Abstract
Perturbation of the endoplasmic reticulum (ER) results in a conserved stress response called the “Unfolded Protein Response” (UPR). Macrophages undergoing a UPR respond to LPS with log-fold increased production of IFN-β, a cytokine with diverse roles in innate and adaptive immunity. In this report, we found that thapsigargin-induced ER stress augmented recruitment of IRF-3, CBP/p300, and transcriptional machinery to the murine ifnb1 promoter during LPS stimulation. Although full synergistic IFN-β production requires XBP-1, this UPR-regulated transcription factor did not appreciably bind the ifnb1 promoter. However, XBP-1 bound a conserved site 6.1kb downstream of ifnb1, along with IRF-3 and CBP only during concomitant UPR and LPS stimulation. XBP-1 physically associates with p300, suggesting a mechanism of multi-molecular assembly at the +6.1kb site. Luciferase reporter assays provide evidence this +6kb region functions as an XBP-1-dependent enhancer of ifnb1 promoter activity. Thus, this study identifies a novel role for an UPR-dependent transcription factor in the regulation of an inflammatory cytokine. Our findings have broader mechanistic implications for the pathogenesis of diseases involving ER stress and type I interferon, including viral infection, ischemia-reperfusion injury, protein-misfolding and inflammatory diseases.
Introduction
Type I interferons (IFN-α/β) play diverse roles in adaptive and innate immune responses. Although they were first noted for their anti-viral properties, type I IFNs also activate macrophages and NK cells, promote T cell survival and dendritic cell maturation, and increase the production of Th1-polarizing cytokines (1). Cells of the innate immune system, such as macrophages and dendritic cells, produce type I IFNs upon detection of pathogens through pattern recognition receptors (PRRs) that include the Toll-like receptors (TLRs) (2). These PRRs bind conserved motifs found on pathogens such as LPS (TLR4), dsRNA (TLR3 and RIG-I) and hypo-methylated CpG DNA (TLR9). TLRs may also mediate responses to “endogenous” products released during tissue necrosis such as hyaluronic acid, heparin sulfate, fibrinogen, and heat shock proteins (3).
IFN-β appears to be the primary cytokine that mediates macrophage type I IFN responses to the TLR4 agonist LPS(4). IFN-β deficient animals were shown to be much more susceptible to lethal sepsis from several strains of pathogenic bacteria, presumably through weakened host inflammatory responses (5). Mice deficient in IFN-β are also more susceptible to particular viral infections, have lower numbers of macrophages and mature B cells, and exhibit reduced bone mass (6–8).
The regulation of IFN-β transcription in the setting of viral infection has been well studied. Briefly, in the uninfected cell, a nucleosome obstructs the 1+ start site, preventing transcription. During infection, a group of transcription factors including NF-κB, AP-1, interferon regulatory factor 7 (IRF-7) and IRF-3 cooperatively assemble over a 55bp stretch of DNA, between −102 to −47 bp upstream of the transcriptional start site (9). This grouping, termed the “enhanceosome” recruits histone acetylases such as CREB binding protein (CBP/p300), a large flexible transcription co-activator that may interact simultaneously with multiple transcription factors (ATF-2, c-Jun, p65 and IRF3/7) (10, 11). CBP-p300 thus acts as a signal integrator. Histone acetylation facilitates the recruitment of chromatin modifiers that slide the nucleosome off the TATA box start site, thus enabling transcription(12, 13). Less is known about the induction of IFN-β transcription following LPS stimulation, although it appears slightly different. For instance, although viral infection induces recruitment of IRF-7 to the enhanceosome, LPS-induced IFN-β appears to depend upon IRF-3 rather than IRF-7 (14–16).
Our previous studies had shown that macrophages undergoing an intracellular stress response called the “Unfolded Protein Response” (UPR) respond to LPS and dsRNA with greatly enhanced IFN-β production (17). The UPR is an adaptive response initiated by environmental stressors (hypoxia, nutrient deprivation, hypoglycemia) or internal derangements (increased protein load, misfolding proteins, calcium gradient deregulation) that disrupt ER function. When ER function is perturbed, excess unfolded protein competes with the ER resident proteins, inositol requiring 1 (IRE-1), protein kinase receptor-like ER kinase (PERK), and activating transcription factor (ATF) 6, for binding of the folding chaperone Ig binding protein (BiP/GRP78). IRE-1 is an endonuclease activated after release of BiP that cleaves a 26bp intron from the X-box binding protein 1 (XBP-1) transcription factor mRNA. This unusual splicing event removes a premature stop-codon through frame shifting the open reading frame, thus allowing for the translation of the full-length XBP-1 transcription factor. Upon release of BiP, PERK transiently inhibits global protein translation apart from select transcripts (e.g. ATF4). Finally, ATF6 leaves the ER and traffics to the Golgi, where it is processed to an active form. UPR target genes aimed at resolving ER stress include folding chaperones and proteins that aid in ER associated protein degradation. If these and other adaptations fail, the UPR results in apoptosis (18).
The UPR appears to play a physiologic role in highly secretory cells such as pancreatic acinar cells, hepatocytes and plasma cells (19). However, the UPR has also been implicated in such diverse pathologic processes as cardiovascular disease, ischemia-reperfusion injury, neurodegenerative diseases, diabetes, viral infections and cancer (20). It is becoming increasingly apparent that the UPR also plays a role in immune function. For instance, the differentiation of B-cells into plasma cells requires splicing of XBP-1(21). XBP-1 deficiency in intestinal epithelial cells leads to spontaneous enteritis and increased susceptibility to Listeria (22). Cholesterol loaded macrophages undergoing a UPR secrete the inflammatory cytokines TNF-α and IL-6(23). ER-stress leads to the proteolytic activation of cyclic-AMP response element binding protein (CREB) H (processed similarly to ATF6), a transcription factor that induces the production of serum amyloid and C-reactive proteins (24).
Understanding how ER stress regulates IFN-β responses may shed light on disease processes in which both UPR and type I IFNs have been implicated such as ischemia-reperfusion injury and viral infections, as well as diseases where they may be related (HLA-B27 associated Spondyloarthritis and inflammatory myopathies) (25–29). Previous work has supported a critical role for the UPR-regulated transcription factor XBP-1 in mediating synergistic IFN-β induction upon TLR stimulation (17). However, the underlying molecular mechanism behind the synergy was not clear. We hypothesized that XBP-1, as a transcription factor, may regulate IFN-β induction by either a direct or epigenetic mechanism during ER stress. In this report we demonstrate binding of XBP-1, CBP and IRF-3 to a DNA region 6.1kb downstream of the ifnb1 gene during conditions of concomitant ER stress and LPS stimulation. Binding of these factors at this +6kb site correlated temporally with increased recruitment of CBP and IRF-3 to the ifnb1 promoter. Finally, the presence of the +6kb site significantly enhanced ifnb1 promoter activity. Together, these data suggest that this newly described region 6kb downstream of the ifnb1 gene is a cis-acting XBP-1-dependent enhancer of IFN-β production that provides a mechanistic link between ER stress and augmented IFN-β induction. As a broader consideration, these findings provide an explanation for how ER stress may drive the pathogenesis of type I IFN-related diseases.
Materials and Methods
Cells, reagents, and stimulations
The RAW264.7 macrophage cell line (ATCC) was maintained in DMEM/high glucose with 4mM L-glutamine, sodium pyruvate (Hyclone), and supplemented with 10%FBS (Hyclone), 100U/mL penicillin, 100µg/mL streptomycin. C57Bl/6 bone marrow macrophages were isolated as previously described (17); briefly, low-density bone marrow cells from C57Bl/6 femurs were isolated on Histopaque 1083 (Sigma) and plated for 3 days in non-tissue culture petri dishes in DMEM (as above) supplemented with 5% M-CSF-containing conditioned supernatant from CMG-14-12 cells(30). Adherent cells were detached by 10mM EDTA and re-plated in tissue culture dishes with the 5% conditioned supernatant 3 more days prior to stimulation. The University of Wisconsin is accredited by AALAC and mouse experiments were performed with IACUC oversight and approval. To induce ER stress, cells were pre-treated with 10µg/mL tunicamycin for 6h, 20mM 2-deoxyglucose for 6h, 10µM A23187 for 4h, 1mM dithiothreitol for 2h, or 1µM thapsigargin for 1h, depending upon time required for maximal XBP-1 mRNA splicing. Splicing was determined by optical density of PCR products separated on a 3–4% agarose gel. S. Enteriditis LPS (Sigma) was used at 100ng/mL. ER stress agents and LPS were from Sigma except DTT (Fisher). The DMSO vehicle for Tpg and A23187 had no effect on IFN-β mRNA induction. Supernatant IFN-β was quantified by ELISA (PBL Interferon source) after 1h Tpg followed by 6h LPS.
XBP-1 knockdown and Immunoblotting
RAW cells were transfected with 200–300nM mXBP-1 stealth siRNA or control “medium GC content” RNAi (Invitrogen, Cat. No 12935–300) by AMAXA nucleofection (kit V). The sequences of the XBP-1 specific sense and antisense strands were: 5’ CAGCGCAGACUGCUCGAGAUAGAAA 3’ and 5’UUUCUAUCUCGAGCAGUCUGCGCUG 3’. 24h post-transfection, cells were stimulated, lysed with RIPA buffer, and whole cell lysates resolved by 4–12% SDS-PAGE (Invitrogen). Nitrocellulose (Whatman) blots were probed with anti-XBP-1 (Santa Cruz) or β-actin (Santa Cruz), horseradish peroxidase conjugated secondary antibody (Bio-Rad), and proteins were visualized by ECL (Amersham)/film.
Quantitative PCR (qPCR)
RNA was purified with Trizol (Invitrogen), treated with DNaseI (Invitrogen), then reverse transcribed using random primers (Promega). Relative cDNA was quantitated by SYBR Green (Bio-Rad), detected by My-IQ (Bio-Rad), and normalized to 18S rRNA. Primers were designed using Beacon design software and are as follows:
18S rRNA: Forward 5’ GGACACGGACAGGATTGACAG 3’ and Reverse 5’ ATCGCTCCACCAACTAAGAACG 3’.
IFN-b: Forward 5’ ACTAGAGGAAAAGCAAGAGGAAAG 3’ and Reverse 5’ CCACCATCCAGGCGTAGC 3’.
ERdj4: Forward 5’ GGCAAAGGACAAAGAGGCAATGG 3’ and Reverse 5’ CCTGGCGTGTGTGGAAGTGG 3’.
IL-1β: Forward 5’ CTCGCAGCAGCACATCAAC and Reverse 5’ ACGGGAAAGACACAGGTAGC.
IL-6: Forward 5’ CTTCCATCCAGTTGCCTTC and Reverse 5’ ATTTCCACGATTTCCCAGAG.
ifnb1 promoter: Forward 5’ AACTGAAAGGGAGAACTGAAAG 3’ and Reverse 5’ GCAAGATGAGGCAAAGGC 3’.
ERdj4 promoter: Forward 5’ AGGGAAGGATGAGGAAATCG 3’ and Reverse 5’ ACTGTTGTTGCCGTTTGG 3’.
+6.0kb site: Forward 5’ CGAAGGGAAAGAGAAATGTG 3’ and Reverse 5’ CTGGAGGTAACTGGTTGC 3’.
XBP-1: Forward 5’ ACACGCTTGGGAATGGACAC 3’ and Reverse 5’ CCATGGGAAGATGTTCTGGG 3’.
Chromatin immunoprecipitation (ChIP)
ChIP was performed as previously described (31); briefly, RAW cells were fixed with 1% formaldehyde, lysed, and sonicated to generate chromatin fragments. After pre-clearing with normal rabbit serum (Covance Research Products), immunoprecipitations were performed with Protein A sepharose (Sigma) coupled to anti-IRF-3, CBP/p300, XBP-1, TFIID/TBP, Pol II (Santa Cruz). DNA-protein complexes were eluted with 0.1M NaHCO3, 1%SDS, cross-links reversed with 0.3M NaCl, and protein degraded with Proteinase K (Promega). Phenol chloroform extracted DNA samples were analyzed by qPCR as above. Percent occupancy was derived by comparison with input chromatin.
Co-Immunoprecipitations
HEK293 T cells were transfected with expression vectors for Flag-tagged XBP1u/s, HA-tagged XBP1s and HA-tagged P300 by Lipofectamine 2000 (Invitrogen) (32, 33). 18–24h later, cells were lysed (150mM NaCl, 1% Triton X-100, 1mM EDTA, 50mM Tris pH7.5, DTT, Protease inhibitor cocktail), and lysates immunoprecipitated with agarose coupled anti-HA or anti-Flag (Sigma). After washing (20mM Tris, 137mM NaCl, 2mM EDTA, 1% Triton-X100, 10% Glycerol, 0.5mM DTT), samples were boiled and proteins resolved on SDS 6–8% gels. Western blots were probed with anti-HA-HRP or anti-Flag-HRP (Sigma), developed with ECL substrate (Pierce), and exposed to film.
Luciferase assays
IFN-β promoter (bp-330 to +9) and +6kb sequences (305bp, 27790207–27790507 (Genbank)/88161592–88161890 (FASTA)) were cloned from RAW264.7 genomic DNA isolated by DNeasy kit (Qiagen). XBP-1 binding sites (IRF-proximal TGCAC (Δ1) or distal TGCA (Δ2)) were deleted by PCR with Pfu DNA polymerase (Stratagene). These segments were inserted into the pGL3 basic luciferase reporter plasmid (Promega) using the KpnI/Xho1and XhoI/BglII sites (respectively) upstream of the luciferase gene. For Tpg stimulations, RAW 264.7 macrophages were transiently transfected with 2 µg luciferase reporter and 0.2 µg Renilla TK (Invitrogen) by AMAXA. 20h later, cells were stimulated for 8h. Luciferase activity was detected by Dual-luciferase reporter assay system (Promega) and read on Synergy Plate reader (BioTeK instruments). Transfection efficiency was normalized to Renilla activity. For comparison of XBP-1s and XBP-1u, RAW cells were transfected with 1 µg pCDNA3.1 vector, XBP-1s or XBP-1u (provided by Dr. Laurie Glimcher, Harvard) plus 1 µg luciferase reporters using TransIT-Neural transfection reagent (Mirus)(21). 16h later, cells were stimulated with LPS for 7h. Luciferase activity was detected using luciferase assay reagent II (Promega) read on a TR717 luminometer (PE applied Biosystems). Results were normalized to total protein (BCA assay, Pierce). All luciferase assay samples were run in duplicate.
Statistics
Statistical differences between groups of data were determined by 2-tailed Student’s T-test. All error bars from combined experiments represent standard errors of the mean. For representative experiments, error bars represent deviations of duplicate determinations.
Results
ER stress augments LPS-induced IFN-β in macrophages
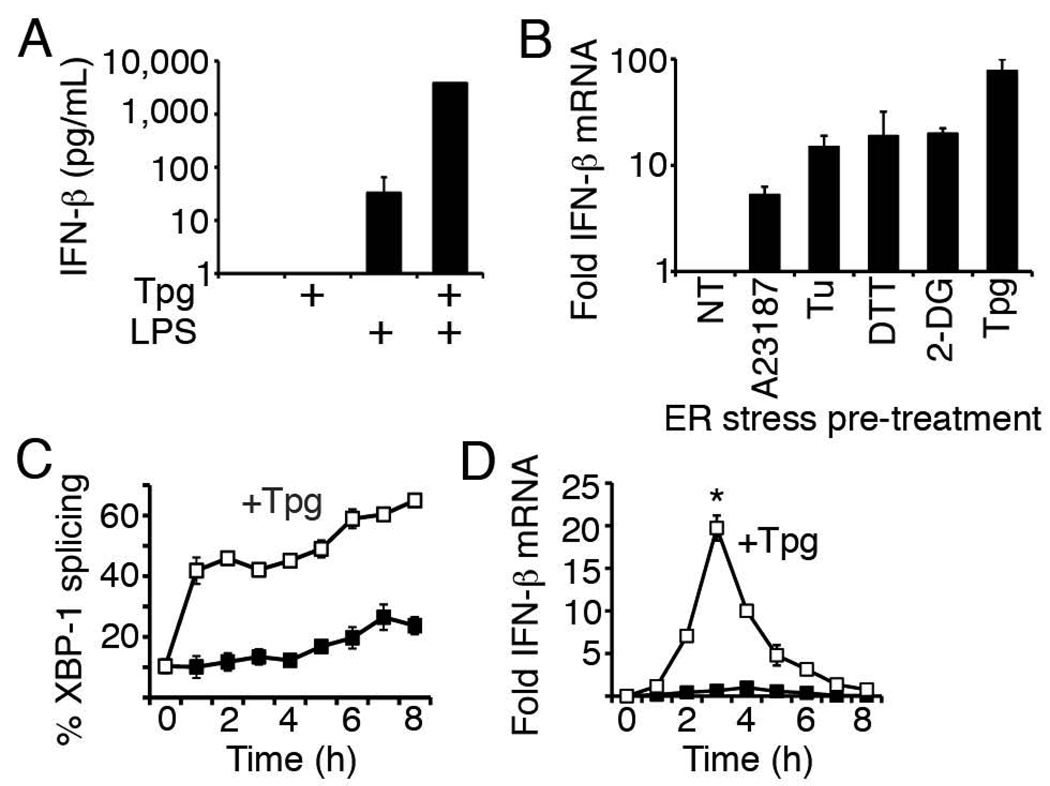
Our previous studies had shown that pre-treatment of primary macrophages with a commonly utilized pharmacologic inducer of ER stress, thapsigargin (Tpg) could significantly augment subsequent LPS stimulated IFN-β transcription. Similarly, macrophages expressing a misfolding protein (HLA-B27) and undergoing a UPR produced more IFN-β mRNA in response to LPS (17). In this study, Tpg-primed bone marrow macrophages responded to LPS with log-fold synergistic induction of IFN-β protein (Figure 1A). Tpg inhibits the SERCA Ca2+pump, which potently and rapidly induces a UPR through disruption of the ER-cytosol calcium gradient(34). Since Tpg could have multiple effects on cell signaling beyond disruption of the ER, we examined the effect of pre-treatment with other pharmacologic UPR inducers on LPS-induced IFN-β transcription (Figure 1B). Pre-treatment with these other UPR-inducing agents also significantly augmented LPS-stimulated IFN-β gene expression in primary bone marrow macrophages. IFN-β induction in the absence of LPS was insignificant. Tpg remained the most potent potentiator of IFN-β transcription. The ~2 log increase in transcript observed with Tpg pre-treatment correlated well with the observed increase in protein secretion. In RAW264.7 macrophages, Tpg rapidly induces XBP-1 splicing (Figure 1C), and pre-treatment for 1h maximally increased LPS-stimulated IFN-β mRNA by 3 hours with a return to baseline by 8 hours (Figure 1D).
Figure 1. Synergistic induction of IFN-β in macrophages by LPS and ER stress.
A) Murine bone marrow macrophages were pre-treated with 1h thapsigargin (Tpg), and then stimulated with 6h LPS. Results were combined from 2 independent experiments. *P=0.001 vs. LPS. B) Murine marrow macrophages were pre-treated with ER stress inducers calcium ionophore (A23187), tunicamycin (Tu), dithiothreitol (DTT), 2-deoxyglucose (2-DG), Tpg, or no ER stress inducer (NT) prior to 3h of LPS. Relative mRNA expression was determined by quantitative PCR (qPCR). Bars represent fold induction of IFN-β mRNA by ER stress pre-treatment plus LPS compared to LPS without ER-stress pre-treatment (NT=1). Results were combined from 3 independent experiments. Except for homocysteine, p=<0.05 for other ER stress inducers vs. NT. (C, D) RAW264.7 macrophages were pretreated with Tpg for 1h and then LPS for up to 8h. Black boxes are LPS only and white boxes are Tpg+LPS. %XBP-1 splicing (C) represents ratio of spliced and spliced+unspliced PCR products. D) For fold induction of IFN-β mRNA, results were combined from 2 independent experiments by normalizing to LPS-induced IFN-β mRNA at 4h (=1). *P=0.003 vs. LPS. Representative experiments for B and D are shown in Supplemental Figure 1.
Tpg amplifies factor recruitment to the ifnb1 promoter
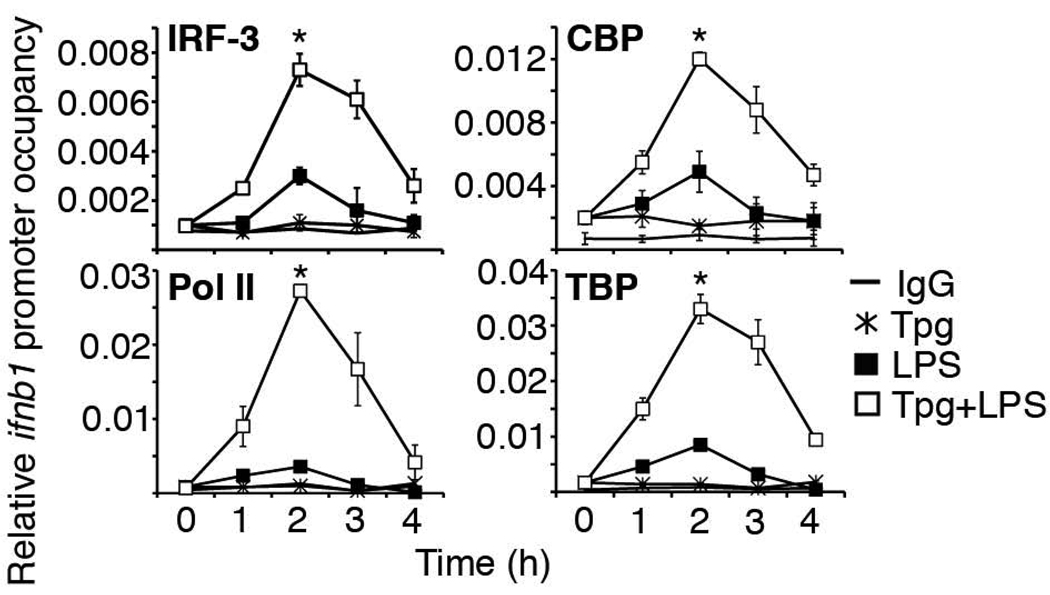
The log-fold increase in IFN-β mRNA and kinetics of synergy suggested that regulation of IFN-β induction by ER stress occurs at the transcriptional level. Preliminary studies with actinomycin D also suggested that ER stress did not prolong IFN-β mRNA half-life (data not shown). To determine how ER stress affected LPS-induced recruitment of transcriptional and other regulatory factors to the ifnb1 gene promoter, occupancy of the ifnb1 promoter was examined by chromatin immunoprecipitation (ChIP). Guided by the kinetics of IFN-β transcriptional synergy (Figure 1D), we examined factor occupancy during the first 4 hours following LPS stimulation. To ensure maximal sensitivity in RAW264.7 macrophages, we utilized the most potent and rapid inducer of ER stress, Tpg. Tpg pre-treatment resulted in a 2–3-fold increase in IRF-3 and CBP occupancy, and a 7–10 fold increase in transcriptional machinery recruitment (RNA Polymerase II (PolII) and TATA-binding protein (TBP)) compared to stimulation with LPS alone (Fig. 2). Preliminary evidence suggests a 2–3-fold increase in NF-κB (p65) occupancy as well at the 2h time point. Maximum occupancy occurred for most factors around 2 hours, with a significant decrease in transcriptional machinery occupancy by 4 hours, correlating with peak IFN-β mRNA kinetics. The enhanced transcriptional machinery recruitment also correlated well with the degree of synergistic mRNA induction typically observed in RAW cells (Figure 1D).
Figure 2. ER stress increases transcription factor and machinery occupancy of the ifnb1 promoter.
RAW 264.7 macrophages were pre-treated with Tpg for 1h and then stimulated with LPS for the times indicated. Binding of IRF-3, CBP, TBP and Pol II to the ifnb1 promoter was detected by ChIP. Relative factor occupancy compared to input chromatin was determined by qPCR. Control IgG results were combined for all stimulation conditions (IgG, no symbol). Results were combined from 4 (IRF-3, TBP), 5 (CBP), and 2 (Pol II) independent experiments. *P<0.001 for LPS vs. Tpg+LPS.
The UPR transcription factor XBP-1 regulates the synergistic induction of IFN-β in macrophages
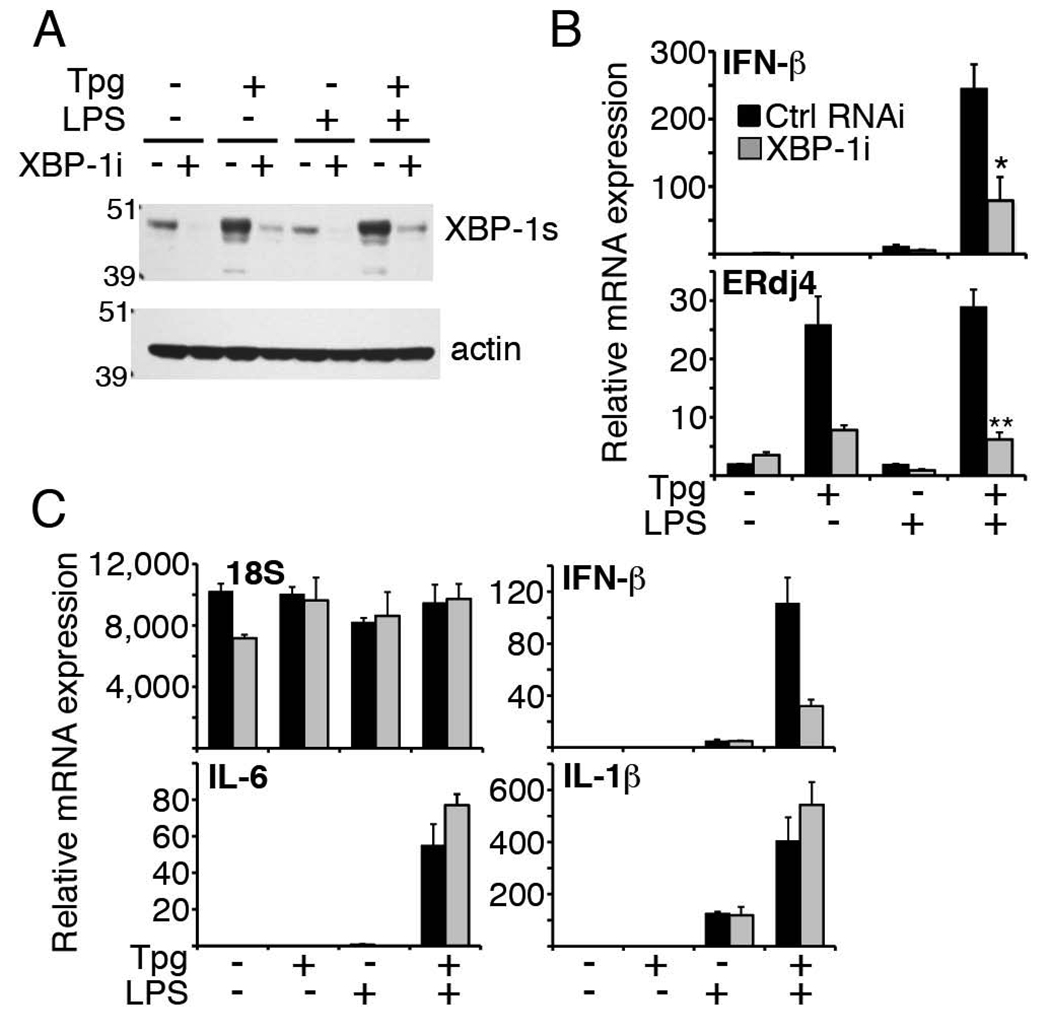
Our previous studies examining the different signaling pathways initiated by the UPR (originating from PERK, IRE-1 and ATF6) had suggested a critical role for the IRE-1-dependent transcription factor XBP-1: Synergy was abrogated in XBP-1 knockout MEFs and by XBP-1 RNAi knockdown in LPS receptor expressing 293 cells (17). In this study, to determine if XBP-1 was required for synergy in macrophages, we used 2 approaches: we initially transfected the RAW cells with a dominant-negative XBP-1 containing the DNA-binding region, but not trans-activating region(35). Interestingly, we were unable to expand macrophages containing this construct, suggesting a role for XBP-1 in macrophage survival. We then knocked down XBP-1 with short hairpin interfering RNA (siRNA). Transiently transfecting RNAi to achieve a 4–5-fold knockdown of XBP-1 mRNA during stimulation did not have an obvious impact on viability or expression of the 18S rRNA housekeeping gene (Figure 3C). XBP-1 RNAi decreased both baseline and Tpg-induced XBP-1 protein (Figure 3A). As can be seen in Fig. 3B, XBP-1 RNAi decreased synergistic induction of IFN-β mRNA by an average of ~70%. The decrease in LPS-induced IFN-β in the absence of Tpg was not statistically significant. By comparison, the induction of ERdj4, a known XBP-1-regulated chaperone, was reduced 80% by XBP-1 RNAi during combined Tpg and LPS stimulation (35). In our transient transfection system, XBP-1 knockdown did not impair induction of IL-1β or IL-6 mRNA (Figure 3C), suggesting relative specificity of XBP-1 regulation for the IFN-β cytokine.
Figure 3. XBP-1 knockdown decreases synergistic induction of IFN-β in macrophages.
A) RAW 264.7 macrophages transfected with 300 nM control or XBP-1 siRNA (XBP-1i) were treated with 1h Tpg and then 3h LPS. XBP-1 (top) or actin (bottom) was detected by Western. Results are representative of 2 separate experiments. B) RAW cells transfected with 300 nM control RNAi or XBP-1i were stimulated as in (A) and relative IFN-β (top) and ERdj4 (bottom) mRNA was determined by qPCR. Results were combined from 2 (ERdj4) and 3 (IFN-β) independent experiments. *P=0.029, **p=0.021. C) RAW cells transfected with 200 nM control RNAi (black) or XBP-1i (gray) were stimulated as in (A) and relative expression of IFN-β, 18S rRNA, and IL-1β, and IL-6 mRNA was determined by qPCR. Results are representative of 3 independent experiments.
XBP-1 binds a site 6.1kb downstream of the ifnb1 gene during concomitant Tpg and LPS stimulation
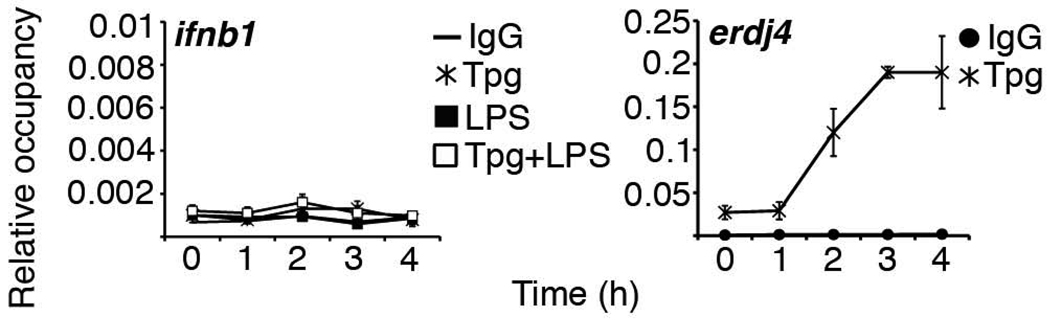
Analysis of the ifnb1 promoter region using TFSEARCH and TRANSFAC on-line databases did not reveal any XBP-1 consensus sites. However, it was possible that XBP-1 recognized a DNA sequence that had not yet been described. By ChIP, XBP-1 did not appear to bind directly to the ifnb1 promoter, although we could detect strong binding of XBP-1 to the ERdj4 promoter (Figure 4).
Figure 4. XBP-1 does not bind the ifnb1 promoter.
RAW 264.7 macrophages were stimulated as described in Figure 2, and then ChIP was performed with anti-XBP-1. Relative occupancy of the ifnb1 (left) and ERdj4 (right) promoters was assessed by qPCR by comparison to input sample. For ERdj4 ChIP, occupancies of control IgG were combined for all stimulation conditions. Results were combined from 4 (ifnb1 promoter) and 2 (Erdj4 promoter) independent experiments.
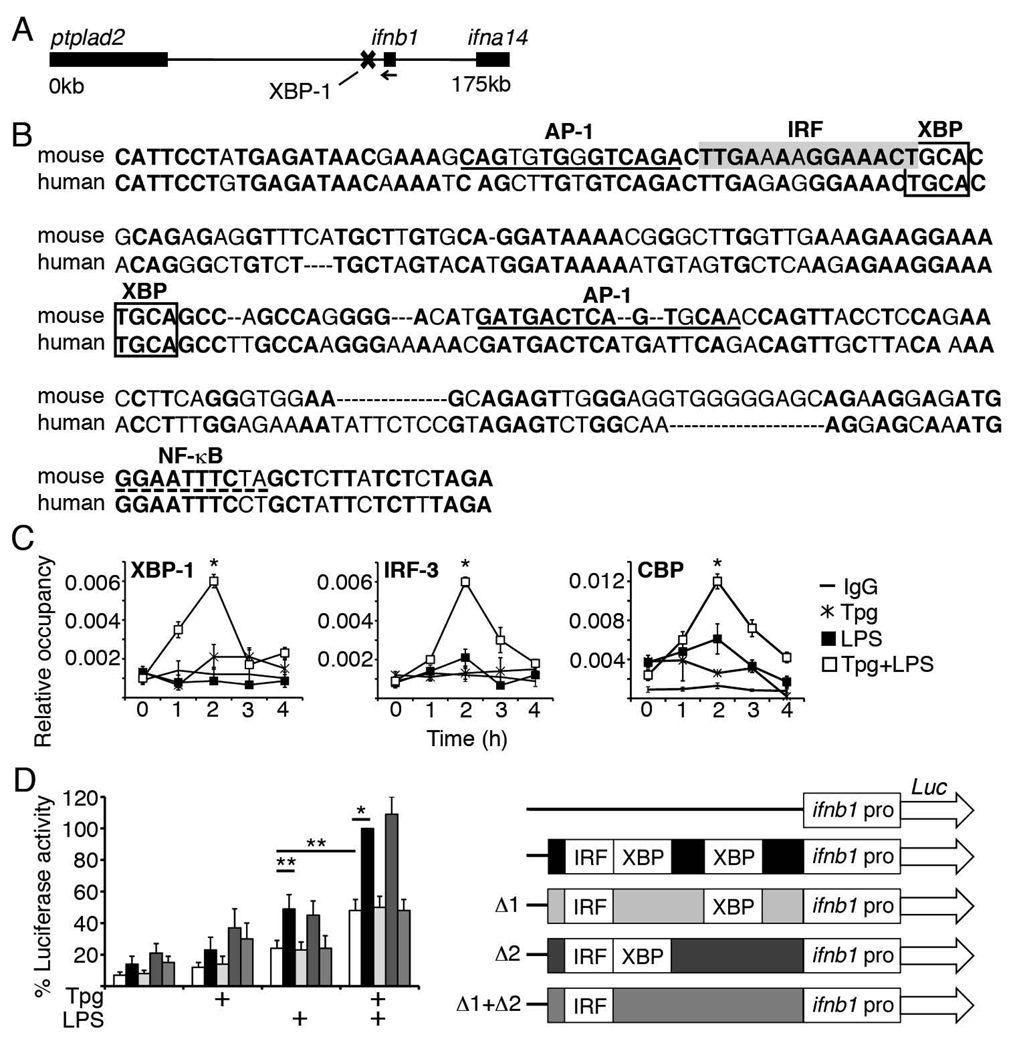
An alternative hypothesis was that XBP-1 binds a regulatory DNA segment near the ifnb1 gene. Therefore, we analyzed the 175kb between the single exon ifnb1 gene and its nearest neighbors, ifna14 and Ptplad2 (Fig 5A). Mouse and human sequences were submitted to Z-picture (zpicture.dcode.org) to search for conserved regions. We then utilized cbrc.jp/research/dg/TFSEARCH to find described XBP-1 consensus binding sites within conserved regions: Using an abbreviated UPRE consensus site (CACG) and core-binding site (ACGT), several candidate regions were identified at 6.1, 11.5, 18.3, 21, 30.6, 43, and 70kb downstream of ifnb1(36, 37). Two sites were predicted to bind multiple other factors relevant to IFN-β transcriptional at 6.1kb (AP-1, IRF, NF-kB) and 18.3kb (IRF, BLIMP-1, HMGI-Y) away. By ChIP, only the +6.1kb site bound XBP-1 unequivocally (5-fold, p=1×10−7) over the IgG immunoprecipitation control. The conserved DNA sequence of this site, predicted transcription factor binding sites and XBP-1 core sites (lower strand is TGCA) are represented in Figure 5B. The DNA sequence surrounding AP-1, IRF and XBP-1 predicted binding sites was relatively highly conserved (78% identity over the first 117 bp) compared to the region as a whole (71% over the whole 234 bp). Interestingly, at the same time the +6kb site bound XBP-1, we observed binding of key ifnb1 enhanceosome components IRF-3 and CBP (Figure 5C). We were unable to detect significant binding of NF-κB (p65) or AP-1 (ATF2) to the +6.1kb site during concomitant thapsigargin and LPS stimulation (data not shown).
Figure 5. Identification of an XBP-1-dependent enhancer site 6.1kb downstream of ifnb1.
A) Genomic region containing ifnb1, XBP-1-binding site, and contiguous genes. Ifnb1 gene: 27796544–27797313 (Genbank)/88168698-88167929 (FASTA). B) Nucleotide sequence of the +6kb site, containing base pairs 27790246–27790479 (Genbank)/88161631–88161864 (FASTA). Conserved nucleotides between mouse and human are bolded. Predicted ifnb1 enhanceosome component binding sites (80–90% consensus identity) are denoted by gray box (IRF) dotted line (NF-κB) and underscore (AP-1). XBP-1 consensus binding sites are in unfilled boxes C) RAW cells were stimulated with 1h Tpg followed by LPS for the times indicated. Factor occupancy of the +6.1kb site was detected by ChIP. Results were combined from 3 (XBP-1) and 4 (CBP, IRF-3) independent experiments. *P<=0.01. D) RAW cells were transfected with luciferase reporters containing the ifnb1 promoter alone or promoter+6kb site with no deletions, deletions of either or both conserved XBP-1 core binding sites (Δ1, Δ2, or Δ1+Δ2). Cells were stimulated with 1h Tpg and/or 7h LPS. Results were normalized to Tpg+LPS stimulation (=100%). Results were combined from 2 (Δ1+Δ2), 3(Δ1, Δ2) and 6 (promoter vs. promoter+6kb enhancer) independent experiments. *P=0.00002, **p<0.04. A sample experiment is shown in Supplemental Figure 2.
The +6kb XBP-1 binding site enhances ifnb1 promoter activity
CBP/p300 occupancy has been proposed as a “gene enhancer signature” (38). Finding CBP bound to the site during concurrent Tpg and LPS stimulation raised the possibility that the +6kb site may be an ER stress sensitive enhancer of IFN-β induction. To determine whether this putative enhancer site had any functional relevance for ifnb1 promoter activity, the ifnb1 promoter and +6kb site were cloned into a vector bearing a luciferase reporter gene. The putative enhancer alone did not induce any luciferase activity over the vector control in the absence of the promoter (data not shown). In the absence of Tpg (LPS only), the +6kb site augmented promoter activity, consistent with baseline presence of spliced XBP-1 in RAW macrophages (Figures 5D, 1C). Tpg treatment augmented promoter activity in the absence of the enhancer, consistent with described induction of NF-κB and MAPkinase signaling by ER stress (23). However, Tpg treatment further increased promoter activity in the presence of the enhancer. Compared to LPS driven promoter activity alone (no ER stress or enhancer), the presence of the enhancer and addition of Tpg pre-treatment augmented activity by ~4 fold in these assays. To determine which conserved XBP-1-binding core site mediated enhancer activity, the IRF consensus-proximal site and more distal site were deleted (Δ1,Δ2, and Δ1+Δ2 respectively, Figure 5D). The Δ1 deletion reduced activity to the level seen with the promoter alone, in both single and double deletion enhancers. Together these data suggest that the IRF consensus-proximal XBP core sequence is critical for enhancer activity of this +6kb site.
XBP-1s physically associates with CBP/p300 and augments ifnb1 promoter activity via the +6kb enhancer
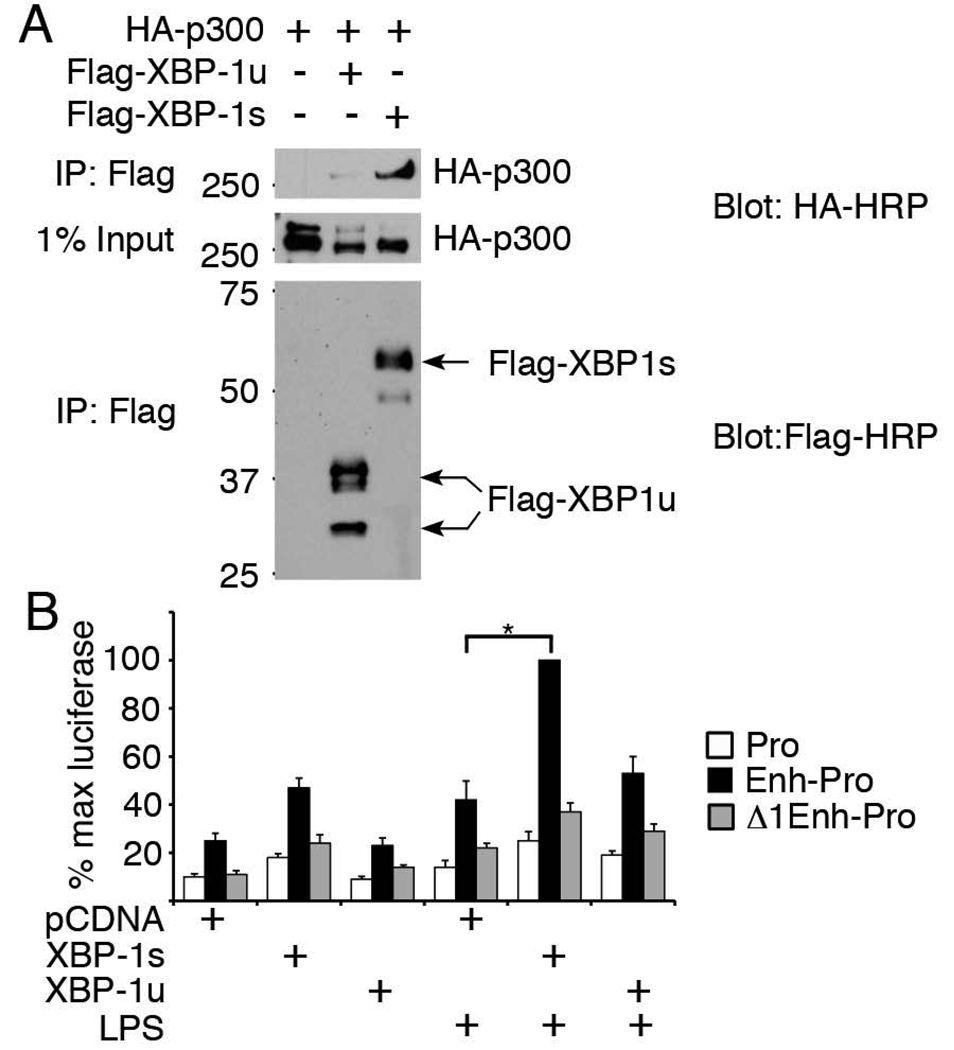
XBP-1 is a CREB family basic leucine zipper transcription factor that can form heterodimers (e.g. with c-fos and ATF6) (39, 40). As a CREB family member, it was possible that XBP-1 interacted with CREB binding protein CBP/p300. CBP/p300 has been shown to associate directly with phosphorylated IRF-3 following viral stimulation (41). Thus, an interaction between XBP-1 and CBP/p300 might explain the increased recruitment of both CBP and IRF-3 to the putative enhancer site in a multi-molecular complex during concomitant ER stress and LPS stimulation. Spliced XBP-1s encodes the 371aa ER stress-induced active transcription factor, whereas the 267aa unspliced XBP-1u has the DNA binding N-terminal domain, but not trans-activating C-terminal domain (18). In over-expression studies (Figure 6A), XBP-1s, but not XBP-1u, co-precipitated with p300. Thus the CBP/p300 co-activator may associate with the active XBP-1 transcription factor during ER stress. The predicted molecular weight of the unspliced XBP-1 is roughly 30 kD, so the higher molecular weight products in lane 2 may represent ubiquitinated protein visualized as a result of the over-expression system (21).
Figure 6. XBP-1s associates with CBP/p300 and enhances ifnb1 promoter activity.
A) Lysates from transfected HEK293 cells were immunoprecipitated with anti-Flag and Western blots probed with anti-HA HRP (top panel) or anti-Flag-HRP (lower panel). 1% of the input lysate was probed with HA-HRP (middle panel). Results are representative of 3 independent experiments. B) RAW 264.7 macrophages co-transfected with pCDNA3.1 vector, XBP-1s, or XBP-1u plus luciferase expression vectors containing either ifnb1 promoter only (Pro), +6kb site (Enh-Pro) or +Δ1 (Δ1 Enh-Pro, Figure 4D) were stimulated with 7h LPS. Results were combined from 4–5 independent experiments by normalization to maximum luciferase activity. P<0.002 for Pro vs. Enh-Pro constructs across all pCDNA3.1, XBP-1s and XBP-1u co-transfections. *P<0.00001. A representative set of experiments is shown in Supplemental Figure 3.
To determine whether XBP-1s or XBP-1u binding regulated enhancer activity, RAW 264.7 macrophages were transfected with XBP-1 expression vectors and the above luciferase reporter constructs. XBP-1s (but not XBP-1u) increased ifnb1 promoter activity in the presence of the +6kb enhancer element (Figure 6B). This increase was abrogated when the IRF-proximal XBP-1 core sequence was deleted (Δ1 Enh-Pro). In the absence of XBP-1, deletion of this core sequence also decreased enhancer-related luciferase activity to the level observed with the promoter alone, suggesting that the background enhancer activity in the unstimulated conditions reflected baseline ER stress and the presence of spliced XBP-1 in RAW cells (Figure 1C). In the LPS stimulated conditions, XBP-1s increased enhancer activity by roughly 2-fold over baseline ER stress, and increased luciferase activity 6–7-fold over the promoter alone. Together these data support a role for the +6.1kb site as an XBP-1-dependent enhancer of ifnb1 promoter function.
Discussion
We have identified an enhancer site 6kb downstream of the ifnb1 gene that exhibits significantly increased binding of XBP-1 only during concomitant Tpg and LPS treatment. Furthermore, enhancer activity of the +6kb site was responsive to the active XBP-1s transcription factor, not XBP-1u and dependent upon a predicted IRF-proximal XBP-1 core binding sequence. This +6kb region also bound the key IFN-β enhanceosome components IRF-3 and CBP/p300 during concomitant LPS stimulation and ER stress. The kinetics of XBP-1, CBP and IRF-3 binding to the enhancer site showed a striking synchronicity with the increased recruitment of CBP and IRF-3 to the ifnb1 gene promoter, with maximal occupancy after 2h LPS. The physical interaction between XBP-1 and CBP/p300 (which is known to associate with phosphorylated IRF-3 following LPS stimulation) provide a mechanistic explanation for the presence of all these factors together at the enhancer site: one could hypothesize the formation of a multi-molecular complex, whereby XBP-1 associates with CREB/p300 that in turn associates with IRF-3, thus allowing for cooperative and synchronous binding of XBP-1 and IRF-3 to the enhancer. This model reflects the need for both ER stress and TLR stimulation to promote simultaneous binding. Precedence for such cooperative assembly may be found at the ifnb1 promoter itself (16).
In previous studies, Tpg pre-treatment had no effect on the induction of the IRF-3 regulated chemokine, RANTES (CCL5) by LPS. Also, XBP-1 RNAi did not affect the induction of the IRF-3-regulated chemokine IL-8 in TLR4 bearing 293 cells (17). These data would suggest that Tpg (and by extension the UPR) does not generally activate all IRF-3 regulated genes and that the effect on IFN-β is more specific. Indeed, by gene expression microarray in primary mouse macrophages, the only chemokine or cytokines showing 10-fold or greater synergy during combined Tpg+LPS stimulation were IFN-β and IL-23 (R.A. Colbert and J. Smith, in press).
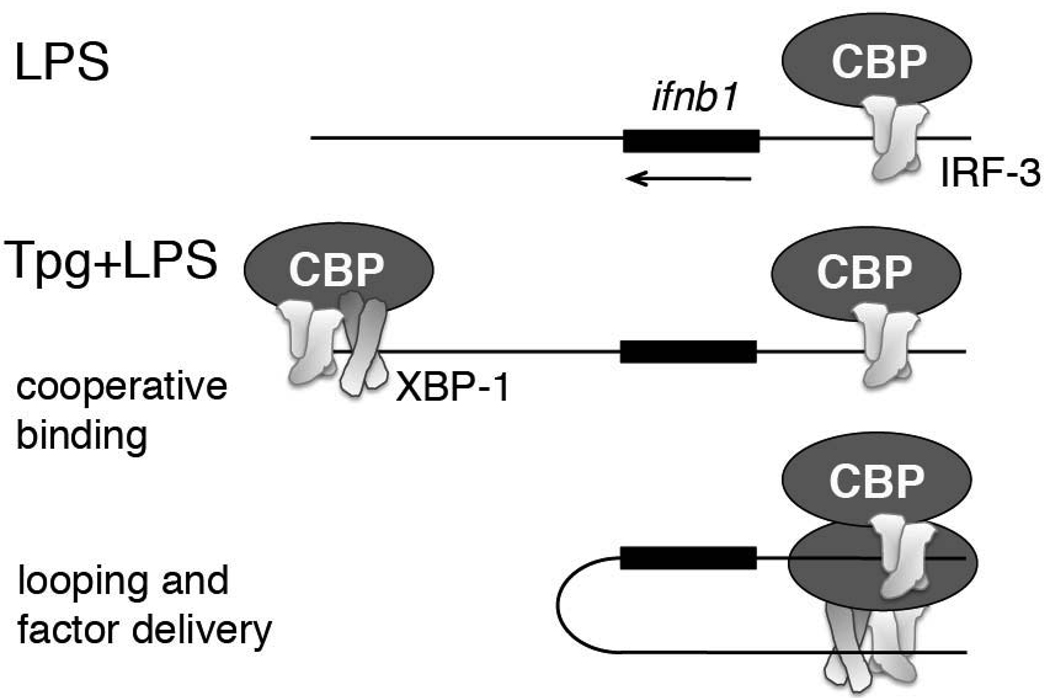
Looping of chromatin has been proposed as a mechanism that brings gene promoters and distal regulatory sites into physical apposition (42). Following synchronous binding of XBP-1, IRF3 and CBP/p300 to the +6kb region during ER stress and LPS stimulation, the enhancer may then loop around to provide increased CBP and IRF-3 delivery to the ifnb1 promoter (Figure 7). Given the cooperative assembly of factors at the ifnb1 promoter, a small amount of “extra” IRF-3 (even 2–3 fold) early in the sequence could be greatly magnified during the successive recruitment of histone modifiers and transcriptional machinery to result in the observed log-fold synergy. According to this looping enhancer theory, the enhancer would only come into play during combined ER stress and LPS stimulation; thus the comparison between LPS-stimulated promoter function and enhancer-promoter function during concomitant Tpg/XBP-1s+LPS would be the most relevant. Since XBP-1 was not detected on the promoter, XBP-1 may dissociate following looping. The compressed kinetics makes it difficult to assess this possibility. Otherwise, the cross-linking may have been insufficient to detect factors indirectly associated with the promoter.
Figure 7. Model for XBP-1 enhanced factor recruitment to the ifnb1 promoter.
In the presence of LPS stimulation alone, IRF-3 and CBP bind to the ifnb1 promoter (top). When macrophages undergoing ER stress (Tpg treatment) are stimulated with LPS, XBP-1, IRF-3, and CBP cooperatively bind the region 6.1kb downstream of the ifnb1 gene. Through chromatin looping, the enhancer bound IRF3 and CBP factors are delivered to the multi-molecular complex at the ifnb1 promoter, ultimately resulting in greater recruitment of transcriptional machinery.
An alternative explanation for the role of XBP-1 in synergy is that XBP-1 induces an unknown factor that binds the ifnb1 promoter. However, the induction of a negative regulator of IFN-β transcription by LPS precluded more direct evaluation of this hypothesis using cyclohexamide (43). The time frame, with 1h Tpg pre-treatment sufficient to detect synergy after 2h of LPS, would argue against the involvement of a newly transcribed XBP-1 gene target.
In this study, the kinetics of promoter occupancy following LPS stimulation was greatly compressed compared to what has been described for viral infection, with a significant decrease in the transcriptional machinery by 4 hours(12). This decreased promoter occupancy correlated well with the disappearance of IFN-β mRNA transcript. Other transcription factors and chromatin modifiers, besides the ones mentioned in this study, have been reported to bind the ifnb1 promoter during viral infection. However, we were unable to detect significant binding (>0.002 occupancy) of IRF-1, IRF-7 or ATF2 transcription factors, general control non-derepressible 5 (GCN5) histone acetyltransferase, or the high mobility group I protein (HMGI-Y) architectural factor (data not shown)(13, 16). The HMGI-Y DNA binding protein has been proposed as a “chaperone” that facilitates and stabilizes assembly of the enhanceosome, although it is not likely to be present in the final structure (9, 44). Acetylation of the HMGI-Y structural protein at lysine-71 by GCN5 promotes association of HMGI-Y with enhanceosome components and protects against destabilization. CBP-mediated acetylation of lysine-65 decreases the affinity of HMGI-Y for DNA and destabilizes the enhanceosome (45). Thus, sequential activity of GCN5 followed by CBP appears to be critical for sustained transcription. In regards to this study, the brief duration of transcription machinery occupancy following LPS stimulation may reflect CBP predominance and insufficient acetylation of HMGI-Y by GCN5 activity.
The luciferase results support a functional role for the newly identified XBP-1 binding enhancer in regulating the ifnb1 promoter. However, the luciferase assay may greatly underestimate the effect of the +6kb enhancer site on promoter activity in situ for the following reasons: 1) The regulation of IFN-β transcription is highly chromatin sensitive: During unstimulated conditions, a nucleosome blocks access of the transcriptional machinery to the TATA box start site. The orchestrated sequential and cooperative recruitment of various factors to the ifnb1 promoter culminates in sliding this nucleosome upstream, thus enabling transcription(13). This event has been described as a regulatory “on-off” switch for ifnb1 transcription. The luciferase construct would not recapitulate this nucleosomal sliding event. 2) The 6kb distance between enhancer and promoter might be required for looping of the chromatin and optimal orientation of enhancer and promoter 3) Finally, there may be a cooperative opening/modification of chromatin by histone acetylation that is simply not captured in a luciferase-bearing vector. All of these issues relating to chromatin structure and regulation would be challenging to recreate in a standard luciferase reporter.
The UPR and type I IFN have been separately implicated in a variety of diseases ranging from viral infections to ischemia-reperfusion injury, and inflammatory myopathies. There is evidence linking the Spondyloarthritis related MHC allele HLA-B27, a molecule shown to misfold and induce a UPR in a rat model and in humans, and type I IFN: Macrophages derived from the HLA-B27 transgenic rat show evidence for both an ongoing UPR and IFN gene signature by microarray(27). Furthermore, macrophages from the transgenic animals produce enhanced levels of IFN-β in response to LPS compared to wild type when undergoing a UPR(17). The data presented here provide a mechanistic link between the UPR and augmented IFN-β. During concomitant ER stress and TLR4 stimulation, XBP-1 binds a potential enhancer element 6kb distal to the ifnb1 gene that may enhance recruitment of IRF-3 and CBP/p300 to the ifnb1 enhanceosome. These findings have significant mechanistic implications for understanding the pathogenesis of protein misfolding and ER stress-related inflammatory diseases.
Acknowledgements
I am indebted to Dr. Emery Bresnick for helpful advice.
This work was supported by the Department of Pediatrics at the University of Wisconsin-Madison, the National Institutes of Health (KL2 grant UL1RR025011 and K08-AI081045), and the American Federation for Aging Research (RAG08061).
References
- 1.Theofilopoulos AN, Baccala R, Beutler B, Kono DH. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu Rev Immunol. 2005;23:307–336. doi: 10.1146/annurev.immunol.23.021704.115843. [DOI] [PubMed] [Google Scholar]
- 2.Honda K, Taniguchi T. Toll-like receptor signaling and IRF transcription factors. IUBMB Life. 2006;58:290–295. doi: 10.1080/15216540600702206. [DOI] [PubMed] [Google Scholar]
- 3.Beg AA. Endogenous ligands of Toll-like receptors: implications for regulating inflammatory and immune responses. Trends Immunol. 2002;23:509–512. doi: 10.1016/s1471-4906(02)02317-7. [DOI] [PubMed] [Google Scholar]
- 4.Thomas KE, Galligan CL, Newman RD, Fish EN, Vogel SN. Contribution of interferon-beta to the murine macrophage response to the toll-like receptor 4 agonist, lipopolysaccharide. J Biol Chem. 2006;281:31119–31130. doi: 10.1074/jbc.M604958200. [DOI] [PubMed] [Google Scholar]
- 5.Mancuso G, Midiri A, Biondo C, Beninati C, Zummo S, Galbo R, Tomasello F, Gambuzza M, Macri G, Ruggeri A, Leanderson T, Teti G. Type I IFN signaling is crucial for host resistance against different species of pathogenic bacteria. J Immunol. 2007;178:3126–3133. doi: 10.4049/jimmunol.178.5.3126. [DOI] [PubMed] [Google Scholar]
- 6.Deonarain R, Verma A, Porter AC, Gewert DR, Platanias LC, Fish EN. Critical roles for IFN-beta in lymphoid development, myelopoiesis, and tumor development: links to tumor necrosis factor alpha. Proc Natl Acad Sci U S A. 2003;100:13453–13458. doi: 10.1073/pnas.2230460100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deonarain R, Cerullo D, Fuse K, Liu PP, Fish EN. Protective role for interferon-beta in coxsackievirus B3 infection. Circulation. 2004;110:3540–3543. doi: 10.1161/01.CIR.0000136824.73458.20. [DOI] [PubMed] [Google Scholar]
- 8.Takayanagi H, Kim S, Matsuo K, Suzuki H, Suzuki T, Sato K, Yokochi T, Oda H, Nakamura K, Ida N, Wagner EF, Taniguchi T. RANKL maintains bone homeostasis through c-Fos-dependent induction of interferon-beta. Nature. 2002;416:744–749. doi: 10.1038/416744a. [DOI] [PubMed] [Google Scholar]
- 9.Panne D, Maniatis T, Harrison SC. An atomic model of the interferon-beta enhanceosome. Cell. 2007;129:1111–1123. doi: 10.1016/j.cell.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merika M, Williams AJ, Chen G, Collins T, Thanos D. Recruitment of CBP/p300 by the IFN beta enhanceosome is required for synergistic activation of transcription. Mol Cell. 1998;1:277–287. doi: 10.1016/s1097-2765(00)80028-3. [DOI] [PubMed] [Google Scholar]
- 11.Yie J, Senger K, Thanos D. Mechanism by which the IFN-beta enhanceosome activates transcription. Proc Natl Acad Sci U S A. 1999;96:13108–13113. doi: 10.1073/pnas.96.23.13108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell. 2000;103:667–678. doi: 10.1016/s0092-8674(00)00169-0. [DOI] [PubMed] [Google Scholar]
- 13.Lomvardas S, Thanos D. Modifying gene expression programs by altering core promoter chromatin architecture. Cell. 2002;110:261–271. doi: 10.1016/s0092-8674(02)00822-x. [DOI] [PubMed] [Google Scholar]
- 14.Sakaguchi S, Negishi H, Asagiri M, Nakajima C, Mizutani T, Takaoka A, Honda K, Taniguchi T. Essential role of IRF-3 in lipopolysaccharide-induced interferon-beta gene expression and endotoxin shock. Biochem Biophys Res Commun. 2003;306:860–866. doi: 10.1016/s0006-291x(03)01049-0. [DOI] [PubMed] [Google Scholar]
- 15.Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, Taniguchi T. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 16.Wathelet MG, Lin CH, Parekh BS, Ronco LV, Howley PM, Maniatis T. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-beta enhancer in vivo. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 17.Smith JA, Turner MJ, DeLay ML, Klenk EI, Sowders DP, Colbert RA. Endoplasmic reticulum stress and the unfolded protein response are linked to synergistic IFN-beta induction via X-box binding protein 1. Eur J Immunol. 2008;38:1194–1203. doi: 10.1002/eji.200737882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 19.Wu J, Kaufman RJ. From acute ER stress to physiological roles of the Unfolded Protein Response. Cell Death Differ. 2006;13:374–384. doi: 10.1038/sj.cdd.4401840. [DOI] [PubMed] [Google Scholar]
- 20.Yoshida H. ER stress and diseases. FEBS J. 2007;274:630–658. doi: 10.1111/j.1742-4658.2007.05639.x. [DOI] [PubMed] [Google Scholar]
- 21.Iwakoshi NN, Lee AH, Vallabhajosyula P, Otipoby KL, Rajewsky K, Glimcher LH. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat Immunol. 2003;4:321–329. doi: 10.1038/ni907. [DOI] [PubMed] [Google Scholar]
- 22.Kaser A, Lee AH, Franke A, Glickman JN, Zeissig S, Tilg H, Nieuwenhuis EE, Higgins DE, Schreiber S, Glimcher LH, Blumberg RS. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Schwabe RF, DeVries-Seimon T, Yao PM, Gerbod-Giannone MC, Tall AR, Davis RJ, Flavell R, Brenner DA, Tabas I. Free cholesterol-loaded macrophages are an abundant source of tumor necrosis factor-alpha and interleukin-6: model of NF-kappaB- and map kinase-dependent inflammation in advanced atherosclerosis. J Biol Chem. 2005;280:21763–21772. doi: 10.1074/jbc.M501759200. [DOI] [PubMed] [Google Scholar]
- 24.Zhang K, Shen X, Wu J, Sakaki K, Saunders T, Rutkowski DT, Back SH, Kaufman RJ. Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell. 2006;124:587–599. doi: 10.1016/j.cell.2005.11.040. [DOI] [PubMed] [Google Scholar]
- 25.Zhai Y, Qiao B, Gao F, Shen X, Vardanian A, Busuttil RW, Kupiec-Weglinski JW. Type I, but not type II, interferon is critical in liver injury induced after ischemia and reperfusion. Hepatology. 2008;47:199–206. doi: 10.1002/hep.21970. [DOI] [PubMed] [Google Scholar]
- 26.He B. Viruses, endoplasmic reticulum stress, and interferon responses. Cell Death Differ. 2006;13:393–403. doi: 10.1038/sj.cdd.4401833. [DOI] [PubMed] [Google Scholar]
- 27.Turner MJ, Sowders DP, DeLay ML, Mohapatra R, Bai S, Smith JA, Brandewie JR, Taurog JD, Colbert RA. HLA-B27 misfolding in transgenic rats is associated with activation of the unfolded protein response. J Immunol. 2005;175:2438–2448. doi: 10.4049/jimmunol.175.4.2438. [DOI] [PubMed] [Google Scholar]
- 28.Nagaraju K, Casciola-Rosen L, Lundberg I, Rawat R, Cutting S, Thapliyal R, Chang J, Dwivedi S, Mitsak M, Chen YW, Plotz P, Rosen A, Hoffman E, Raben N. Activation of the endoplasmic reticulum stress response in autoimmune myositis: potential role in muscle fiber damage and dysfunction. Arthritis Rheum. 2005;52:1824–1835. doi: 10.1002/art.21103. [DOI] [PubMed] [Google Scholar]
- 29.Somani AK, Swick AR, Cooper KD, McCormick TS. Severe dermatomyositis triggered by interferon beta-1a therapy and associated with enhanced type I interferon signaling. Arch Dermatol. 2008;144:1341–1349. doi: 10.1001/archderm.144.10.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takeshita S, Kaji K, Kudo A. Identification and characterization of the new osteoclast progenitor with macrophage phenotypes being able to differentiate into mature osteoclasts. J Bone Miner Res. 2000;15:1477–1488. doi: 10.1359/jbmr.2000.15.8.1477. [DOI] [PubMed] [Google Scholar]
- 31.Im H, Grass JA, Johnson KD, Boyer ME, Wu J, Bresnick EH. Measurement of protein-DNA interactions in vivo by chromatin immunoprecipitation. Methods Mol Biol. 2004;284:129–146. doi: 10.1385/1-59259-816-1:129. [DOI] [PubMed] [Google Scholar]
- 32.Sha H, He Y, Chen H, Wang C, Zenno A, Shi H, Yang X, Zhang X, Qi L. The IRE1alpha-XBP1 pathway of the unfolded protein response is required for adipogenesis. Cell Metab. 2009;9:556–564. doi: 10.1016/j.cmet.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen H, Qi L. SUMO Modification Regulates Transcriptional Activity of XBP1. Biochem J. 2010 doi: 10.1042/BJ20100193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Denmeade SR, Isaacs JT. The SERCA pump as a therapeutic target: making a "smart bomb" for prostate cancer. Cancer Biol Ther. 2005;4:14–22. doi: 10.4161/cbt.4.1.1505. [DOI] [PubMed] [Google Scholar]
- 35.Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanemoto S, Kondo S, Ogata M, Murakami T, Urano F, Imaizumi K. XBP1 activates the transcription of its target genes via an ACGT core sequence under ER stress. Biochem Biophys Res Commun. 2005;331:1146–1153. doi: 10.1016/j.bbrc.2005.04.039. [DOI] [PubMed] [Google Scholar]
- 37.Yamamoto K, Yoshida H, Kokame K, Kaufman RJ, Mori K. Differential contributions of ATF6 and XBP1 to the activation of endoplasmic reticulum stress-responsive cis-acting elements ERSE, UPRE and ERSE-II. J Biochem. 2004;136:343–350. doi: 10.1093/jb/mvh122. [DOI] [PubMed] [Google Scholar]
- 38.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 39.Ono SJ, Liou HC, Davidon R, Strominger JL, Glimcher LH. Human X-box-binding protein 1 is required for the transcription of a subset of human class II major histocompatibility genes and forms a heterodimer with c-fos. Proc Natl Acad Sci U S A. 1991;88:4309–4312. doi: 10.1073/pnas.88.10.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Acharya A, Rishi V, Moll J, Vinson C. Experimental identification of homodimerizing B-ZIP families in Homo sapiens. J Struct Biol. 2006;155:130–139. doi: 10.1016/j.jsb.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 41.Hiscott J, Pitha P, Genin P, Nguyen H, Heylbroeck C, Mamane Y, Algarte M, Lin R. Triggering the interferon response: the role of IRF-3 transcription factor. J Interferon Cytokine Res. 1999;19:1–13. doi: 10.1089/107999099314360. [DOI] [PubMed] [Google Scholar]
- 42.Li Q, Barkess G, Qian H. Chromatin looping and the probability of transcription. Trends Genet. 2006;22:197–202. doi: 10.1016/j.tig.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 43.Whittemore LA, Maniatis T. Postinduction turnoff of beta-interferon gene expression. Mol Cell Biol. 1990;10:1329–1337. doi: 10.1128/mcb.10.4.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Merika M, Thanos D. Enhanceosomes. Curr Opin Genet Dev. 2001;11:205–208. doi: 10.1016/s0959-437x(00)00180-5. [DOI] [PubMed] [Google Scholar]
- 45.Munshi N, Agalioti T, Lomvardas S, Merika M, Chen G, Thanos D. Coordination of a transcriptional switch by HMGI(Y) acetylation. Science. 2001;293:1133–1136. doi: 10.1126/science.293.5532.1133. [DOI] [PubMed] [Google Scholar]