Abstract
Helicases of the superfamily (SF) 1 and 2 are involved in virtually all aspects of RNA and DNA metabolism. SF1 and SF2 helicases share a catalytic core with high structural similarity, but different enzymes even within each superfamily perform a wide spectrum of distinct functions on diverse substrates. To rationalize similarities and differences between these helicases, we outline a classification based on protein families that are characterized by typical sequence, structural and mechanistic features. This classification complements and extends existing SF1 and SF2 helicase categorizations and highlights major structural and functional themes for these proteins. We discuss recent data in the context of this unifying view of SF1 and SF2 helicases.
Introduction
Helicases use ATP to bind or remodel nucleic acids, nucleic acid-protein complexes, or both [1–4]. All forms of cellular life and many viruses encode helicases, which constitute one of the largest class of enzymes [5,6]. Virtually all biological processes involving DNA or RNA employ one or more helicases [4,7–9]. Defects in their function as well as deregulated expression of these proteins have been linked to numerous diseases including cancers, developmental defects, and neurodegenerative diseases [10,11].
Two distinct types of helicases exist, those forming toroidal, predominantly hexameric structures, and those that do not [3]. Pioneering sequence analysis by Gorbalenya and Koonin and more recent comparative structural and functional analysis by Wigley and co-workers showed that helicases can be classified in several superfamilies (SFs) [3,12]. The toroidal enzymes comprise SFs3 to 6, and the non-ring forming ones comprise SFs1 and 2 [3].
The catalytic cores of SF1 and SF2 helicases share almost identical folds and extensive structural similarity [3]. Within each superfamily, the level of structural conservation is still higher [3,7]. Notwithstanding, different helicases of each superfamily perform a wide spectrum of functions on diverse substrates ranging from chromosomal DNA over ribosome pre-cursors to small, non-coding RNAs [2,13,14]. Classical helicase function, i.e., ATP-driven duplex unwinding, is not always the primary physiological function of SF1 and SF2 enzymes [2,7,15,16], and even for helicases that unwind duplexes, there are significant mechanistic differences. Unwinding can be based on translocation on the nucleic acid [2–4], but there is also translocation without unwinding [17–20], and unwinding without translocation [21–24].
To rationalize functional differences and structural similarity of SF1 and SF2 helicases, classifications have been proposed based on sequence, structural and mechanistic features [3,12]. However, none of these classifications extended to all members of the SF1 and 2, because gaps in structural and functional information precluded a comprehensive and systematic view that integrated sequence, structure, and mechanism. Some of these knowledge gaps have been narrowed by recent data and there is renewed interest in a systematic and inclusive SF1/SF2 classification. Here we outline such a classification, based on distinct protein families that are characterized by typical sequence, structural and mechanistic features. We briefly describe how this classification highlights major structural and functional themes of SF1 and 2 helicases, and we discuss recent data in the context of this unifying view of these enzymes.
SF1 and SF2 Helicase Families
The hallmark of SF1 and SF2 helicases is a conserved helicase core consisting of two similar protein domains that resemble the fold of the recombination protein RecA [3]. This helicase core contains characteristic sequence motifs, based on which the original subdivision of helicases in superfamilies was accomplished [12]. It has long been noted that both SF1 and SF2 encompass defined protein families with distinct sequence, structural, and mechanistic features, such as the DEAD-box and the Swi/Snf families in SF2, or the Pif1-like family in SF1 [20,25,26]. Yet, no comprehensive classification of these families has been reported for either superfamily.
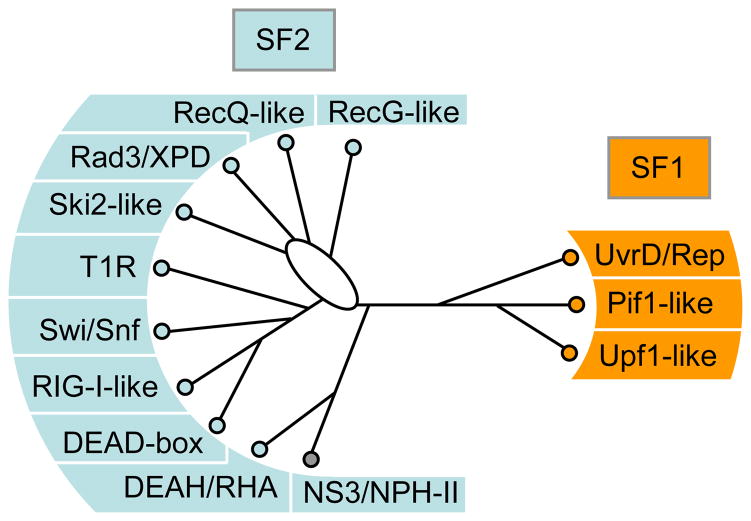
To devise a family classification for the entirety of both superfamilies, we aligned the sequences of the helicase cores of all SF1 and SF2 proteins from human, Saccharomyces cerevisiae, E.coli, and select viral proteins (Suppl. Fig. S1). We then performed phylogenetic analysis of this alignment. Consistent clustering of proteins with similar sequence characteristics was found (Fig. 1, Suppl. Fig. S2). We identified several well defined clusters of different sizes in both superfamilies. Some of these clusters encompass proteins that have long been viewed as family, such as the DEAD-box and the RecQ-like proteins [26,27]. Extrapolating from this concept, we suggest calling clusters with more than three proteins from a single organism (e.g., DEAD-box proteins) a family, and clusters with two proteins from a single organism a group. No specific term appears necessary for proteins that are encoded by a single ORF in a given organism, even though these proteins may be highly conserved throughout evolution (e.g. Suv3). The distinction between groups and families seems useful because further substructure (subfamilies) can exist in a family with 3 or more proteins, but not in a group with two proteins. We suggest an exception for viral SF2 helicases related to NS3/NPH-II, which we propose to view as a group, even though these proteins are usually encoded only by a single ORF per virus. However, these proteins cluster despite originating from very different viruses.
Figure 1. The Families of the SF1 and SF2 helicases.
Schematic, unrooted cladogram showing the three identified families of the SF1 (right), and the 9 families and one group of the SF2 (left). A comprehensive list of proteins in each family in human, S.cerevisiae, and E.coli is given in the Supplementary Tables ST1 and ST2. Families were identified by a combination of phylogenetic analysis of the alignment of all SF1 and SF2 proteins from human, S.cerevisiae, E.coli, and selected viruses (Suppl. Fig. S1), and scoring for presence or absence of distinct sequence features and characteristic domain organization (for more detail, see Suppl. Figs. S1–3) and corresponding captions). Assignment of proteins to a family was highly robust. The phylogenetic relationships between the families were more ambiguous and tree topologies varied for some branches according to method and parameters used for generating the trees (Suppl. Fig. S2). The cladogram shown represents an average of multiple trees generated by different means (Suppl. Fig. S2). Branch lengths are not to scale. The oval indicates significant uncertainty in the tree topology in this region. Families were named according to names in use, or according to prominent members. Families named after a single protein were termed – like (i.e., Ski2-like). Non-standard abbreviations are as follows: T1R – type 1 restriction enzymes, RHA – RNA helicase A.
According to this family/group definition, we identified 9 families and one group in the SF2 and three families in the SF1 (Fig. 1). Several proteins (e.g., Suv3) do not fall into a family or group, despite a high level of conservation in eukaryotes or bacteria (Tabs. ST1, ST2). The families were named according to terms already in use (e.g. DEAD-box), or according to a prominent member (e.g., Ski2-like). Our analysis did not include a comprehensive cross-section of viral SF1 and SF2 proteins [9,28] (see Suppl. Fig. S1 for explanation). It is very likely that further viral proteins constitute additional groups, analogous to the NS3/NPH-II group. Moreover, future surveys of SF1 and SF2 proteins from more organisms may reveal additional groups or families. In this sense, the presented family structure should be viewed as minimal and basic, although it is comprehensive for human, S.cerevisiae, and E.coli. (Fig. 1).
The identified families of both SF1 and SF2 correspond remarkably well to the classification based on translocation and unwinding polarity proposed by Wigley and coworkers [3], (Tab. 1). The family-based system now extends this functional classification by including all SF2 proteins and by adding a sequence dimension. The presented family structure is further consistent with previous phylogenetic analyses of subsets of SF2 proteins [25,29–31], and with the original sequence analysis and classification by Gorbalenya and Koonin [12].
Table 1.
Mechanistic characteristics of the SF1 and SF2 families.
| Number of Proteins(a) | Nucleic Acid Preference(b) | NTP Preference(c) | Unwinding Polarity(d) | Functional Classification(e) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H | Y | E.c. | D | R | A | G | C | U/T | 3′ 5′ | 5′ 3′ | ||
| DEAD-box | 37 | 26 | 4 | + | + | + | + | n/a (f) | ||||
| DEAH/RHA | 15 | 7 | 2 | + | + | + | + | + | + | + | (+) (g) | SF2Aα (g) |
| NS3/NPH-II | − | − | − | + | + | + | + | + | + | + | SF2Aα | |
| Ski2-like | 7 | 5 | 2 | + | + | + | + | SF2Aα | ||||
| RIG-I-like | 5 | 1 | − | + | + | + | + | SF2Aα/β(h) | ||||
| RecQ-like | 5 | 2 | 1 | + | + | + | * (i) | SF2Aα | ||||
| RecG-like | − | − | 3 | + | + | + | SF2Aβ | |||||
| Swi/Snf | 28 | 16 | 1 | + | + | n/a | SF2β | |||||
| T1R | − | − | 3 | + | + | n/a | SF2β | |||||
| Rad3/XPD | 5 | 2 | 2 | + | + | + | SF2Bα | |||||
| UvrD/Rep | 2 | 2 | 4 | + | + | + | SF1Aα | |||||
| Pif1-like | 2 | 2 | 2 | + | + | + | SF1Bα | |||||
| Upf1-like | 11 | 5 | 2 | + | + | + | + | SF1Bα | ||||
The number of proteins in each family was determined by mining the Swissprot and the Genbank databases (for more detail, see Suppl. Fig. S1, and Tab. TS1, TS2). (H – human, Y – S.cerevisiae, E.c. – E.coli).
Nucleic acid preference indicates the ability to unwind either DNA (D) or RNA (R). In families with both DNA and RNA helicases, individual proteins mostly function either as RNA or DNA helicase. Only some viral and some Upf1-like proteins have been shown to unwind both, DNA and RNA duplexes.
NTP preference indicates the ability of family members to hydrolyze all nucleotides with similar efficiency, or to be specific for adenosine triphosphates.
The unwinding polarity indicates whether proteins from a given family require duplex substrates with a single straded overhang 3′ (3′ → 5′) or 5′ (5′ → 3′) to the duplex region.
Functional classification is assigned according to the system proposed by Wigley and co-workers [3].
DEAD-box proteins, which do not unwind duplexes with defined polarity [23,24], are not classified in this system.
Several DEAH proteins have been shown to unwind with dual polarities, although 3′ → 5′ is generally preferred.
RIG-I has been shown to perform both polar unwinding (SF2Aα) [78] and translocation on duplex RNA (SF2β) [19].
Several RecQ-like proteins are specific for distinct DNA structures, and it is not clear whether all family members display unique polarity on these substrates.
Proteins within each of the identified families or groups share high sequence conservation and family/group-typical sequence characteristics (Suppl. Figs. S1, S3), as well as distinct structural features, as discussed below. In addition, proteins within a family or group display family/group-typical mechanistic features, including promiscuous NTP usage vs. specificity for adenosine triphosphates, the ability to unwind duplexes, and unwinding polarity (Tab. 1). The notion of family-typical mechanistic features is important, because it might guide specific experimental approaches for the characterization of SF1 or 2 helicases for which only sequence information is available.
Characteristics of the Helicase Core
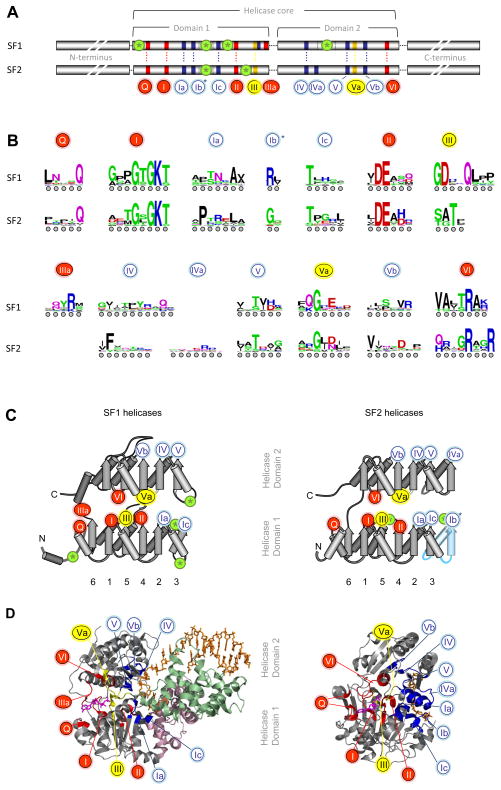
One of the most notable SF1 and SF2 helicase signatures are the characteristic sequence motifs, nine of which had been previously identified [7,12]. Closer inspection of the sequences reveals at least 12 characteristic sequence motifs shared by both superfamilies (Fig. 2A). However, not all motifs are present in each family (Suppl. Fig. S3). As noted previously [12], sequence conservation in the characteristic motifs is high within each family (Suppl. Fig. S1). The level of conservation in most motifs decreases throughout the respective superfamilies, and only limited sequence conservation remains across both superfamilies (Fig. 2B).
Figure 2. Sequence and structural organization of the helicase core of SF1 and SF2 proteins.
(A) Sequence organization of the helicase core in SF1 and SF2. Characteristic sequence motifs are colored according to their predominant biochemical function: red, ATP binding and hydrolysis; yellow, coordination between nucleic acid and NTP binding sites; blue, nucleic acid binding. Green circled asterisks mark insertions of additional domains. The lengths of the blocks and the distance between the conserved domains is not to scale. Characteristic motifs were identified from the alignment of all SF1 and SF2 proteins from human, S.cerevisiae, E.coli and selected viruses (Suppl. Fig. S1). Considering numbering schemes already in use [7], motifs were numbered consecutively. Motifs in SF1 and SF2 proteins are located at identical positions in the RecA-fold (panel B, below). Motif IIIa in SF1 has been occasionally marked as motif IV. The Q-motif is equivalent to motif 0 in RecQ proteins. Motif IVa in SF2 proteins is frequently marked QxxR, motif Ic often TPGR. The asterisk on motif Ib indicates that in some proteins this motif is replaced by an additional domain. (B) Sequence conservation within the characteristic helicase motifs. The height of the amino acids reflects the level of conservation at a given position, tall letters indicate higher conservation. The universally conserved E in motif II corresponds to 4 bits. Coloring marks the chemical properties of a given amino acid position: green - polar, blue - basic, red – acidic, and black - hydrophobic. Sequence logos were created from the alignment of SF1 and SF2 proteins (Suppl. Fig. S1) according to reference [91]. Circles under the letters are for visual guidance. For sequence logos of the characteristic motifs of the individual families see Suppl. Fig. S3 (C) Position of the characteristic motifs in the RecA-like folds of the helicase core domains. The β-strands are indicated by arrows, α-helices by cylinders. The β-strands of the first RecA-like domain are numbered according to their position in the primary structure. The position of the characteristic motifs is indicated by numbered circles, colored as in panel A. The position of inserted domains are marked by green circled asterisks, as in panel A. Blue coloring of the rightmost β-strand and α-helix in the SF2 representation indicates the absence of this part in several SF2 protein families. (D) Position of the characteristic motifs in three-dimensional structures of SF1 and SF2 proteins. Structures of the SF1 helicase UvrD (left, UvrD/Rep family) and the SF2 helicase Vasa (right, DEAD-box family) [40,92]. The bound ATP analog is colored margenta, the nucleic acid is colored wheat. Conserved sequence motifs are colored as in panel (A), inserted domains 1B and 2B of UvrD are light-pink and light-green.
The highest level of sequence conservation across both superfamilies is seen in the residues that coordinate binding and hydrolysis of the triphosphate (motifs I, II, VI, Fig. 2). These residues are located in the cleft between the two conserved RecA-like helicase domains (Fig. 2C,D). The spatial arrangement of functionalities presented by these residues is highly conserved in other P-loop NTPases, probably reflecting significant evolutionary constraints in the active site for phosphoester hydrolysis [5].
The Q-motif, which coordinates the adenine base, is somewhat less conserved across both superfamilies [32]. This motif is absent in the DEAH/RHA and viral DExH proteins (SF2), and these enzymes are not specific for adenosine triphosphates (Suppl. Figs. S1, S2, Tab. 1). Motif IIIa is seen in SF1 proteins (frequently annotated as motif IV) and also appears to be present in some members of the Swi/Snf family (Suppl. Figs. S1, S4). This motif seems to supply a stacking platform (conserved tyrosine) for the adenine base, a functionality that in other SF2 proteins is located on the opposite site of the bound adenine base, preceding the Q-motif (Suppl. Fig. S4).
The motifs primarily implied in the coordination between NTP and nucleic acid binding site (motifs III, Va) are highly conserved within each superfamily, but not across both (Fig. 2B, Suppl. Fig. S3). Mutations in the conserved residues in these motifs usually impair the coupling of NTP hydrolysis to nucleic acid binding and unwinding (e.g., refs. [33–35]), perhaps by affecting the ability to properly align both helicase domains in response to NTP binding, or by failing to coordinate the assembly of the NTP active site in response to nucleic acid binding [35,36]. Sequence differences in motifs III and Va between both superfamilies suggests that communication between NTP and nucleic acid binding site may vary between SF1 and 2. Such potential disparities are perhaps most obvious in motif III, which is followed by a nucleic acid binding site in many SF1 [37], but not in SF2 helicases. While it is not well understood how exactly NTP and nucleic acid binding sites communicate in either superfamily, overarching themes begin to emerge, such as arrangements resembling the “glutamate switch” in AAA+ proteins [38].
For some helicases, including PcrA, Rep, UvrD (UvrD/Rep family, [37,39,40]), RecD2 (Pif1-like family, [41]), Dengue virus NS3 and HCV NS3 (NS3/NPH-II group, [42,43]), and Mss116p (DEAD-box family, [44]), structural data at various ATP-hydrolysis states have been integrated into models of conformational changes during unwinding and ATP-dependent nucleic acid binding. These models highlight common themes such as domain closure upon ATP binding, but also significant differences, such as the involvement of non-conserved, accessory domains in the unwinding mechanisms. For the UvrD/Rep and Pif1-like SF1 families, motifs Ia and III have been proposed to play particularly important roles in defining translocation polarity [41].
The motifs contacting nucleic acid (motifs Ia,b,c, IV, IVa, V, Vb) are located on the face opposite the ATP binding site on both helicase domains (Fig. 2A, B). The arrangement of nucleic acid binding motifs in the secondary structure of both RecA-like helicase domains is very similar for both domains (Fig. 2C). Besides binding to the nucleic acid, these motifs are likely to also participate in the communication between nucleic acid and NTP binding sites, as recently shown for DEAD-box proteins [45]. The nucleic acid binding motifs are well conserved within the protein families (Suppl. Fig. S3), but much less across both superfamilies (Fig. 2B). Only two residues, two Ts (occasionally substituted by S) in motif Ic and V, are widely conserved in both superfamilies (Fig. 2B). The two conserved T residues have been assigned critical roles in unwinding mechanisms of helicases that separate duplexes by translocation [46,47]. However, the two Ts are absent in Rad3/XPD proteins (Suppl. Fig. S3), which unwind duplexes with 5′ to 3′ polarity (Tab. 1).
Contacts to the nucleic acid by residues of the conserved sequence motifs are primarily made to the phosphate-sugar backbone. Additional base contacts, which are critical for unwinding by several helicases, are usually established by residues that are either located in the helicase core outside of the conserved motifs, or in different domains altogether [41,43]. Several contacts to the nucleic acid involve only the peptide backbone, explaining the occasionally low sequence conservation of the nucleic acid binding motifs (e.g., motif IVa, Fig. 2A).
Interestingly, some families encompass both RNA and DNA helicases, other families are comprised solely of DNA helicases (Tab. 1), and only the DEAD-box family appears to contain exclusively RNA helicases. Notwithstanding, it has been shown that even some DEAD-box proteins can bind DNA, although the binding cannot be modulated by ATP in the way RNA binding is modulated [23]. Several helicases, including viral proteins of the NS3/NPH-II group and Upf1-like proteins have been shown to work on both DNA and RNA [48–50]. The absence of a clear correlation between the helicase families and specificity for RNA or DNA suggests that discrimination between RNA and DNA may not have been a predominant evolutionary force for the differentiation of the families. Mechanistic features of proteins from the respective families may be utilized in both RNA and DNA-related processes, and DNA and RNA specificity may have developed after the families were established. Which sequences or structural features dictate specificity for DNA or RNA remains to be elucidated.
In addition to the family-typical sequence domains, some helicase families harbor characteristic structural features. For example, structures of Ski2-like, DEAH/RHA and NS3/NPH-II proteins revealed a prominent β-hairpin between motifs Va and VI that is not present in the other SF2 families [51–53] (Suppl. Fig. S5). Structural and mechanistic studies of the Ski2-like Hel308 helicase indicate that this hairpin functions as a “pin” to separate duplex strands at the junction [52]. It is assumed that this “pin” plays similar roles in viral DExH and DEAH/RHA proteins [53], and it is intriguing to note that the presence of this β-hairpin correlates with polar 3′ to 5′ unwinding activity (Tab. 1). Moreover, this β-hairpin might be functionally similar to the strand separating “pin” in SF1 helicases [54], and perhaps to a potential “pin” in RecQ proteins [55]. These pins, however, are localized at different positions in the RecA-fold, or outside the helicase core (Suppl. Fig. S5).
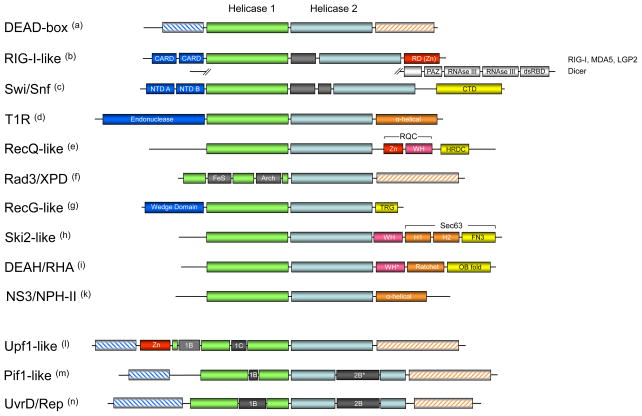
A further, generally family-typical structural feature is the presence of occasionally large inserts within the helicase core domains. Inserts are seen in all SF1 families [37,41,56,57] and in the Rad3/XPD family [58–60]. The Swi/Snf and the RIG-I-like families contain inserts between the two helicase domains [61–63] (Fig. 3). The position of these inserts is usually conserved within a family. The inserts adopt independent folds, which in most cases have only small or virtually no effects on the RecA-fold of the helicase core domains. Where tested, the inserted domains were found to impact the function of these proteins, both in the cell and in vitro (e.g. refs. [54,64]). A few individual proteins in other helicase families also feature inserts, such as the DEAD-box protein DDX1 [65] and the RecG-like PriA [66], but these inserts are not typical for the families.
Figure 3. Domain organization of SF2 and SF1 helicase families and groups.
Domains are not to scale. (a) C- and N-termini of DEAD-box proteins include RRMs, Zn-fingers, tudor domains and others [26]. (b) The family-typical domain inserted between the helicase domains is shown in grey. RIG-I-like proteins vary in their terminal domains [70]. Prominent RIG-I-like proteins are shown, Mph1p/FancM-related proteins are not shown. (CARD – caspase recruitment domain, RD – regulatory domain, a Zn-binding domain [67], PAZ – PIWI, Argonaute, Zwille, dsRBD – double strand RNA binding domain) (c) Domain organization of bacterial RapA [93]. Inserted domain between the helicase core domains is family-typical (Suppl. Fig. 1) (NTD – N-terminal domain, CTD – C terminal domain). (d) Domain organization of EcoR124 [80]. (e) RecQ-like proteins feature multiple C and N-terminal domains including exonuclease domains. Shown are the most conserved features of the RecQ-like family [94]. (Zn – Zn finger domain, WH – winged helix domain, HRDC – Helicase and RNAseD-like C terminal domain, RQC – RecQ C-terminal domain) (f) Rad3/XPD proteins also feature diverse C and N-termini. The domains inserted in helicase domain 1 are family-typical (FeS – iron-sulfur cluster, Arch – Arch domain, a structural domain, see Fig. 4) (g) Domain organization in RecG-like protein is largely conserved, but PriA has a non-typical Zn finger inserted in the helicase domain 2 [66] (TRG – translocation by RecG) (h) Domain organization of Hel 308 [52]. The organization of the C-terminal domains is conserved in Brr2 [95,96]. (WH – winged helix, H1 – helical 1, H2 – helical 2, FN3 – fibronectin 3) (i) DEAH/RHA proteins have varying N-terminal domains, but show a very high degree of conservation in their C-termini, especially among the spliceosomal DEAH proteins. It is possible that DEAH proteins and perhaps even most DEAH/RHA proteins show a conserved domain organization of their C-termini. Shown is the domain organization of Prp43 [53]. The domain organization of the C-terminus, with the exception of the OB-fold domain, resembles that of Ski2-like proteins [52,95,96]. (WH* - degenerated winged helix, Ratchet corresponds to H1 and H1 in the Ski2-like proteins) (k) NS3/NPH-II proteins have pronounced C- and N-terminal domains, but with the exception of the shown helical C-terminus of NS3 proteins from flaviviridae [51], no further information about these domains is available. (l) Upf1-like proteins feature variable termini. The location of domains 1B and 1C is conserved in the family. Shown is the domain organization of Upf1. (m) Pif1-like proteins feature variable termini, but location of domains 1B is conserved in the family. Domain 2B varies in size from about a dozen residues (RecD1) to more than 100 amino acids (Pif1p). (n) UvrD/Rep proteins also have variable termini. The location of domains 1B and 2B is largely conserved, but domain 2B is absent in LBA1 and HelD. LBA1 also features 3 ankyrin repeats inserted in the helicase domain 1 before domain 1B.
Terminal accessory domains
In most SF1 and SF2 helicases the helicase core is surrounded by C and N-terminal domains, which often exceed the helicase core in size. In many cases, these domains adopt defined folds with specific functions such as nucleases, RNA or DNA binding domains (e.g., Zn-fingers, OB-folds, dsRBDs [53,67–69]), or domains engaged in protein-protein interactions (e.g., CARD-domains [70]) (Fig. 3). In many proteins, multiple defined C- and N-terminal domains are seen (e.g., Dicer [71] Fig. 3). It is clear that these regions influence or even define the function of a helicase, not only through additional enzymatic activities, such as nuclease function, but also, for example, by promoting oligomerization [72]. In addition, C- and N-terminal domains are thought to be critical for the physiological specificity of helicases. Terminal domains have been demonstrated to direct recruitment to certain complexes, to promote interactions with other proteins, or to facilitate recognition of specific nucleic acid regions [70,73–76]. Consistent with critical roles in establishing physiological specificity for individual enzymes, C- and N-terminal accessory domains are usually not conserved within a family. I.e., a family-typical helicase core is surrounded by variable accessory domains. Notwithstanding, recent studies have revealed some degree of structural conservation of the C-terminal domains in the Ski2-like and the DEAH/RHA families [52,53] (Fig. 3).
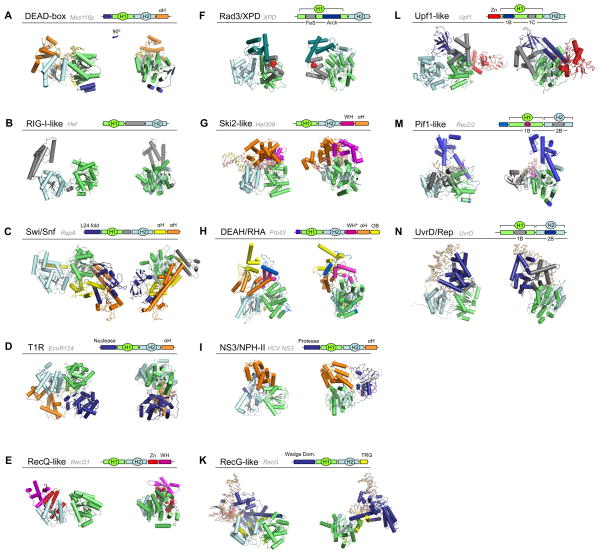
Structures for members of all SF1 and SF2 families and groups have been reported, and a broad view of the domain architecture of the different families is now possible (Fig. 4). It is important to note that architectures within a given family might differ, depending on the various C and N-terminal domains. It is not currently possible to systematically assess such intra-family variability, because too few structures are available for helicases within each given family that encompass the helicase core and significant or all parts of C- and N-terminal domains. Nonetheless, it is intriguing to compare the available information. Most helicases that unwind duplexes with defined polarity, either 3′ to 5′, or 5′ to 3′ (Rad3/XPD, Ski2-like, DEAH/RHA, NS3/NPH-II, all SF1 families) have functionally important accessory domains located on top of the nucleic acid binding site on the helicase core (Fig. 4). This arrangement encloses the bound nucleic acid strand to some extent, possibly facilitating directional translocation upon which polar unwinding is based for many helicases [3,4]. Polar unwinding has also been shown for some RecQ-like helicases [77] and for RIG-I [78], but structures of proteins from these families do not feature domains on top of the nucleic acid binding site (Fig. 4E,B). However, currently available structures do not represent full-length proteins, and it remains to be seen whether these families ultimately have domains located on top of the helicase domain, or if these proteins accomplish polar unwinding through a distinct architecture, or by other means, such as oligomerization [79].
Figure 4. Domain architecture of SF2 and SF1 helicase families and groups.
A representative structure of one protein from each family is shown, as indicated. The schematic cartoons show the domain organization of the displayed structure. In all structures, the helicase domain 1 (H1) is green, helicase domain 2 (H2) cyan, and nucleic acid, where present, beige. Terminal and inserted accessory domains are colored according to their respective folds, as indicated. All structures are oriented in a similar fashion, right panels show the structures rotated by roughly 90°, as indicated in panel (A). (Abbreviations: αH - helical domain, L24 fold – fold resembling the ribosomal protein L24, Zn – Zinc binding domain, WH – winged helix domain, FeS – iron-sulphur cluster, OB – OB fold, TRG – translocation by RecG) 1B,C and 2B mark the inserts in SF1 proteins. The following structures are shown: (A) Mss116p (S.cerevisiae) [44], (B) Hef (Pyrococcus furiosis) [63], (C) RapA (E.coli) [93], (D) EcoR124I (E.coli) [80], (E) RecQ1 (H.sapiens) [55], (F) XPD (Sulfolobus acidocaldarius) [58], (G) Hel308 (Archaeoglobus fulgidus) [52], (H) Prp43p (S.cerevisiae) [53], (I) HCV NS3 (Hepatitis C virus) [97], (K) RecG (Thermotoga maritima) [98], (L) Upf1 (H.sapiens) [99], (M) RecD2 (D.radiourans) [41], (N) UvrD (E.coli) [40].
Helicases that do not unwind duplexes (Swi/Snf, T1R) do not have domains on top of the nucleic acid binding site (Fig. 4C,D). This architecture is consistent with the notion that these enzymes need to bind duplex DNA [18,20,80]. The RIG-I-like helicase Hef also does not feature a domain on top of its RNA binding site in the helicase core, although RIG-I-like proteins unwind duplexes in a polar fashion [78,81] (Tab. 1). However, RIG-I and related proteins also act on duplex RNA [19,70,82,83]. Available structures for DEAD-box proteins also do not show domains on top of the RNA binding site on the helicase core [44,84,85] (Fig. 4A). This arrangement is consistent with the observation that the distinct unwinding mechanism of these enzymes involves direct binding to the duplex region [21,23,24,86,87]. However, conclusive inferences for both, the RIG-I-like and the DEAD-box families cannot be made until full-length structures for proteins with large C- or N-terminal domains are available.
Conclusions and future challenges
Significant progress has been made in recent years towards understanding structure-function relations of SF1 and SF2 helicases. It has become clear that despite a highly conserved core fold, these enzymes perform diverse functions on nucleic acids both in vitro and in the cell. The presented family-based classification may be a helpful step towards rationalizing common themes as well as differences between individual helicases.
Notwithstanding the impressive progress, much remains to be learned. For helicases where sophisticated structural models explain complex reactions, such as unwinding, focus for further inquiry may be on elucidating the communication between NTP and nucleic acid binding sites. For other helicases, it is critical to obtain structural data of full-length proteins, bound to nucleic acids and preferably to ATP analogs representing different states of the ATPase cycle. Structural information needs to be complemented by mechanistic studies addressing how the different states of the ATPase cycle are coupled to conformational work on nucleic acid substrates.
In addition to investigating connections between structural and mechanistic aspects of helicase functions, it is most critical to elucidate how helicase mechanisms are manifested in physiological contexts. Although not discussed in this review, enormous progress has been made over the last years in structural and functional analysis of helicases bound to other proteins and in authentic functional complexes. Examining helicase function in complexes amenable to biochemical and structural analysis is likely to come into even greater focus over the next years.
For eukaryotic RNA helicases, deeper understanding of physiological substrates and contexts remained elusive for many years, but significant hurdles have now been taken. Studies of specific systems such as the DEAH/RHA helicase Prp22 [88], and the DEAD-box protein eIF4A-III [89], as well as the application of novel, genome wide survey techniques [90] have started to reveal the interaction of RNA helicases with their physiological substrates on a physical level. Integration of biological studies with structural and mechanistic analyses of RNA helicases will be bound to illuminate many aspects of post-transcriptional regulation of gene expression.
Supplementary Material
Acknowledgments
We apologize to all colleagues whose work could not be discussed or cited here, due to space limitations. Among the cited articles, too many contributions are of particular significance, and we therefore refrained from highlighting selected references. We thank Dr. Patrick Linder (Geneva) for the many fruitful discussions that were most instrumental in shaping our view on SF1 and SF2 helicases. Research in the author’s laboratory is supported by the Burroughs Wellcome Fund and the NIH (GM067700). UPG is supported by a DFG postdoctoral fellowship (GU-1146).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Abdel-Monem M, Hoffman-Berling H. Enzymic Unwinding of DNA 2. Chain separtion by an ATP-dependent DNA unwinding enyzme. Eur J Biochem. 1976;65:441–449. doi: 10.1111/j.1432-1033.1976.tb10359.x. [DOI] [PubMed] [Google Scholar]
- 2.Lohman TM, Tomko EJ, Wu CG. Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat Rev Cell Biol. 2008;9:391–401. doi: 10.1038/nrm2394. [DOI] [PubMed] [Google Scholar]
- 3.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Ann Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 4.Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Ann Rev Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- 5.Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol. 2002;317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 6.Anantharaman V, Koonin EV, Aravind L. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res. 2002;30:1427–1464. doi: 10.1093/nar/30.7.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jankowsky E, Fairman M. RNA helicases - one fold for many functions. Curr Opin Struct Biol. 2007;17:316–324. doi: 10.1016/j.sbi.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 8.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 9.Kadare G, Haenni AL. Virus-encoded RNA helicases. J Virol. 1997:2583–2590. doi: 10.1128/jvi.71.4.2583-2590.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abdelhaleem M. Do human RNA helicases have a role in cancer? Biochim Biophys Acta. 2004;1704:37–46. doi: 10.1016/j.bbcan.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 11.Hanada K, Hickson ID. Molecular genetics of RecQ helicase disorders. Cell Mol Life Sci. 2007;64:2306–2322. doi: 10.1007/s00018-007-7121-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorbalenya AE, Koonin EV. Helicases: amino acid comparisons and structure-function relationships. Curr Opin Struct Biol. 1993;3:419–429. [Google Scholar]
- 13.Linder P, Owttrim GW. Plant RNA helicases: linking aberrant and silencing RNA. Trends Plant Sci. 2009;14:344–352. doi: 10.1016/j.tplants.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 14.Strunk BS, Karbstein K. Powering through ribosome assembly. RNA. 2009;15:2083–2104. doi: 10.1261/rna.1792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mayas RM, Staley JP. DEAD on. Nat Struct Mol Biol. 2006;13:954–955. doi: 10.1038/nsmb1106-954. [DOI] [PubMed] [Google Scholar]
- 16.Jankowsky E, Bowers H. Remodeling of ribonucleoprotein complexes with DExH/D RNA helicases. Nucleic Acids Res. 2006;34:4181–4188. doi: 10.1093/nar/gkl410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soultanas P, Dillingham MS, Wiley P, Webb MR, Wigley DB. Uncoupling DNA translocation and helicase activity in PcrA: direct evidence for an active mechanism. EMBO J. 2000;19:3799–3810. doi: 10.1093/emboj/19.14.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seidel R, Bloom JG, Dekker C, Szczelkun MD. Motor step size and ATP coupling efficiency of the dsDNA translocase EcoR124I. EMBO J. 2008;27:1388–1398. doi: 10.1038/emboj.2008.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Myong S, Cui S, Cornish PV, Kirchhofer A, Gack MU, Jung JU, Hopfner KP, Ha T. Cytosolic viral sensor RIG-I is a 5′-triphosphate-dependent translocase on double-stranded RNA. Science. 2009;323:1070–1074. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Durr H, Flaus A, Owen-Hughes T, Hopfner KP. Snf2 family ATPases and DExx box helicases: differences and unifying concepts from high-resolution crystal structures. Nucleic Acids Res. 2006;34:4160–4167. doi: 10.1093/nar/gkl540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tijerina P, Bhaskaran H, Russell R. Nonspecific binding to structured RNA and preferential unwinding of an exposed helix by the CYT-19 protein, a DEAD-box RNA chaperone. Proc Natl Acad Sci U S A. 2006;103:16698–16703. doi: 10.1073/pnas.0603127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bizebard T, Ferlenghi I, Iost I, Dreyfus M. Studies on three E. coli DEAD-box helicases point to an unwinding mechanism different from that of model DNA helicases. Biochemistry. 2004;43:7857–7866. doi: 10.1021/bi049852s. [DOI] [PubMed] [Google Scholar]
- 23.Yang Q, Jankowsky E. The DEAD-box protein Ded1 unwinds RNA duplexes by a mode distinct from translocating helicases. Nat Struct Mol Biol. 2006;13:981–986. doi: 10.1038/nsmb1165. [DOI] [PubMed] [Google Scholar]
- 24.Yang Q, Del Campo M, Lambowitz AM, Jankowsky E. DEAD-box proteins unwind duplexes by local strand separation. Mol Cell. 2007;28:253–263. doi: 10.1016/j.molcel.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 25.Bessler JB, Torredagger JZ, Zakian VA. The Pif1p subfamily of helicases: region-specific DNA helicases? Trends Cell Biol. 2001;11:60–65. doi: 10.1016/s0962-8924(00)01877-8. [DOI] [PubMed] [Google Scholar]
- 26.Linder P. Dead-box proteins: a family affair--active and passive players in RNP-remodeling. Nucleic Acids Res. 2006;34:4168–4180. doi: 10.1093/nar/gkl468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chu WK, Hickson ID. RecQ helicases: multifunctional genome caretakers. Nature Rev Cancer. 2009;9:644–654. doi: 10.1038/nrc2682. [DOI] [PubMed] [Google Scholar]
- 28.Frick DN, Lam AM. Understanding helicases as a means of virus control. Curr Pharm Des. 2006;12:1315–1338. doi: 10.2174/138161206776361147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanjuán R, Marín I. Tracing the origin of the compensasome: evolutionary history of DEAH helicase and MYST acetyltransferase gene families. Mol Biol Evol. 2001;18:330–343. doi: 10.1093/oxfordjournals.molbev.a003809. [DOI] [PubMed] [Google Scholar]
- 30.Sarkar D, Desalle R, Fisher PB. Evolution of MDA-5/RIG-I-dependent innate immunity: independent evolution by domain grafting. Proc Natl Acad Sci USA. 2008;105:17040–17045. doi: 10.1073/pnas.0804956105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pang PS, Jankowsky E, Planet P, Pyle AM. The hepatitis C viral NS3 protein is a processive DNA helicase with co-factor enhanced RNA unwinding. EMBO J. 2002;21:1168–1176. doi: 10.1093/emboj/21.5.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanner NK, Cordin O, Banroques J, Doere M, Linder P. The Q motif: a newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol Cell. 2003;11:127–138. doi: 10.1016/s1097-2765(03)00006-6. [DOI] [PubMed] [Google Scholar]
- 33.Dillingham MS, PS, Wigley DB. Site-directed mutagenesis of motif III in PcrA helicase reveals a role in coupling ATP hydrolysis to strand separation. Nucleic Acids Res. 1999;27:3310–3317. doi: 10.1093/nar/27.16.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanaka N, Schwer B. Mutations in PRP43 that uncouple RNA-dependent NTPase activity and pre-mRNA splicing function. Biochemistry. 2006;45:6110–6121. doi: 10.1021/bi052656g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Banroques J, Doère M, Dreyfus M, Linder P, Tanner NK. Motif III in Superfamily 2 “Helicases” Helps Convert the Binding Energy of ATP into a High-Affinity RNA Binding Site in the Yeast DEAD-Box Protein Ded1. J Mol Biol. 2010 doi: 10.1016/j.jmb.2009.12.025. epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 36.Karow AR, Klostermeier D. A conformational change in the helicase core is necessary but not sufficient for RNA unwinding by the DEAD box helicase YxiN. Nucleic Acids Res. 2009;37:4464–4471. doi: 10.1093/nar/gkp397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 38.Zhang X, Wigley DB. The ‘glutamate switch’ provides a link between ATPase activity and ligand binding in AAA+ proteins. Nature Struct Biol. 2008;15:1223–1227. doi: 10.1038/nsmb.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hsieh J, Moore KJ, Lohman TM. A two-site kinetic mechanism for ATP binding and hydrolysis by E. coli Rep helicase dimer bound to a single-stranded oligodeoxynucleotide. J Mol Biol. 1999;288:255–274. doi: 10.1006/jmbi.1999.2666. [DOI] [PubMed] [Google Scholar]
- 40.Lee JY, Yang W. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell. 2006;127:1349–1360. doi: 10.1016/j.cell.2006.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saikrishnan K, Powell B, Cook NJ, Webb MR, Wigley DB. Mechanistic basis of 5′-3′ translocation in SF1B helicases. Cell. 2009;137:849–859. doi: 10.1016/j.cell.2009.03.036. [DOI] [PubMed] [Google Scholar]
- 42.Luo D, Xu T, Watson RP, Scherer-Becker D, Sampath A, Jahnke W, Yeong SS, Wang CH, Lim SP, Strongin A, et al. Insights into RNA unwinding and ATP hydrolysis by the flavivirus NS3 protein. EMBO J. 2008;27:3209–3219. doi: 10.1038/emboj.2008.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gu M, Rice CM. Three conformational snapshots of the hepatitis C virus NS3 helicase reveal a ratchet translocation mechanism. Proc Natl Acad Sci USA. 2010;107:521–528. doi: 10.1073/pnas.0913380107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Del Campo M, Lambowitz AM. Structure of the Yeast DEAD box protein Mss116p reveals two wedges that crimp RNA. Mol Cell. 2009;35:598–609. doi: 10.1016/j.molcel.2009.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Banroques J, Cordin O, Doère M, Linder P, Tanner NK. A conserved phenylalanine of motif IV in superfamily 2 helicases is required for cooperative, ATP-dependent binding of RNA substrates in DEAD-box proteins. Mol Cell Biol. 2008;28:3359–3371. doi: 10.1128/MCB.01555-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Myong S, Bruno MM, Pyle AM, Ha T. Spring-loaded mechanism of DNA unwinding by hepatitis C virus NS3 helicase. Science. 2007;317:513–516. doi: 10.1126/science.1144130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Myong S, Ha T. Stepwise translocation of nucleic acid motors. Curr Opin Struct Biol. 2010 doi: 10.1016/j.sbi.2009.12.008. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tackett AJ, Wei L, Cameron CE, Raney KD. Unwinding of nucleic acids by HCV NS3 helicase is sensitive to the structure of the duplex. Nucleic Acids Res. 2001;29:565–672. doi: 10.1093/nar/29.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taylor SD, Solem A, Kawaoka J, Pyle AM. The NPH-II helicase displays efficient DNA-RNA helicase activity and a pronounced purine sequence bias. J Biol Chem. 2010 doi: 10.1074/jbc.M109.088559. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guenther UP, Handoko L, Laggerbauer B, Jablonka S, Chari A, Alzheimer M, Ohmer J, Plöttner O, Gehring N, Sickmann A, et al. IGHMBP2 is a ribosome-associated helicase inactive in the neuromuscular disorder distal SMA type 1 (DSMA1) Hum Mol Genet. 2009;18:1288–1300. doi: 10.1093/hmg/ddp028. [DOI] [PubMed] [Google Scholar]
- 51.Kim JL, Morgenstern KA, Griffith JP, Dwyer MD, Thomson JA, Murcko MA, Lin C, Caron PR. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure. 1998;6:89–100. doi: 10.1016/s0969-2126(98)00010-0. [DOI] [PubMed] [Google Scholar]
- 52.Büttner K, Nehring S, Hopfner KP. Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat Struct Biol. 2007;14:647–352. doi: 10.1038/nsmb1246. [DOI] [PubMed] [Google Scholar]
- 53.He Y, Andersen GR, Nielsen KH. Structural basis for the function of DEAH helicases. EMBO Rep. 2010 doi: 10.1038/embor.2010.11. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saikrishnan K, Griffiths SP, Cook N, Court R, Wigley DB. DNA binding to RecD: role of the 1B domain in SF1B helicase activity. EMBO J. 2008;27:2222–2229. doi: 10.1038/emboj.2008.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pike AC, Shrestha B, Popuri V, Burgess-Brown N, Muzzolini L, Costantini S, Vindigni A, Gileadi O. Structure of the human RECQ1 helicase reveals a putative strand-separation pin. Proc Natl Acad Sci USA. 2009;106:1039–1044. doi: 10.1073/pnas.0806908106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheng Z, Muhlrad D, Lim MK, Parker R, Song H. Structural and functional insights into the human Upf1 helicase core. EMBO J. 2007;26:253–264. doi: 10.1038/sj.emboj.7601464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Singleton MR, Dillingham MS, Gaudier M, Kowalczykowski SC, Wigley DB. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature. 2004;432:187–193. doi: 10.1038/nature02988. [DOI] [PubMed] [Google Scholar]
- 58.Fan L, Fuss JO, Cheng QJ, Arvai AS, Hammel M, Roberts VA, Cooper PK, Tainer JA. XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations. Cell. 2008;133:789–800. doi: 10.1016/j.cell.2008.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu H, Rudolf J, Johnson KA, McMahon SA, Oke M, Carter L, McRobbie AM, Brown SE, Naismith JH, White MF. Structure of the DNA repair helicase XPD. Cell. 2008:133. doi: 10.1016/j.cell.2008.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wolski SC, Kuper J, Hänzelmann P, Truglio JJ, Croteau DL, Van Houten B, Kisker C. Crystal structure of the FeS cluster-containing nucleotide excision repair helicase XPD. PLOS Biol. 2008;6:e149. doi: 10.1371/journal.pbio.0060149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dürr H, Körner C, Müller M, Hickmann V, Hopfner KP. X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell. 2005;121:363–373. doi: 10.1016/j.cell.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 62.Thomä NH, Czyzewski BK, Alexeev AA, Mazin AV, Kowalczykowski SC, Pavletich NP. Structure of the SWI2/SNF2 chromatin-remodeling domain of eukaryotic Rad54. Nature Struct Biol. 2005;12:350–356. doi: 10.1038/nsmb919. [DOI] [PubMed] [Google Scholar]
- 63.Nishino T, Komori K, Tsuchiya D, Ishino Y, Morikawa K. Crystal structure and functional implications of Pyrococcus furiosus hef helicase domain involved in branched DNA processing. Structure. 2005;13:143–153. doi: 10.1016/j.str.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 64.Brendza KM, Cheng W, Fischer CJ, Chesnik MA, Niedziela-Majka A, Lohman TM. Autoinhibition of Escherichia coli Rep monomer helicase activity by its 2B subdomain. Proc Natl Acad Sci USA. 2005;102:10076–100781. doi: 10.1073/pnas.0502886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Godbout R, Packer M, Bie W. Overexpression of a DEAD box protein (DDX1) in neuroblastoma and retinoblastoma cell lines. J Biol Chem. 1998;273:21161–21168. doi: 10.1074/jbc.273.33.21161. [DOI] [PubMed] [Google Scholar]
- 66.Gabbai CB, Marians KJ. Recruitment to stalled replication forks of the PriA DNA helicase and replisome-loading activities is essential for survival. DNA Repair. 2010 doi: 10.1016/j.dnarep.2009.12.009. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cui S, Eisenächer K, Kirchhofer A, Brzózka K, Lammens A, Lammens K, Fujita T, Conzelmann KK, Krug A, Hopfner KP. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol Cell. 2008;29:169–179. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 68.Bernstein DA, Keck JL. Domain mapping of Escherichia coli RecQ defines the roles of conserved N- and C-terminal regions in the RecQ family. Nucleic Acids Res. 2003;31:2778–2785. doi: 10.1093/nar/gkg376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang S, Grosse F. Multiple functions of nuclear DNA helicase II (RNA helicase A) in nucleic acid metabolism. Acta Biochim Biophys Sin. 2004;36:177–183. doi: 10.1093/abbs/36.3.177. [DOI] [PubMed] [Google Scholar]
- 70.Yoneyama M, Fujita T. Structural mechanism of RNA recognition by the RIG-I-like receptors. Immunity. 2008;29:178–181. doi: 10.1016/j.immuni.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 71.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 72.Klostermeier D, Rudolph MG. A novel dimerization motif in the C-terminal domain of the Thermus thermophilus DEAD box helicase Hera confers substantial flexibility. Nucleic Acids Res. 2009;37:421–430. doi: 10.1093/nar/gkn947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shereda RD, Reiter NJ, Butcher SE, Keck JL. Identification of the SSB binding site on E. coli RecQ reveals a conserved surface for binding SSB’s C terminus. J Mol Biol. 2009;386:612–625. doi: 10.1016/j.jmb.2008.12.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Trubetskoy D, Proux F, Allemand F, Dreyfus M, Iost I. SrmB, a DEAD-box helicase involved in Escherichia coli ribosome assembly, is specifically targeted to 23S rRNA in vivo. Nucleic Acids Res. 2009:6540–6549. doi: 10.1093/nar/gkp685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Karginov FV, Caruthers JM, Hu Y, McKay DB, Uhlenbeck OC. YxiN is a modular protein combining a DEx(D/H) core and a specific RNA-binding domain. J Biol Chem. 2005;280:35499–35505. doi: 10.1074/jbc.M506815200. [DOI] [PubMed] [Google Scholar]
- 76.Killoran MP, Keck JL. Structure and function of the regulatory C-terminal HRDC domain from Deinococcus radiodurans RecQ. Nucleic Acids Res. 2008;36:3139–3149. doi: 10.1093/nar/gkn143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Capp C, Wu J, Hsieh TS. Drosophila RecQ4 has a 3′-5′ DNA helicase activity that is essential for viability. J Biol Chem. 2009;284:30845–30852. doi: 10.1074/jbc.M109.008052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Takahasi K, Yoneyama M, Nishihori T, Hirai R, Kumeta H, Narita R, Gale M, Inagaki F, Fujita T. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol Cell. 2008;29:428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 79.Li N, Henry E, Guiot E, Rigolet P, Brochon JC, Xi XG, Deprez E. Multiple Escherichia coli RecQ helicase monomers cooperate to unwind long DNA Substrates: A fluorescence cross-correlation spectroscopy study. J Biol Chem. 2010 doi: 10.1074/jbc.M109.069286. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lapkouski M, Panjikar S, Janscak P, Smatanova IK, Carey J, Ettrich R, Csefalvay E. Structure of the motor subunit of type I restriction-modification complex EcoR124I. Nature Struct Mol Biol. 2009;16:94–95. doi: 10.1038/nsmb.1523. [DOI] [PubMed] [Google Scholar]
- 81.Prakash R, Krejci L, Van Komen S, Schürer AK, Kramer W, Sung P. Saccharomyces cerevisiae MPH1 gene, required for homologous recombination-mediated mutation avoidance, encodes a 3′ to 5′ DNA helicase. J Biol Chem. 2005;280:7854–7860. doi: 10.1074/jbc.M413898200. [DOI] [PubMed] [Google Scholar]
- 82.Schlee M, Roth A, Hornung V, Hagmann CA, Wimmenauer V, Barchet W, Coch C, Janke M, Mihailovic A, Wardle G, et al. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schmidt A, Schwerd T, Hamm W, Hellmuth JC, Cui S, Wenzel M, Hoffmann FS, Michallet MC, Besch R, Hopfner KP, et al. 5′-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc Natl Acad Sci USA. 2009;106:12067–12072. doi: 10.1073/pnas.0900971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Andersen CB, Ballut L, Johansen JS, Chamieh H, Nielsen KH, Oliveira CL, Pedersen JS, Seraphin B, Le Hir H, Andersen GR. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- 85.Bono F, Ebert J, Lorentzen E, Conti E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell. 2006;126:713–725. doi: 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 86.Liu F, Putnam A, Jankowsky E. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc Natl Acad Sci U S A. 2008;105:20209–20214. doi: 10.1073/pnas.0811115106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen Y, Potratz JP, Tijerina P, Del Campo M, Lambowitz AM, Russell R. The DEAD-box Protein CYT-19 Uses a Single ATP to Completely Separate a Short RNA Duplex. Proc Natl Acad Sci U S A. 2008;105:20203–20208. doi: 10.1073/pnas.0811075106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schwer B. A conformational rearrangement in the spliceosome sets the stage for Prp22-dependent mRNA release. Mol Cell. 2008;30:743–754. doi: 10.1016/j.molcel.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Le Hir H, Andersen GR. Structural insights into the exon junction complex. Curr Opin Struct Biol. 2008;18:112–119. doi: 10.1016/j.sbi.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 90.Bohnsack MT, Martin R, Granneman S, Ruprecht M, Schleiff E, Tollervey D. Prp43 bound at different sites on the pre-rRNA performs distinct functions in ribosome synthesis. Mol Cell. 2009;36:583–592. doi: 10.1016/j.molcel.2009.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 93.Shaw G, Gan J, Zhou YN, Zhi H, Subburaman P, Zhang R, Joachimiak A, Jin DJ, Ji X. Structure of RapA, a Swi2/Snf2 protein that recycles RNA polymerase during transcription. Structure. 2008;16:1417–1427. doi: 10.1016/j.str.2008.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vindigni A, Hickson ID. RecQ helicases: multiple structures for multiple functions? HFSP J. 2009;3:153–164. doi: 10.2976/1.3079540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang L, Xu T, Maeder C, Bud LO, Shanks J, Nix J, Guthrie C, Pleiss JA, Zhao R. Structural evidence for consecutive Hel308-like modules in the spliceosomal ATPase Brr2. Nature Struct Mol Biol. 2009;16:731–739. doi: 10.1038/nsmb.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pena V, Jovin SM, Fabrizio P, Orlowski J, Bujnicki JM, Lührmann R, Wahl MC. Common design principles in the spliceosomal RNA helicase Brr2 and in the Hel308 DNA helicase. Mol Cell. 2009;35:454–466. doi: 10.1016/j.molcel.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 97.Yao N, Reichert P, Taremi SS, Prosise WW, Weber PC. Molecular views of viral polyprotein processing revealed by the crystal structure of the hepatitis C virus bifunctional protease-helicase. Structure. 1999;15:1353–1363. doi: 10.1016/s0969-2126(00)80025-8. [DOI] [PubMed] [Google Scholar]
- 98.Singleton MR, Scaife S, Wigley DB. Structural analysis of DNA replication fork reversal by RecG. Cell. 2001;107:79–89. doi: 10.1016/s0092-8674(01)00501-3. [DOI] [PubMed] [Google Scholar]
- 99.Clerici M, Mourão A, Gutsche I, Gehring NH, Hentze MW, Kulozik A, Kadlec J, Sattler M, Cusack S. Unusual bipartite mode of interaction between the nonsense-mediated decay factors, UPF1 and UPF2. EMBO J. 2009;28:2293–2306. doi: 10.1038/emboj.2009.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.