042/bst0300945. [DOI] [PubMed] [Google Scholar]
- 27.Lai WS, Carballo E, Strum JR, Kennington EA, Phillips RS, Blackshear PJ. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol Cell Biol. 1999;19:4311–4323. doi: 10.1128/mcb.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brewer BY, Malicka J, Blackshear PJ, Wilson GM. RNA sequence elements required for high affinity binding by the zinc finger domain of tristetraprolin: conformational changes coupled to the bipartite nature of Au-rich MRNA-destabilizing motifs. J Biol Chem. 2004;279:27870–27877. doi: 10.1074/jbc.M402551200. [DOI] [PubMed] [Google Scholar]
- 29.Mello CC, Draper BW, Krause M, Weintraub H, Priess JR. The pie-1 and mex-1 genes and maternal control of blastomere identity in early C. elegans embryos. Cell. 1992;70:163–176. doi: 10.1016/0092-8674(92)90542-k. [DOI] [PubMed] [Google Scholar]
- 30.Cuenca AA, Schetter A, Aceto D, Kemphues K, Seydoux G. Polarization of the C. elegans zygote proceeds via distinct establishment and maintenance phases. Development. 2003;130:1255–1265. doi: 10.1242/dev.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reese KJ, Dunn MA, Waddle JA, Seydoux G. Asymmetric segregation of PIE-1 in C. elegans is mediated by two complementary mechanisms that act through separate PIE-1 protein domains. Mol Cell. 2000;6:445–455. doi: 10.1016/s1097-2765(00)00043-5. [DOI] [PubMed] [Google Scholar]
- 32.Pinkston-Gosse J, Kenyon C. DAF-16/FOXO targets genes that regulate tumor growth in Caenorhabditis elegans. Nat Genet. 2007;39:1403–1409. doi: 10.1038/ng.2007.1. [DOI] [PubMed] [Google Scholar]
- 33.Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat Struct Mol Biol. 2004;11:257–264. doi: 10.1038/nsmb738. [DOI] [PubMed] [Google Scholar]
- 34.Farley B, Pagano JM, Ryder S. RNA target specificity of the embryonic cell fate determinant. POS-1 RNA. 2008;14:2685–2697. doi: 10.1261/rna.1256708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pagano JM, Farley B, McCoig LM, Ryder S. Molecular basis of RNA recognition by the embryonic polarity determinant MEX-5. J Biol Chem. 2007;282:8883–8894. doi: 10.1074/jbc.M700079200. [DOI] [PubMed] [Google Scholar]
- 36.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 38.Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- 39.Sijen T, Steiner FA, Thijssen KL, Plasterk RH. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007;315:244–247. doi: 10.1126/science.1136699. [DOI] [PubMed] [Google Scholar]
- 40.Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- 41.Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 42.Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, et al. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell. 2008;31:67–78. doi: 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gu W, Shirayama M, Conte DJ, Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ, et al. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell. 2009;36:231–244. doi: 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••44.Claycomb JM, Batista PJ, Pang KM, Gu W, Vasale JJ, van Wolfswinkel JC, Chaves DA, Shirayama M, Mitani S, Ketting RF, et al. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell. 2009;139:123–134. doi: 10.1016/j.cell.2009.09.014. The biological role of CSR-1, an interesting WAGO with a function in chromosome segregation, is described in this article. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Wolfswinkel JC, Claycomb JM, Batista PJ, Mello CC, Berezikov E, Ketting RF. CDE-1 affects chromosome segregation through uridylation of CSR-1-bound siRNAs. Cell. 2009;139:135–148. doi: 10.1016/j.cell.2009.09.012. [DOI] [PubMed] [Google Scholar]
- ••46.Wang Y, Juranek S, Li H, Sheng G, Wardle GS, Tuschl T, Patel DJ. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009;461:754–761. doi: 10.1038/nature08434. The structural basis for Argonaute guide–target recognition and evidence for the two-state binding model is presented in this paper. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 48.Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- 49.Aoki K, Moriguchi H, Yoshioka T, Okawa K, Tabara H. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J. 2007;26:5007–5019. doi: 10.1038/sj.emboj.7601910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Farley B, Ryder S. Regulation of maternal mRNAs in early development. Critical Reviews in Biochemistry and Molecular Biology. 2008;43:135–162. doi: 10.1080/10409230801921338. [DOI] [PubMed] [Google Scholar]