Abstract
Rod cGMP phosphodiesterase 6 (PDE6) is a key enzyme of the phototransduction cascade, consisting of PDE6α, PDE6β and two regulatory PDE6γ subunits. PDE6 is membrane-associated through isoprenyl membrane anchors attached to the C-termini of PDE6α and PDE6β and can form a complex with PrBP/δ, an isoprenyl-binding protein highly expressed in photoreceptors. The stoichiometry of PrBP/δ to PDE6 binding and how the PDE6-PrBP/δ complex assembles is not fully characterized, and the location of the regulatory PDE6γ subunits within the protein assembly has not been elucidated. To clarify these questions, we have developed a rapid purification method for PDE6-PrBP/δ from bovine rod outer segments (ROS) utilizing recombinant PrBP/δ. Transmission electron microscopy of negatively stained samples revealed the location of PrBP/δ and thus where the carboxyl-termini of PDE6α and PDE6β must be located. The three-dimensional structure of the PDE6αβγ complex was determined to 18 Å resolution from single particle projections, and was interpreted by model building to identify the probable location of isoprenylation, PDE6γ subunits and catalytic sites.
Keywords: Photoreceptor cells, phosphodiesterase 6, 3D electron microscopy, phototransduction, vision
Introduction
Rod-specific cGMP phosphodiesterase 6 (PDE6)1 is a key effector enzyme in vertebrate visual signal transduction. PDE6 belongs to the PDE superfamily, whose members (PDE1–11) regulate cellular concentrations of the second messengers cAMP and cGMP 1; 2. PDEs exist as homo- or heterodimeric proteins that comprise a highly conserved C-terminal catalytic domain, and a regulatory N-terminus, often consisting of an N-terminal GAF-A and an intermediate GAF-B domain 1; 2; 3; 4. In contrast to other PDE family members, PDE6 utilizes an additional small regulatory protein and is thus made of three subunits, PDE6α (~99 kDa), PDE6β (~99 kDa) and PDE6γ (~10 kDa) that form a heterotetramer in a molar ratio of 1:1:2 5; 6; 7. Although PDE6γ inhibits the catalytic domains of both PDE6α and PDE6β 8; 9, PDEαβγ2 still has basal cGMP hydrolytic activity 6.
Both catalytic subunits PDE6α and PDE6β carry a CAAX motif at their C-termini for post-translational isoprenylation and carboxymethylation; PDE6α is farnesylated, whereas PDE6β is geranylgeranylated 10. These modifications serve as membrane anchors for attachment of PDE6 to ROS membranes 5; 10; 11. The 17 kDa prenyl-binding protein PrBP/δ 12; 13; 14 is able to extract PDE6 from ROS membranes by binding to the isoprenylated carboxyl-terminus of the catalytic subunits 15. Using fluorescence resonance energy transfer techniques, Zhang et al. 13 showed that PrBP/δ specifically binds geranylgeranyl and farnesyl moieties with a Kd of 19.06 μM and 0.70 μM, respectively. PrBP/δ is evolutionarily highly conserved and expressed throughout the animal kingdom. The homology among bovine, human and mouse PrBP/δ polypeptides is ~99% and falls to 69% between C. elegans and human 16; 17. In mammals, PrBP/δ is primarily localized to the retina, but is also present in non-retinal tissues 12. Deletion of PrBP/δ in mouse has profound effects on trafficking of farnesylated GRK1 and PDE6 to the outer segments 18 suggesting that PrBP/δ functions as a chaperone in transport of a subset of prenylated proteins.
A wealth of structural information on PDEs exists; the recently determined nearly full-length structure of PDE2 provides the first structure of an intact phosphodiesterase and insights into its regulation, domain arrangement and function 19. The structure of the cone GAF-A domain of chicken PDE6 has been solved 20. NMR measurements indicate a conformational change upon cGMP binding, and unfolding experiments revealed that that cGMP-free GAF-A is thermodynamically less stable than the cGMP-bound form 20. Neither the structure of the PDE6α or PDE6β GAF-B domains nor their functional implications are known, but the structure of the catalytic domain of a PDE5/6 chimeric protein complexed with the inhibitory PDE6γ peptide has been elucidated, giving insight into the regulatory mechanism of PDE6γ 21. A co-crystal of the transducin α subunit (Gtα) complexed with the core of the PDE6γ specifies the region of interaction between these entities 22. Moreover, the crystal structure of PrBP/δ revealed that it has an immunoglobulin-like β-sandwich fold that forms a hydrophobic isoprenyl-binding pocket 18. Finally, 3D electron microscopy of negatively stained samples unveiled the overall architecture of PDE6 at 30 Å resolution 23; 24.
Herein we describe a novel method for purification of PDE6 in complex with PrBP/δ. We present the biochemical characterizations of both PDE6αβγ2 and PDE6αβγ2-PrBP/δ. By comparing the projection structures of negatively stained PDE6αβγ2 and PDE6αβγ2-PrBP/δ samples the PrBP/δ binding site/isoprenylation site was identified. In addition, we present the 3D structure of the PDE6 complex at 18 Å resolution generated from single particle images by weighted back projection 25. Model building has been performed to fully interpret this structure.
Results
Purification of PDE6 and PDE6-PrBP/δ
PDE6 complexes were purified as described in the Materials and Methods section. PDE6 was extracted from ROS membranes with hypotonic buffer and further purified by propyl-agarose affinity chromatography. Fractions containing PDE6 were pooled, concentrated, and subjected to gel filtration chromatography using a Superdex 200 column (Fig. 1A). PDE6-PrBP/δ-GST was isolated by extraction of PDE6 from ROS membranes with recombinant GST-PrBP/δ and purified on a GSTrap column. This purification step allowed the separation of PDE6-PrBP/δ-GST from contaminating proteins (Fig. S1). Fractions containing PDE6-PrBP/δ-GST were pooled, digested with thrombin, and separated by gel filtration (Fig. 1B). Gel filtration profiles showed effective separation of PDE6 from transducin subunits (Gt in Fig. 1A) and PDE6-PrBP/δ from GST, and an excess of the PrBP/δ protein (Fig. 1B). Purified PDE6 elutes from the gel filtration column with apparent molecular weight of 220 kDa whereas PDE6-PrBP/δ elutes at 250 kDa.
Figure 1. Purification of PDE6 and PDE6-PrBP/δ by FPLC gel filtration and functional analysis.
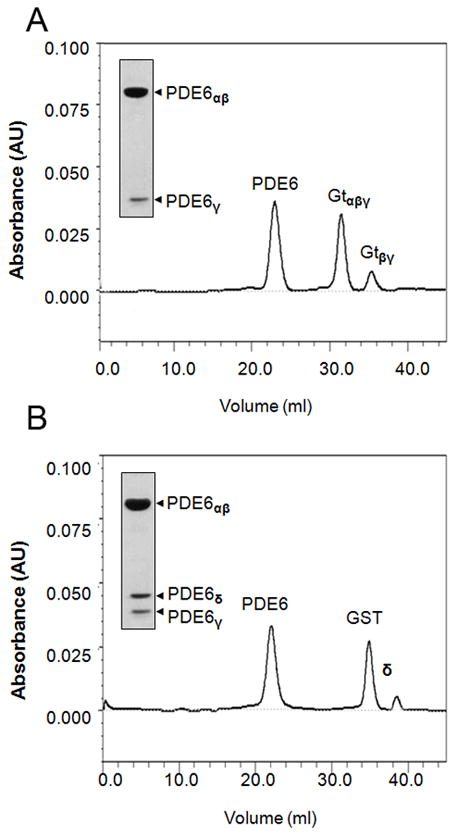
A. Propyl-agarose chromatographic purification of PDE6 released from ROS membranes. The peak fractions contain PDE6αβγ and Gtαβγ and Gtβγ, respectively. Inset shows PDE6 peak after gel-filtration. B. Elution profile of concentrated pooled PDE6-PrBP/δ-GST fractions after thrombin digestion. Peak fractions are PDE6-PrBP/δ, GST and free (excess) PrBP/δ. PDE6-PrBP/δ fractions were collected and used for further biochemical, biophysical and microscopic experiments. Inset, 12% SDS-PAGE separation of PDE6αβγ2-PrBP/δ (3 μg).PDE6-PrBP/δ.
Stochiometry of purified PDE6 and PDE6-PrBP/δ
Subunits of PDE6 and PDE6-PrBP/δ preparations were separated by SDS-PAGE and the molar ratios of PDE6α, PDE6β, PDE6γ was confirmed to be 1:1:2 for PDE6, in agreement with published results 7; 26, whereas the molar ratio was 1:1:2:2 for PDE6α:β:γ:PrBP/δ, in contrast to 1:1:2:1 as previously published 12. Native electrophoresis followed by immunoblotting showed the single bands of PDE6αβγ2 and PrBP/δ-PDE6, respectively, suggesting that the hetero-oligomers formed stable complexes (data not shown).
Projection maps of PDE6, PDE6-PrBP/δ and PDE6-PrBP/δ-GST
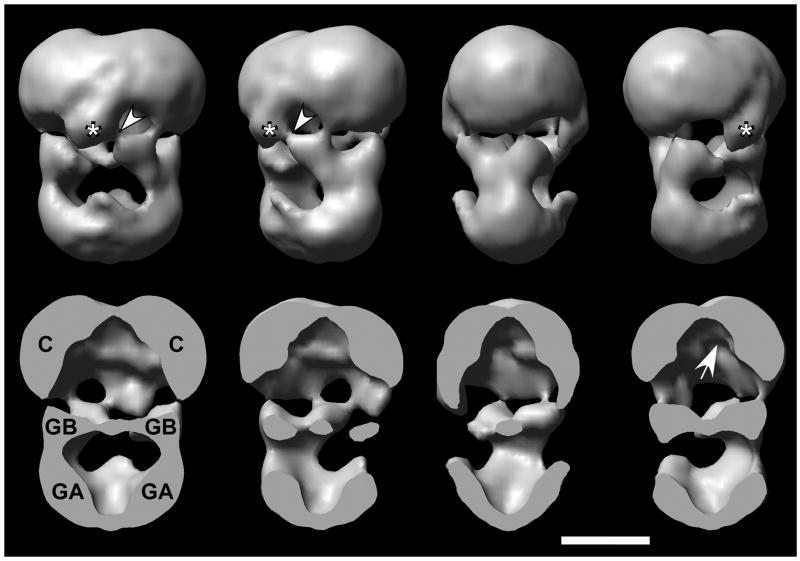
Negatively stained PDE6 and PDE6-PrBP/δ preparations exhibit particles having a length of 132±7 Å and a width of 100±3 Å (Figs. 2A and 2B). They have a wide and a narrow end, the latter being identified previously as the N-terminus 23. Upon initial observation, these particles appear identical. At closer inspection a minor fraction (<10%) of the PDE6-PrBP/δ complexes showed an additional small domain protruding from the catalytic domains, and an even smaller fraction of particles having two additional protruding domains (arrowheads in Fig. 2B). These protrusions have a size that is compatible with the mass of PrBP/δ (~17 kDa), and are most clearly discerned in <10% of the class averages obtained by multivariate statistical analysis (inset Fig. 2B). A few class averages also showed the existence of two protrusions and document the presence of two binding sites for the PrBP/δ. PDE6-PrBP/δ-GST complexes have a length of 175±8 Å and roughly the same width as the PDE6 particles. Because GST (~25 kDa) significantly increases the mass of the protrusions attached to the isoprenylation sites at the catalytic domains, the additional domain is well visible (Fig. 2C, arrowheads). The class averages shown in the top row of the inset exhibit a domain that corresponds to two δ-GST moieties, whereas the bottom row shows averages of PDE6 complexes that have a single PrBP/δ-GST construct attached. Visible connections to the catalytic domains are mostly only on one side – notably, all of them linking the PrBP/δ-GST protein to the same location as where the PrBP/δ subunits are found (Fig. 2B inset). To further establish the location of PrBP/δ, ten class averages of PDE6 and of PrBP/δ, the corresponding standard deviation (SD) maps, and finally the difference map PDE6-PrBP/δ – PDE6 were calculated. As illustrated in Fig. 2D, most of the statistical fluctuations between PDE6 class averages occur at the periphery of the complex as result of variations of the negative stain depth (Fig. 2D2). In spite of the disorder of the PrBP/δ, two clear yet smeared-out additional blobs are visible in the average from the ten PDE6-PrBP/δ class averages in inset of B (Fig. 2D3). The corresponding SD map exhibits a strong signal at the putative location of PrBP/δ (Fig. 2D4). Finally, the difference map (Fig. 2D5) reveals clear signals due to two PrBP/δ’s in addition to stain variations mainly at the periphery of the PDE6-PrBP/δ complex.
Figure 2. Projection maps of PDE6, PDE6-PrBP/δ and PDE6-PrBP/δ-GST complexes.
. Negatively stained PDE6 complexes exhibit an elongated shape with a wide top and a narrower end. Reproducible substructure is enhanced in the class averages shown in the inset, revealing two stain-filled cavities. B. Similar particles are observed in micrographs of PDE6-PrBP/δ preparations. Upon closer inspection, ~10% of these complexes show an additional domain protruding from the wider end (arrowheads). Even less frequently, two such domains are found to be attached. The selected class averages (inset) show that these protruding domains are emerging from a well-defined site, which is the putative location of isoprenylation in the catalytic domain (see also Figure 5). C. PDE6-PrBP/δ-GST complexes are significantly longer as result of the PrBP/δ-GST chimeric protein attached to the wider end (arrowhead). Class averages in the top row of the inset display PDE6-(PrBP/δ-GST)2 complexes, whereas the bottom row shows PDE6-PrBP/δ-GST complexes. D. Averages, standard deviation (SD) maps and difference maps from PDE6 and PDE6-PrBP/δ complexes. D1: PDE6; D2: SD of PDE6; D3: PDE6-PrBP/δ; D4: SD of PDE6-PrBP/δ; D5: PDE6-PrBP/δ – PDE6. The scale bars represent 150 Å; boxes of the insets have a width of 290 Å, and those in D are 250 Å wide.
3D structure of the PDE6 complex
As illustrated in Figs. 2 and S2C, the elongated particles tended to adsorb to the carbon film in a preferential orientation. Therefore, we recorded additional images from both the PDE6 and the PDE6-PrBP/δ grid preparations with the grid tilted at 45°. Because the population of complexes with one or two PrBP/δ proteins bound was <10% for the PDE6-PrBP/δ preparation, both data sets were merged to determine the 3D structure of PDE6. The complete data set comprising 14,180 single particle projections allowed us to calculate a 3D map using a simple model for bootstrapping the reconstruction and refinement procedure. The model selected comprised 2 Gaussian blobs having a diameter corresponding to the approximate mass of GAF-A, 2 blobs for the GAF-B and 2 larger blobs for the catalytic domains. The centers of these spherical objects were selected to match the shape of the most prominent class averages of particles acquired at 0° tilt. Because bovine PDE6α and PDE6β share a sequence homology of ~91% and exhibit almost the same mass (98,331 vs 99,341 Da), and the holoenzyme PDE6αβ is associated with two PDE6γ subunits, we imposed two-fold symmetry during the refinement process; calculation of maps without the two-fold averaging results in poorer, noisy maps which are also two-fold symmetric. The 3D maps converged rapidly when this model was used: within a few cycles the characteristic shape of the PDE6 complex we observed became visible and a stable map was obtained within 15 cycles (Fig. S2A). According to the 50% criterion, the Fourier shell correlation (FSC) function documents a resolution of 18 Å, with additional information above the theoretical three-sigma level out to 13 Å (Fig. S2B). Thus the final 3D map was bandwidth-limited to 13 Å.
The structure of the PDE6 complex roughly resembles a skull (Figs. 3, 4 and S4) and shows considerably more detail than maps previously produced by essentially the same approach 23; 24. Notably, the PDE6 complex appears to have two pronounced cavities, the larger being formed by the catalytic domains at its top and the GAF-B domains at its bottom. This C-terminal cavity is partially closed by an oblate 30 Å wide and 15 Å thick domain (Fig. 3, denoted with an *) that is linked to the GAF-B domain by a thin connector (Fig. 3, arrowheads). The N-terminal cavity is more open and is closed off at its bottom by the GAF-A domains. Cross-sections in the bottom row of Fig. 3 demonstrate that the catalytic domains are hemi-spherical, that the GAF-B domains exhibit a sheet-like density separating the upper and lower cavities, and that the GAF-A domains are connected by a narrow interface.
Figure 3. 3D structure of PDE6 at 18 Å resolution.
A. The skull-like shape of the PDE6 complex houses two major cavities, one enclosed at the top by the catalytic domains (C) and an oblate 30 Å wide density (*), the other by the GAF-A domains at the bottom (GA). The oblate density is connected to the GAF-B domain (GB) by a slender linker (arrowheads). GAF-B forms a shelf-like structure separating the two cavities. The cut-away representations show the hemispherical shape of the catalytic domains. This shape was decisive to fit the x-ray structure of the PDE5/6 chimeric protein into the map, resulting in our assignment of the active site being on the interior surface of the top cavity (arrow). The map was rendered at 100% mass, and each view is rotated wrt to its left neighbor by 45° in counter-clock direction. Scale bar: 50Å.
Figure 4. PDE6 exhibits an entangled topology.
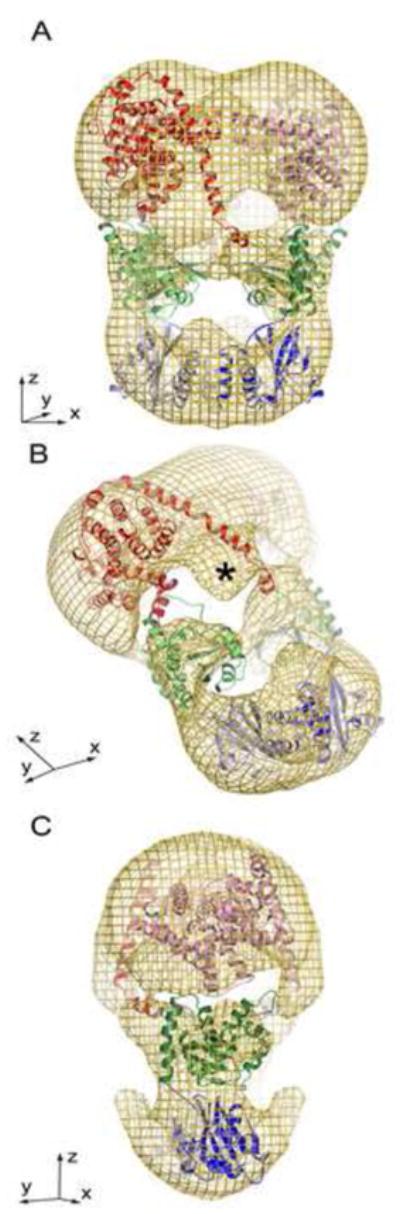
A homology model for PDE6 was generated by docking structures of the closest in sequence of individual subdomains into the 18 Å map. Crystal contacts from subdomain structures were also examined to understand likely interaction surfaces between the individual PDE protamer subdomains (for details see text). A. The PDEα and PDEβ subunits wrap around one another exhibiting an entangled topology. Catalytic domains are shown in red/pink, GAF-B domains are shown in green/light green and GAF-A domains are shown in blue/light blue. B. Model has been rotated to show the unmodeled density (*), which comprises the putative location of the central region of PDE6γ. C. View of A rotated 90° about the y-axis.
Modeling
To better understand our EM map we used all the structural information available and constructed a chimeric structure from the GAF-A domain of PDE6C (PDB ID: 3DBA), a homology model of the GAF-B domain and the PDE5/6 chimeric catalytic domain (PDB ID: 3JWQ). Attempts to fit the structure of PDE2A (PDB ID: 3IBJ) failed, because this structure was ~16 Å longer than the PDE6 map (Fig. S3). Therefore, we began by fitting the GAF-A dimer into the lower lobe with minor disruption of the dimer present in the crystal structure. As there is no structure of a closely related homolog for the GAF-B domain, we utilized SWISSMODEL 27 to build a homology model based on GAF-B from PDE2A and fit it into the middle lobe of the map. The PDE5/6 catalytic domain has roughly the shape of a half-sphere, having the catalytic cleft on its rather flat surface, and the last residue on the spherical surface. Our observation that PrBP/δ binds to the spherical side of the catalytic domain supports the placement of the catalytic domain into the half-sphere density with the flat side facing the cavity. Therefore, the catalytic cleft 21 must open toward the interior of the upper cavity of PDE6, as marked in Fig. 3 (bottom row) with an arrow. Finally, the connection between GAF-B and the catalytic domain predicted to be mostly helical needed to accommodate ~50 residues. These constraints were best satisfied with an entangled arrangement of the PDE6α and PDE6β subunits, assigning the thin connection between the GAF-B and catalytic domains to a long, connecting helix. After placing the domains into the map employing the features of CHIMERA, connecting helices were built and the model was refined using COOT. Once one entire monomer was fit, we generated the dimer by applying the 2-fold symmetry imposed on the map obtained by electron microscopy (Fig. 4).
Discussion
In this study, we present a purification of free PDE6 and the complexes formed by PDE6 and PrBP/δ in form of PDE6-PrBP/δ-GST and PDE6-PrBP/δ. We compare the biochemical properties of PDE6 and PDE6-PrBP/δ, and elucidate their structure by electron microscopy and single particle processing of negatively stained PDE6 samples.
Complex formation between the PDE6 and PrBP/δ subunits
Previous isolation protocols for PDE6-PrBP/δ resulted in a complex containing one PrBP/δ per PDE6 oligomer, although the possibility of additional PrBP/δ subunits was not excluded 12. Our attempts to reconstitute the PDE6-PrBP/δ complex in solution from PDE6 (purified on propyl-agarose resin after extraction under hypotonic conditions) and recombinant PrBP/δ by size exclusion chromatography failed to yield a homogenous complex with a well-defined ratio between PDE6 and PrBP/δ. For that reason, we elected to purify PDE6-PrBP/δ complex directly from ROS membranes, where binding of PDE6 and PrBP/δ occurs and PDE6 is properly presented on the surface of the lipid bilayer. The stochiometry estimates by Coomassie Blue stained SDS-PAGE for the PrBP/δ subunits suggested that ~two PrBP/δ are bound per PDE6 oligomer, indicating that both isoprenyl anchors (farnesyl and geranylgeranyl) bind PrBP/δ. Since our primary goal was simply to identify the binding site of PrBP/δ, we did not attempt to determine the stoichiometry with a more quantitative method.
We emphasize that excess PrBP/δ-GST does not result in unspecific binding to PDE6. Firstly, gel filtration chromatograms exhibit a symmetric, sharp peak of PDE6-PrBP/δ, with complete separation from free unbound PrBP/δ subunits (Fig. 1B). Secondly, single particle projections document that PrBP/δ-GST always bound the catalytic domain where the isoprenylation is located (Figs. 2C). If PrBP/δ-GST proteins were to bind nonspecifically, evidence of this would be seen in the images of negatively stained complexes. All of our biochemical experiments and functional analyses (data not shown) revealed that after purification both PDE6 and PDE6-PrBP/δ are active and stable in biochemical assays. Moreover, we could detect no difference in catalytic efficiency, consistent with results published by other groups 28.
The differences between projections of PDE6 and PDE6-PrBP/δ complexes
To recognize a single 17 kDa protein within a complex of roughly 220 kDa mass by negative stain transmission electron microscopy is an ambitious goal. However, as a result of the preferential adsorption of PDE6 complexes with the long axis parallel to the carbon film and the peripheral location of the isoprelynation sites, the PrBP/δ protein could be seen as a protruding domain at the wider end of the PDE6 complex (Fig. 2B). During sample transfer between two laboratories and negative staining procedure, a large fraction of the PDE6-PrBP/δ complexes dissociated. The low occupancy (<10% for a single protruding domain/PDE6; <2% for two protruding domains/PDE6) and the conformational variation of these protrusions suggested that they might not be identified by single particle analysis. However, multivariate statistical classification and averaging of projections from negatively stained PDE6-PrBP/δ complexes reveal 10% of the class averages had one or even less frequently, two protrusions located at the periphery of the catalytic domains (Fig. 2B, inset). Comparative analyses of top-view class averages of both PDE6 and PDE6-PrBP/δ complexes showed variations in PDE6 projections mainly at their periphery, which reflects variations of the stain thickness (Figs. 2D1 and D2). For PDE6-PrBP/δ, the standard deviation map (Fig. 2D4) exhibits maxima that correlate with the locations of PrBP/δ, which are distinct in the PDE6-PrBP/δ average (Fig, D3). This signal and the one of the staining variation are prominent in the difference map displayed in Fig. D5. The location of PrBP/δ is confirmed by class averages of PDE6-PrBP/δ-GST complexes (Fig. 2C). It is interesting to note that class averages of PDE6-PrBP/δ-GST particles unveil more often only one connection to the catalytic domain than two connections, even when two PrBP/δ-GSTs are bound (Fig. 2C inset, top row) possibly reflecting the difference in affinity of PrBP/δ for geranylgeranyl and farnesyl moieties 13.
3D map of PDE6
We selected a total of 14,180 single particle projections from negatively stained PDE6 and PDE6-PrBP/δ preparations, with about 40% acquired from grids inclined at 45° with respect to the optical axis of the electron microscope. This helped to improve the coverage of the tilt angle space (Fig. S2), a problem often found with particles adsorbed in a preferential orientation to the carbon film. We calculated 3D maps without enforcing the 2-fold symmetry expected from biochemical data to check for deviations from this symmetry. Such maps were noisier, but clearly symmetric and prompted us to implement the 2-fold symmetry constraint throughout the refinement. The final map shown in Fig. 3 comprised 11481 projections, 81% of all particles picked. All projections were corrected for effects of the contrast transfer function – an effort usually not invested for processing images of negatively stained samples. This and the large data pool helped to significantly improve the resolution compared to previous reports 23; 24. The pertinent features of the PDE6 3D map are the overall skull-like shape and upper and lower cavities. Starting from a crude model composed of six spherical densities, the open lower cavity and a rather enclosed upper cavity emerged after four refinement cycles, and the map converged after 15 cycles to where the FSC function indicated a resolution of 18 Å (Fig. S2B). Densities partially closing the top cavity are not visible in the lower resolution maps previously published 23; 24, and represent new features in the 3D map presented here.
The challenge was to interpret this 18 Å resolution map. We exploited the available structural information to the fullest extent, starting from the most complete structure of a PDE family member, that of PDE2A 19. However, this structure with a length of ~150 Å could not be accommodated into the skull-like PDE6 structure of ~130 Å length (Fig. S3). Moreover, the dimer interface between the linear extended PDA2 monomers, which forms the center and extends over the entire length of the complex is not compatible with the two distinct, large cavities we observe in PDE6. In contrast, the good fit of the GAF-A structure of PDE6C (PDB ID: 3DBA) to the narrower end of our PDE6 map provided the basis to begin model building and fitting. The information on the isoprenylation site (Figs. 2B and C) and the hemispherical shape of the density allocated to the catalytic domain gave the constraints with which to fit the structure of the PDE5/6 catalytic domain (PDB ID: 3JWQ) into the top of the skull. Accordingly, the catalytic cleft formed by the H-loop and M-loops 21 was found to open into the upper, rather secluded cavity. This result is of profound functional significance, as sequestration of the catalytic site from the bulk cytoplasm might have implications for the regulation of activity through access to substrate.
Because the GAF-B sequence of PDE6 exhibits low sequence homology to any GAF structure available, we resorted to the GAF-B homology model built by SWISSMODEL 27, based on the PDE2A GAF-B domain structure 29, and the 20% sequence homology to the corresponding PDE6 sequence. The GAF-B homology model fits rather well to the shelf-like shape of the density between GAF-A and catalytic domains, requiring a short helical connection to the GAF-A domain and providing the starting location of the long helical connection linking GAF-B and catalytic domain. The map and the necessity to incorporate a 50 residue long helical segment gave sufficient constraints to adjust the position of the catalytic domain. Taking into account the ~91% homology of the PDE6α and PDE6β subunits, the model of PDE6αβ was obtained by applying the 2-fold symmetry imposed for calculating the 18 Å map. This chimeric model is shown in Fig. 4, which illustrates the entangled fold of PDE6αβ. An alternative fit would have exhibited a right-handed entanglement rather than the left-handed one presented here, but the functional implications of both models would be similar.
It is important to note that PDE6 is the only phosphodiesterase that comprises an additional regulatory subunit, the inhibitory PDE6γ. Therefore, PDE6 is expected to possess a distinctly different structure than the PDE2A dimer, whose active sites are buried in the dimeric interface. Here, cGMP binding to the GAF-B domain leads to a conformational change disrupting the dimer interface between the catalytic domains, allowing substrate to access the active site 19. In contrast, PDE6 regulation is mediated by binding of Gtα to PDE6γ, whereas roles and mechanisms of PDE6 GAF domains in regulation of the enzyme remain largely obscure 30.
Putative location of the inhibitory PDE6γ subunit
A wealth of information has accumulated on the PDE6γ-PDE6αβ interactions: PDE6γ N-terminal (residues 16–30) has been reported to interact with GAF-A, the central domain (residues 24–46) to enhance the affinity of PDE6γ for PDE6αβ, and the inhibitory C-terminal domain (residues 73–87) to block the catalytic sites of PDE6γ-PDE6αβ 5; 8; 31; 32; 33; 34; 35; 36. The recent structure of the PDE5/6 chimeric catalytic domain showed the inhibitory PDE6γ-peptide (residues 70–87) to bind to the H–M loops that form the entrance to the catalytic site 21, thus confirming previous biochemical analyses. While the polycationic, GAF domain-interacting region of PDE6γ (residues 24–45) was reported to interact with transducin 37, the structure of the C-terminal half (residues 50–87) that interacts with the Gtα subunit has been solved 22. Finally, most recent data demonstrate the importance of the interaction of Gtα with the N-terminal half of PDE6γ for allowing catalysis to occur 36. Thus, the emerging picture of PDE6γ is that of an elongated adaptable protein with a central domain that binds to GAF-B, a C-terminus that interacts with the catalytic domain, and an unstructured N-terminal extension that binds to GAF-A 31.
Our 18 Å map contains a distinct, oblate 30 Å wide density that does not result from any part of the model and cannot readily be explained by residues lacking in the X-ray structures or homology models used (denoted with an * in Fig. 3, 4, and indicated in red shades in Fig. S4). We interpret this density to represent part of the PDE6γ subunit, because it is at a location, which would allow the C-terminal extension to reach the catalytic cleft 21, and the N-terminus to interact with the GAF-A domain 31; 32. This density also would be accessible to Gtα, which must bind to the central region of PDE6γ in order to dislodge the inhibitory C-terminal domain buried in the upper cavity of PDE6. It is not known whether the Gtα-PDE6γ complex remains bound to PDE6, but it is conceivable that the density we interpret as the central region of PDE6γ would be displaced upon binding to Gtα, thereby not only removing the inhibitory PDE6γ from, but also improving access to the catalytic site.
A working model for PDE6 regulation
The model of PDE6 shown in Fig. 4 and S4 together with the wealth of biochemical and structural information available fosters our hypothesis on the molecular events in the activation of PDE6. Upon initiating the visual signaling cascade by a photon, many Gtα subunits are activated and released from transducin to relieve inhibition of PDE6. Our localization of PrBP/δ (depicted in pale blue in Fig. S4) indicates how PDE6 is attached to the membrane. Because both Gtα and PDE6 are anchored in the membrane, they are also optimally aligned for interaction 28; 38. Previous biochemical and structural studies implicate the C-terminus of PDE6γ in directly binding residues surrounding the catalytic site 21. Through its interaction with the central region of PDE6γ (shaded in red in Fig. S4) Gtα is able to extract PDE6γ from the PDE6αβγ2 heterotetramer, and thus to displace the inhibitory C-terminal domain. This provides access to the previously blocked PDE6 catalytic cleft that faces the interior of the top enclosed cavity.
Materials and Methods
Expression and purification PrBP/δ Proteins
PrBP/δ-GST plasmid in the pGex-2T expression vector and purification was performed as previously described 13. The PrBP/δ protein was purified from an overnight digest of PrBP/δ-GST with biotinylated thrombin followed by 30 min incubation with streptavidin agarose to remove thrombin. The PrBP/δ was then separated from GST by Superdex 200 10/300 gel filtration chromatography.
Purification of PDE6
Bovine ROS membranes were prepared from 100 frozen retinas (W.L. Lawson Co., Lincoln, NE) under dim red light according to the Papermaster procedure 39, and PDE6 was purified according to the procedure described in Goc et al. 40. Fractions containing PDE6 were pooled and concentrated to 2–4 mg/ml with a 30 K MWCO Centricon (Millipore, Billerca, MA). SDS-PAGE analysis indicated that PDE6 was purified to homogeneity, and typically 100 bovine retinas yielded ~1–2 mg of PDE6 by this methodology. All protein concentrations were assessed utilizing Bradford reagent 41.
Purification of PDE6-PrBP/δ
ROS membranes were incubated for 30 min at RT with PrBP/δ-GST at a ratio of 10:1 mg total protein and washed with 10 ml of isotonic buffer (20 mM Hepes, pH 7.5, 5 mM MgCl2, 1 mM DTT, and 100 mM NaCl) followed by centrifugation at 25,000g at 4°C for 30 min. This extraction was performed three times and the combined supernatants were cleared of ROS membrane particles by an additional centrifugation at 25,000g at 4°C for 60 min followed by an overnight dialysis at 4°C against PBS containing 1 mM DTT. This solution was applied at 5 ml/min onto a 5 ml GSTrap column and washed with 3 column volumes of equilibrating buffer. Bound proteins were eluted with 20 ml of 50 mM Tris-HCl, pH 8.0, containing 10 mM glutathione and 1 mM DTT Fractions containing PrBP/δ-GST-PDE6 were pooled, dialyzed at 4°C against 10 mM Hepes, pH 7.5, containing 2 mM MgCl2, 1 mM DTT, and 100 mM NaCl, and digested with thrombin. The sample was then concentrated in a 10 K MWCO Centricon (Millipore, Billerica, MA) to a final volume of 0.25 ml and separated by gel filtration. Fractions containing PDE6 were again pooled and concentrated to 2–4 mg/ml. PDE6 was purified to homogeneity as assessed by SDS-PAGE and immunoblotting. Typical yield of PDE6-PrBP/δ was 0.4 – 0.6 mg / 100 retinas.
Chemicals and Reagents
TPCK-treated trypsin and soybean trypsin inhibitors were purchased from Worthington (Lakewood, NJ). All chemicals used for SDS-PAGE and immunoblotting were electrophoresis grade from Bio-Rad (Hercules, CA), except BCIP/NBT, which was from Promega (Madison, WI). Biotinylated thrombin and streptavidin agarose were from Novagen (Gibbstown, NJ). Other chemicals were reagent-grade from Sigma (St. Louis, MO). The anti-PDEβ subunit (PDEβ), the anti-PDE6γ subunit (PDE6γ), and anti-PrBP/δ polyclonal antibodies were purchased from Abcam Inc. (Cambridge, MA.). Alkaline phosphatase-conjugated goat anti-rabbit IgG from Promega was used as a secondary antibody. Propyl-agarose resin was prepared by coupling propylamine to CNBr-activated agarose (Santa Cruze Biotechnology Inc.). Superdex 200 and GSTrap columns were purchased from GE Healthcare Bio-Sciences (Piscataway, NJ).
Quantification of the stochiometry of PDE6α, PDE6β, PDE6γ subunits and PrBP/δ protein in purified PDE6 and PDE6-PrBP/δ
Samples of PDE6 were separated by 12% SDS-PAGE and stained with Coomassie Blue. Ratios between the PDE6β, PDE6γ subunits and PrBP/δ protein were scanned and then quantified by densitometric analysis in Image J (http://rsb.info.nih.gov/ij/).
Electron microscopy
PDE6 samples were diluted as required in the above dialysis buffer and adsorbed for 1 min to glow discharged 400 mesh carbon-coated grids. These were washed and negatively stained with 2% (w/v) uranyl acetate and imaged either with a CM 100 operated at 80 kV or a CM 200 FEG transmission electron microscope operated at 200 kV (Philips, Eindhoven, Netherlands). Electron micrographs were recorded at a nominal magnification of 52,000 x on Kodak So-263 film, at 0° as well as at a 45° tilt. Micrographs were digitized on a Primescan D 7100 drum scanner (Heidelberger Druckmaschinen AG) at a pixel size corresponding to 3.3 Å or 3.7 Å depending on the microscope used.
Image analysis
All image processing was achieved with the EMAN software package 42. Particles were manually selected for single-particle 3D reconstruction using BOXER. CTFIT achieved the phase-flip required for images of tilted samples acquired with the CM 200 FEG. Refine2d.py calculated class averages from data sets collected from PDE6, PDE6-PrBP/δ and PDE6-PrBP/δ-GST samples. Class averages were compared and used to appropriately scale projections recorded with different microscopes. Class averages of the magnification-refined and CTF-corrected particle projections were then taken to produce an initial 3D map of PDE6. A rough model comprising 2x3 Gaussian blobs that correspond to the GAF-A, GAF-B, and C-terminal domains of the PDE6α subunit was used to assign initial Euler angles to the class averages from a data set comprising ~8,500 projections taken at 0° tilt and ~5,700 projections taken at 45° tilt. The emerging maps were refined until they converged and finally submitted to the EOTEST function provided by EMAN, to compute the FSC function for determining the resolution of the map.
Model building
Model was built as described in the text. Briefly, Homology models (GAF-A based on PDE6C structure, PDB: 3DBA; GAF-B from PDE2A, PDB 3LBJ; and the structure of the PDE5/6 chimera: PDB, 3JWQ) were fit by hand as rigid bodies utilizing COOT. Where appropriate connecting helices were built to connect the individual protein domains, utilizing the distantly related full length PDE2A structure as a guide.
Supplementary Material
Acknowledgments
This research was supported by NIH grants EY008061, EY019478, T32EY007157, GM079191 and P30EY11373, the Foundation Fighting Blindness, the Swiss National Science Foundation, the Swiss National Center of Competence in Structural Biology, and the Maurice E. Müller Foundation, Switzerland. KP is a Senior Fellow of the American Asthma and Sandler Program for Asthma Research.
Footnotes
Abbreviations used: BCIP/NBT, 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium; cGMP, guanosine cyclic 3′,5′-monophosphate; DTT, dithiothreitol; EDTA, ethylenediaminetetraacetic acid; FSC, Fourier Shell Correlation, GMP, guanosine 5′-monophosphate; GST, glutathione S-transferase; Gt, rod photoreceptor G protein (transducin); guanosine 5′-triphosphate; HEPES, 4-(2-hydroxyethyl)-1-piperazinethanesulfonic acid; HPLC, high-pressure liquid chromatography; PBS, phosphate buffered saline; PDE6, rod-specific cGMP phosphodiesterase 6; PrBP/δ, prenyl-binding protein δ; PrBP/δ-GST, PrBP/δ glutathione S-transferase fusion protein; Rho, rhodopsin; Rho*, photoactivated rhodopsin; ROS, rod outer segment(s); TPCK, L-(tosylamido-2-phenyl) ethyl chloromethyl ketone.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Conti M, Beavo J. Biochemistry and physiology of cyclic nucleotide phosphodiesterases: essential components in cyclic nucleotide signaling. Annu Rev Biochem. 2007;76:481–511. doi: 10.1146/annurev.biochem.76.060305.150444. [DOI] [PubMed] [Google Scholar]
- 2.Lugnier C. Cyclic nucleotide phosphodiesterase (PDE) superfamily: a new target for the development of specific therapeutic agents. Pharmacol Ther. 2006;109:366–98. doi: 10.1016/j.pharmthera.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 3.Aravind L, Ponting CP. The GAF domain: an evolutionary link between diverse phototransducing proteins. Trends Biochem Sci. 1997;22:458–9. doi: 10.1016/s0968-0004(97)01148-1. [DOI] [PubMed] [Google Scholar]
- 4.Martinez SE, Beavo JA, Hol WG. GAF domains: two-billion-year-old molecular switches that bind cyclic nucleotides. Mol Interv. 2002;2:317–23. doi: 10.1124/mi.2.5.317. [DOI] [PubMed] [Google Scholar]
- 5.Artemyev NO, Surendran R, Lee JC, Hamm HE. Subunit structure of rod cGMP-phosphodiesterase. J Biol Chem. 1996;271:25382–8. doi: 10.1074/jbc.271.41.25382. [DOI] [PubMed] [Google Scholar]
- 6.Baehr W, Devlin MJ, Applebury ML. Isolation and characterization of cGMP phosphodiesterase from bovine rod outer segments. J Biol Chem. 1979;254:11669–77. [PubMed] [Google Scholar]
- 7.Deterre P, Bigay J, Forquet F, Robert M, Chabre M. cGMP phosphodiesterase of retinal rods is regulated by two inhibitory subunits. Proc Natl Acad Sci U S A. 1988;85:2424–8. doi: 10.1073/pnas.85.8.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Artemyev NO, Hamm HE. Two-site high-affinity interaction between inhibitory and catalytic subunits of rod cyclic GMP phosphodiesterase. Biochem J. 1992;283 ( Pt 1):273–9. doi: 10.1042/bj2830273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hurley JB, Stryer L. Purification and characterization of the gamma regulatory subunit of the cyclic GMP phosphodiesterase from retinal rod outer segments. J Biol Chem. 1982;257:11094–9. [PubMed] [Google Scholar]
- 10.Anant JS, Ong OC, Xie HY, Clarke S, O'Brien PJ, Fung BK. In vivo differential prenylation of retinal cyclic GMP phosphodiesterase catalytic subunits. J Biol Chem. 1992;267:687–90. [PubMed] [Google Scholar]
- 11.Qin N, Baehr W. Expression and mutagenesis of mouse rod photoreceptor cGMP phosphodiesterase. J Biol Chem. 1994;269:3265–71. [PubMed] [Google Scholar]
- 12.Gillespie PG, Prusti RK, Apel ED, Beavo JA. A soluble form of bovine rod photoreceptor phosphodiesterase has a novel 15-kDa subunit. J Biol Chem. 1989;264:12187–93. [PubMed] [Google Scholar]
- 13.Zhang H, Liu XH, Zhang K, Chen CK, Frederick JM, Prestwich GD, Baehr W. Photoreceptor cGMP phosphodiesterase delta subunit (PDEdelta) functions as a prenyl-binding protein. J Biol Chem. 2004;279:407–13. doi: 10.1074/jbc.M306559200. [DOI] [PubMed] [Google Scholar]
- 14.Norton AW, Hosier S, Terew JM, Li N, Dhingra A, Vardi N, Baehr W, Cote RH. Evaluation of the 17-kDa prenyl-binding protein as a regulatory protein for phototransduction in retinal photoreceptors. J Biol Chem. 2005;280:1248–56. doi: 10.1074/jbc.M410475200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cook TA, Ghomashchi F, Gelb MH, Florio SK, Beavo JA. Binding of the delta subunit to rod phosphodiesterase catalytic subunits requires methylated, prenylated C-termini of the catalytic subunits. Biochemistry. 2000;39:13516–23. doi: 10.1021/bi001070l. [DOI] [PubMed] [Google Scholar]
- 16.Li N, Baehr W. Expression and characterization of human PDEdelta and its Caenorhabditis elegans ortholog CEdelta. FEBS Lett. 1998;440:454–7. doi: 10.1016/s0014-5793(98)01501-4. [DOI] [PubMed] [Google Scholar]
- 17.Li N, Florio SK, Pettenati MJ, Rao PN, Beavo JA, Baehr W. Characterization of human and mouse rod cGMP phosphodiesterase delta subunit (PDE6D) and chromosomal localization of the human gene. Genomics. 1998;49:76–82. doi: 10.1006/geno.1998.5210. [DOI] [PubMed] [Google Scholar]
- 18.Hanzal-Bayer M, Renault L, Roversi P, Wittinghofer A, Hillig RC. The complex of Arl2-GTP and PDE delta: from structure to function. EMBO J. 2002;21:2095–106. doi: 10.1093/emboj/21.9.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pandit J, Forman MD, Fennell KF, Dillman KS, Menniti FS. Mechanism for the allosteric regulation of phosphodiesterase 2A deduced from the X-ray structure of a near full-length construct. Proc Natl Acad Sci U S A. 2009;106:18225–30. doi: 10.1073/pnas.0907635106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez SE, Heikaus CC, Klevit RE, Beavo JA. The structure of the GAF A domain from phosphodiesterase 6C reveals determinants of cGMP binding, a conserved binding surface, and a large cGMP-dependent conformational change. J Biol Chem. 2008;283:25913–9. doi: 10.1074/jbc.M802891200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barren B, Gakhar L, Muradov H, Boyd KK, Ramaswamy S, Artemyev NO. Structural basis of phosphodiesterase 6 inhibition by the C-terminal region of the gamma-subunit. EMBO J. 2009;28:3613–22. doi: 10.1038/emboj.2009.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Slep KC, Kercher MA, He W, Cowan CW, Wensel TG, Sigler PB. Structural determinants for regulation of phosphodiesterase by a G protein at 2.0 A. Nature. 2001;409:1071–7. doi: 10.1038/35059138. [DOI] [PubMed] [Google Scholar]
- 23.Kameni Tcheudji JF, Lebeau L, Virmaux N, Maftei CG, Cote RH, Lugnier C, Schultz P. Molecular organization of bovine rod cGMP-phosphodiesterase 6. J Mol Biol. 2001;310:781–91. doi: 10.1006/jmbi.2001.4813. [DOI] [PubMed] [Google Scholar]
- 24.Kajimura N, Yamazaki M, Morikawa K, Yamazaki A, Mayanagi K. Three-dimensional structure of non-activated cGMP phosphodiesterase 6 and comparison of its image with those of activated forms. J Struct Biol. 2002;139:27–38. doi: 10.1016/s1047-8477(02)00502-6. [DOI] [PubMed] [Google Scholar]
- 25.Frank J, Wagenknecht T, McEwen BF, Marko M, Hsieh CE, Mannella CA. Three-dimensional imaging of biological complexity. J Struct Biol. 2002;138:85–91. doi: 10.1016/s1047-8477(02)00019-9. [DOI] [PubMed] [Google Scholar]
- 26.Fung BK, Young JH, Yamane HK, Griswold-Prenner I. Subunit stoichiometry of retinal rod cGMP phosphodiesterase. Biochemistry. 1990;29:2657–64. doi: 10.1021/bi00463a006. [DOI] [PubMed] [Google Scholar]
- 27.Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 2009;37:D387–92. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu YT, Matte SL, Corbin JD, Francis SH, Cote RH. Probing the catalytic sites and activation mechanism of photoreceptor phosphodiesterase using radiolabeled phosphodiesterase inhibitors. J Biol Chem. 2009;284:31541–7. doi: 10.1074/jbc.M109.018606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez SE, Wu AY, Glavas NA, Tang XB, Turley S, Hol WG, Beavo JA. The two GAF domains in phosphodiesterase 2A have distinct roles in dimerization and in cGMP binding. Proc Natl Acad Sci U S A. 2002;99:13260–5. doi: 10.1073/pnas.192374899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muradov H, Boyd KK, Artemyev NO. Structural determinants of the PDE6 GAF A domain for binding the inhibitory gamma-subunit and noncatalytic cGMP. Vision Res. 2004;44:2437–44. doi: 10.1016/j.visres.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 31.Guo LW, Muradov H, Hajipour AR, Sievert MK, Artemyev NO, Ruoho AE. The inhibitory gamma subunit of the rod cGMP phosphodiesterase binds the catalytic subunits in an extended linear structure. J Biol Chem. 2006;281:15412–22. doi: 10.1074/jbc.M600595200. [DOI] [PubMed] [Google Scholar]
- 32.Mou H, Cote RH. The catalytic and GAF domains of the rod cGMP phosphodiesterase (PDE6) heterodimer are regulated by distinct regions of its inhibitory gamma subunit. J Biol Chem. 2001;276:27527–34. doi: 10.1074/jbc.M103316200. [DOI] [PubMed] [Google Scholar]
- 33.Muradov H, Boyd KK, Artemyev NO. Analysis of PDE6 function using chimeric PDE5/6 catalytic domains. Vision Res. 2006;46:860–8. doi: 10.1016/j.visres.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 34.Oppert B, Takemoto DJ. Identification of the gamma-subunit interaction sites in the retinal cyclic-GMP phosphodiesterase beta-subunit. Biochem Biophys Res Commun. 1991;178:474–9. doi: 10.1016/0006-291x(91)90131-p. [DOI] [PubMed] [Google Scholar]
- 35.Paglia MJ, Mou H, Cote RH. Regulation of photoreceptor phosphodiesterase (PDE6) by phosphorylation of its inhibitory gamma subunit re-evaluated. J Biol Chem. 2002;277:5017–23. doi: 10.1074/jbc.M106328200. [DOI] [PubMed] [Google Scholar]
- 36.Zhang XJ, Skiba NP, Cote RH. Structural Requirements of the Photoreceptor Phosphodiesterase {gamma}-Subunit for Inhibition of Rod PDE6 Holoenzyme and for Its Activation by Transducin. J Biol Chem. 2010;285:4455–63. doi: 10.1074/jbc.M109.057406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Artemyev NO, Rarick HM, Mills JS, Skiba NP, Hamm HE. Sites of interaction between rod G-protein alpha-subunit and cGMP-phosphodiesterase gamma-subunit. Implications for the phosphodiesterase activation mechanism. J Biol Chem. 1992;267:25067–72. [PubMed] [Google Scholar]
- 38.Wensel TG. Signal transducing membrane complexes of photoreceptor outer segments. Vision Res. 2008;48:2052–61. doi: 10.1016/j.visres.2008.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Papermaster DS. Preparation of retinal rod outer segments. Methods Enzymol. 1982;81:48–52. doi: 10.1016/s0076-6879(82)81010-0. [DOI] [PubMed] [Google Scholar]
- 40.Goc A, Angel TE, Jastrzebska B, Wang B, Wintrode PL, Palczewski K. Different Properties of the Native and Reconstituted Heterotrimeric G Protein Transducin. Biochemistry. 2008 doi: 10.1021/bi8015444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 42.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.