Abstract
It is generally accepted that glioma develops through accumulation of genetic alterations. We hypothesized that polymorphisms of candidate genes involved in the DNA repair pathways may contribute to susceptibility to glioma. To address this possibility, we conducted a study of 373 Caucasian glioma cases and 365 cancer-free Caucasian controls to assess associations between glioma risk and 18 functional SNPs in DNA repair genes. We evaluated potential gene-gene and gene-environment interactions using a multi-analytic strategy combining logistic regression, multifactor dimensionality reduction (MDR), and classification and regression tree (CART) approaches. In the single-locus analysis, six SNPs (ERCC1 3’ UTR, XRCC1 R399Q, APEX1 E148D, PARP1 A762V, MGMT F84L, and LIG1 5’UTR) showed a significant association with glioma risk. In the analysis of cumulative genetic risk of multiple SNPs, a significant gene-dosage effect was found for increased glioma risk with increasing numbers of adverse genotypes involving the above-mentioned six SNPs (P trend = 0.0004). Further, both the MDR and CART analyses identified MGMT F84L as the predominant risk factor for glioma, and revealed strong interactions among ionizing radiation (IR) exposure, PARP1 A762V, MGMT F84L and APEX1 E148D. Interestingly, the risk for glioma was dramatically increased in IR exposure individuals who had the wild-type genotypes of both MGMT F84L and PARP1 A762V [adjusted odds ratios (OR), 5.95; 95% confidence intervals (CI), 2.21–16.65]. Taken together, these results suggest that polymorphisms in DNA repair genes may act individually or together to contribute to glioma risk.
INTRODUCTION
Malignant gliomas are the most common primary brain tumor in adults. Few factors have so far been conclusively shown to affect glioma risk namely, family history, rare genetic syndromes and exposure to high doses of ionizing radiation (IR). However, these factors only account for a small proportion of cases (1–4). Therefore, the most generally accepted model of carcinogenesis postulates that glioma develops through accumulation of genetic alterations that allow the cells to escape normal growth-regulatory mechanisms (5).
It is increasingly clear that genetic susceptibility to cancer is complex because of interactions between and among genes and environmental factors. Such interactions are ubiquitous to complex genetic diseases such as cancer (6), and association studies are designed to address this complexity. Molecular epidemiological studies have moved from the evaluation of a single candidate gene to the consideration of a pathway comprising dozens of genes and their environmental factors, and even to multiple pathways that form complex networks.
DNA is continually subjected to a variety of assaults, both as a result of internal, cellular metabolic processes and exposure to genotoxic or clastogenic agents. Efficient and proficient DNA repair is thus required for the effective maintenance of genome integrity. Such DNA damage requires the concerted actions of a number of DNA repair genes to restore genomic integrity, including the nucleotide excision repair (NER), base excision repair (BER), DSB repair (DSBR), and mismatch repair (MMR) pathways, as well as direct reversal of damage. Thus, common polymorphisms of DNA repair genes are plausible candidates that may contribute to susceptibility to glioma. While several studies have investigated the role of single nucleotide polymorphisms (SNPs) in DNA repair genes and susceptibility to glioma, and the results are encouraging (7–11), few have systematically examined glioma risk in the context of SNPs in the different DNA repair pathways together (12, 13). Hence, we tested the hypothesis that polymorphisms of candidate genes involved in the DNA repair pathway genes may contribute to susceptibility to glioma. In this study, we used a candidate pathway-based approach to investigate 18 potential functional polymorphisms in 12 key genes in the different DNA repair pathways including NER pathway, BER pathway, DSBR pathway and direct reversal of damage. Further, based on the interaction of these genes at the molecular level and the possibility that these interactions might be modified by specific environmental factors, we also tested the hypothesis that gene-gene, gene-environment interactions contribute to glioma susceptibility using a multi-analytic strategy that combined traditional statistical methods with novel computational algorithms.
MATERIALS AND METHODS
Study Population
Cases were Caucasian adults (18 years or older) with newly diagnosed gliomas (International Classification of Diseases, ICD-9 codes 9380–9481) residing in 15-counties surrounding Harris country Texas (Austin, Brazoria, Chambers, Colorado, Fort Bend, Galveston, Harris, Jefferson, Liberty, Montgomery, Orange, San Jacinto, Walker, Waller, and Wharton), and recruited into the study between January 2001 and January 2006. A pathology specimen was obtained for all patients, and glioma diagnosis was confirmed by a neuropathologist. Glioma cases with prior diagnosis of cancers were excluded. Patients signed an informed consent and were interviewed to obtain the information described below. Proxies were interviewed if the patients were unable to participate because of cognitive disability. We exclude minority patients for the analysis since only a small number of glioma patients were reached (15 African-American and 34 Hispanic). Controls were frequency-matched to cases on age (± 5 years), gender, and race/ethnicity. Controls were all English-speaking residents of the same Houston area counties, and were recruited through random-digit dialing through a contracting company using standard methods (14, 15). Controls must have not been previously diagnosed with cancer at the time of enrollment. Eligible controls were contacted to obtain informed consent and schedule an interview. The participation rates for cases and controls were 77% and 83%, respectively.
Both cases and controls underwent a detailed interview, either in person or over the telephone. The following information was collected: demographic variables, cigarette smoking and alcohol consumption, family history of brain tumor (first-degree) or other cancers and IR exposure histories (medical and occupational). The reported exposures to IR are likely to cover a wide range of doses. Medical IR exposure history was assessed by collecting information on diagnostic (i.e., routine chest X-rays), and any radiotherapy for a medical problem (such as acne or hyperthyroidism). For the diagnostic exposure, details such as the dates, site of body and reasons for exposure were obtained. For therapeutic exposure, details such as the nature of the disease requiring radiation, the number of treatment cycles, area of the body that received irradiation and age at initiation and at the end of the treatments. Only exposures that had occurred at least two years prior to diagnosis (cases) or reference date (interview for the controls) were included as “exposed.” Occupational IR exposure included work as a pilot, flight attendant, astronaut, uranium miner, workers in the nuclear power industries, radiologists or X-ray medical worker, dentist or dental hygienist and other participants who self-reported IR exposure in the workplace. We only included occupational exposures of at least one year in duration for a minimum of two years before the reference date (diagnosis for cases; interview for controls). A 20-mL blood specimen was obtained from all patients and control subjects. The research protocol was approved by The University of Texas M. D. Anderson Institutional Review Board.
Selection of Candidate SNPs and Genotyping
Candidate SNPs were selected from the best evidence from published studies, and to represent more commonly occurring variants (minor allele frequencies >10%), in order to gain statistical power to detect interactions. We selected a total of 18 literature-defined putative functional polymorphisms in 12 key genes (16–20), six SNPs in the NER pathway (XPC V499A rs2228000, XPC Q939K rs2228001, XPD/ERCC2 R156R rs238406, XPD Q751K rs13181, XPG/ERCC5 H1104D rs17655, ERCC1 3’ UTR rs3212986), five SNPs in the BER pathway (XRCC1 W194R rs1799782, XRCC1 R399Q rs25487, OGG1 C326S rs1052133, PARP1 A762V rs1136410, APEX1 E148D rs3136819), three SNPs in direct reversal of damage pathway (MGMT F84L rs12917, V143I rs2308321and R178K rs2308327), two SNPs in DSBR pathway (XRCC3 T241M rs861539 and NBS Q185E rs1805794), and two SNPs in the other repair genes (LIG1 5’UTR rs20579 and LIG1 A170A rs20580),
Genomic DNA was extracted from peripheral blood lymphocytes using the Qiagen Blood Kit (Qiagen Inc., Valencia, CA). Genotyping was performed using the Sequenom MassARRAY iPLEX™ platform according to the manufacturer's instructions (http://www.sequenom.com/seq.genotyping.html). The quality control analysis included the genotyping of internal positive control samples, the use of no template controls, and the use of replicates for 10% of the samples.
Statistical Analysis
Goodness of fit to the Hardy-Weinberg equilibrium (HWE) expectation in control subjects was assessed by the χ2 test for each SNP. Genotype frequencies in cases and control subjects were compared using the χ2 test. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated by unconditional logistic regression analysis with adjustment for age and gender. The Akaike's information criterion (AIC) was used to determine the best genetic model for each SNP (21). Besides the permutation testing (1000 times), we also adopted a powerful bootstrapping method to reduce the potential for spurious findings due to multiple testing and to validate the results in our sample (22). The bootstrap approach selects random samples of size N (cases + controls) with replacement from the original data (23, 24). In our study, within each bootstrap replicate (1000 replications), we chose a random sample (each random sample comprised 373 cases and 365 controls) which allows for any one participant to be chosen once, more than once, or not at all for each replicate. We then calculated the OR for each replicate and constructed an empirical distribution for the OR. We calculated the mean OR across all replicates and obtained 2.5% and 97.5% percentiles of the empirical distribution to construct a 95% CI for the OR. All statistical tests were two-sided.
We examined linkage disequilibrium (LD) among the polymorphisms using Lewontin’s standardized coefficient D’ and the LD coefficient r2 (25). We used the HAPLO.STATS package developed by Schaid et al. (http://www.mayo.edu/hsr/Sfunc.html) for the haplotype analysis (26). This method, which is based on the generalized linear model framework, allows for the adjustment of possible confounding variables and provides both global and haplotype-specific tests. Haplotypes with a frequency of less than 0.03 were pooled into a combined group. Empirical P values, based on 1000 simulations, were computed for the global score test and each of the haplotype-specific score tests.
We used a multiphase strategy for analyzing gene-gene and gene-environment interactions. First, using multivariate logistic regression, global effect interactions by the number of adverse genotypes identified from the single SNPs analysis were examined. Then, we applied the multifactor dimensionality reduction (MDR) method to identify interaction models. The nonparametric and genetic model-free MDR analysis was performed using version 0.5.1 of the open-source MDR software package that is available online (http://www.epistasis.org/software.html). The MDR approach is described in detail by Ritchie et al. (27) and reviewed by Moore et al. (6, 28). In this study, the MDR analysis was conducted using the dichotomous groupings of the polymorphisms selected by the AIC, and we used 100-fold cross-validation consistency (CVC) and 1000-fold permutation testing. MDR results were considered statistically significant at the 0.05 level. To better confirm and visualize the interaction models identified by MDR, we further built an entropy-based interaction dendrogram (29, 30). This would enable the attributes (i.e., SNPs) that strongly interact to appear close together at the leaves of the tree, with those not interacting appearing distant from one another. Lastly, we conducted a classification and regression tree (CART) analysis to detect and characterize the high-order interactions using the Helix-Tree Genetics Analysis software (version 4.1.0; Golden Helix, Bozeman, MT). CART is a binary recursive partitioning method that produces a decision tree to identify subgroups of subjects at higher risk (31). Specifically, the recursive partitioning algorithm in Helix-Tree starts at the root node (with the entire data set) and uses a statistical hypothesis testing method, i.e., formal inference-based recursive modeling, to determine the first locally optimal split and each subsequent split of the data set, with multiplicity-adjusted P values to control tree growth (P < 0.05). This process continues until the terminal nodes have no subsequent statistically significant splits or the terminal nodes reach a pre-specified minimum size (at least 10 subjects for each terminal node). Subgroups of individuals with differential risk associations were identified in the different nodes of the tree, indicating the presence of interactions. In our analyses, the SNP variables were considered as two categories according to their AIC criterion selected best effect model. Referent group is the least percentage of cases. OR and 95% CI were adjusted by age and gender.
RESULTS
Characteristics of Study Subjects
Our analysis included 373 Caucasian glioma cases, including 33 (4%) proxy-reports, and 365 cancer-free Caucasian controls. The characteristics of cases and control subjects are summarized in Table 1. The case group had more males than the control group (56.8% versus 43.6%), and were more likely than the controls to report a family history of brain tumor (4.0% versus 2.7%) in their first-degree relatives. With regard to IR exposure, 35 glioma cases (8 medical exposures, 27 occupationally exposed) and 21 cancer-free controls (2 medical exposures, 19 occupationally exposed) reported a history of IR exposure (9.4% versus 5.9%). For participants undergoing medical radiation treatment, the diagnoses were “severe acne” (2 cases and 1 control), “hyperthyroid” (2 cases), “birth mark” (1 case and 1 control), “thick sinus mucus” (1 case), “bell's palsy” (1 case) and “tonsillitis” (1 case). More than two-thirds of the 46 occupationally exposed participants were in the medical field (physicians, radiologists and nurses). Amongst the exposed professions were also pilots and engineers. Of the 373 cases, 214 were glioblastoma, categorized as high-grade glioma; 77 were anaplastic astrocytoma, medium-grade glioma; and 82 were other low-grade glioma, which included oligodendroglioma, not-otherwise-specified astrocytoma, and mixed glioma.
Table 1.
Frequency distribution of selected characteristics of study subjects by the case-control status
| Cases (n=373) | Controls (n=365) | |
|---|---|---|
| Variable | No. (%)* | No. (%) * |
| Age (years) | ||
| ≤45 | 142 (38.1) | 143 (39.2) |
| >45 | 231 (61.9) | 222 (60.8) |
| Gender | ||
| Male | 212 (56.8) | 159 (43.6) |
| Female | 161 (43.2) | 206 (56.4) |
| Smoking status | ||
| Never | 222 (59.7) | 205 (56.2) |
| Ever | 150 (40.3) | 160 (43.8) |
| Family history of brain tumor | ||
| No | 339 (96.0) | 326 (97.3) |
| Yes | 14 (4.0) | 9 (2.7) |
| IR exposure history | ||
| No | 337 (90.6) | 337 (94.1) |
| Yes | 35 (9.4) | 21 (5.9) |
| Histology type† | ||
| High grade | 214 (57.4) | |
| Medium grade | 77 (20.6) | |
| Low grade | 82 (22.0) | |
Numbers do not add up to the column totals due to missing values.
High grade glioma (glioblastoma); medium grade glioma (anaplastic astrocytoma); low grade glioma (oligodendroglioma, not-otherwise-specified astrocytoma, and mixed glioma).
Individual SNP Association Analysis
The SNP IDs, locations, and allele frequencies are given in Supplementary Table S1. The genotype distributions of 18 selected SNPs in the cases and controls are summarized in Table 2. All genotype distributions in the controls were consistent with those expected from the HWE test (Supplementary Table S1). The allele frequencies of three SNPs (i.e. MGMT F84L, LIG1 5’UTR and XRCC3 T241M) were significantly different between the cases and the controls, even after 1000 permutations (P = 0.028, 0.044, and 0.044, respectively). Marginal differences in distribution were observed for XPD Q751K, XRCC1 R399Q, and PARP1 A762V after 1000 permutations (P = 0.059, 0.066, and 0.064, respectively).
Table 2.
Genotype frequencies of 18 SNPs among cases and controls and their associations with risk of glioma
| Pathway | Gene & SNP | Genotype | No. (frequency) |
Logistic regression OR (95%CI)* |
Bootstrap OR (95%CI)† |
|||
|---|---|---|---|---|---|---|---|---|
| Cases | Controls | |||||||
| NER | ||||||||
| DM | XPC V499A | CC | 220 | (59.9%) | 206 | (56.6%) | 1.00 (reference) | 1.00 (reference) |
| rs2228000 | CT/TT | 147 | (40.1%) | 158 | (43.4%) | 0.85 (0.63–1.16) | 0.87 (0.63–1.18) | |
| XPD R156R | GG | 123 | (33.0%) | 101 | (27.7%) | 1.00 (reference) | 1.00 (reference) | |
| rs238406 | GT/TT | 250 | (67.0%) | 263 | (72.3%) | 0.77 (0.56–1.05) | 0.78 (0.56–1.05) | |
| XPD Q751K | TT | 139 | (37.9%) | 161 | (44.5%) | 1.00 (reference) | 1.00 (reference) | |
| rs13181 | TG/GG | 228 | (62.1%) | 201 | (55.5%) | 1.30 (0.96–1.75) | 1.31 (0.97–1.74) | |
| RM | XPC Q939K | AA/AC | 315 | (84.9%) | 297 | (81.4%) | 1.00 (reference) | 1.00 (reference) |
| rs2228001 | CC | 56 | (15.1%) | 68 | (18.6%) | 0.75 (0.50–1.11) | 0.76 (0.49–1.10) | |
| XPG H1104D | CC/CG | 353 | (94.6%) | 351 | (96.4%) | 1.00 (reference) | 1.00 (reference) | |
| rs17655 | GG | 20 | (5.4%) | 13 | (3.6%) | 1.47 (0.71–3.05) | 1.61 (0.69–3.36) | |
| ERCC1 3’ UTR | GG/GT | 338 | (91.6%) | 345 | (95.3%) | 1.00 (reference) | 1.00 (reference) | |
| rs3212986 | TT | 31 | (8.4%) | 17 | (4.7%) | 1.86 (1.01–3.46) | 1.97 (1.01–3.73) | |
| BER | ||||||||
| DM | XRCC1 R399Q | GG | 149 | (39.9%) | 169 | (46.4%) | 1.00 (reference) | 1.00 (reference) |
| rs25487 | GA/AA | 224 | (60.1%) | 195 | (53.6%) | 1.43 (1.05–1.92) | 1.44 (1.06–1.92) | |
| PARP1 A762V | TT | 267 | (71.8%) | 236 | (64.7%) | 1.00 (reference) | 1.00 (reference) | |
| rs1136410 | TC/CC | 105 | (28.2%) | 129 | (35.3%) | 0.71 (0.52–0.97) | 0.72 (0.52–0.99) | |
| RM | XRCC1 W194R | CC/CT | 209 | (99.7%) | 362 | (99.2%) | 1.00 (reference) | 1.00 (reference) |
| rs1799782 | TT | 1 | (0.3%) | 3 | (0.8%) | 0.52 (0.07–5.26) | N.A‡ | |
| OGG1 C326S | CC/CG | 355 | (95.4%) | 345 | (94.5%) | 1.00 (reference) | 1.00 (reference) | |
| rs1052133 | GG | 17 | (4.6%) | 20 | (5.5%) | 0.85 (0.44–1.65) | 0.92 (0.40–1.84) | |
| APEX1 E148D | CC/CT | 289 | (78.1%) | 262 | (72.2%) | 1.00 (reference) | 1.00 (reference) | |
| rs3136819 | TT | 81 | (21.9%) | 101 | (27.8%) | 0.68 (0.48–0.97) | 0.70 (0.50–0.98) | |
| Direct repair | ||||||||
| DM | MGMT F84L | CC | 299 | (81.0%) | 267 | (73.6%) | 1.00 (reference) | 1.00 (reference) |
| rs12917 | CT/TT | 70 | (19.0%) | 96 | (26.4%) | 0.67 (0.45–0.95) | 0.69 (0.47–0.97) | |
| RM | MGMT V143I | AA/AG | 379 | (97.9%) | 369 | (98.8%) | 1.00 (reference) | 1.00 (reference) |
| rs2308321 | GG | 8 | (2.1%) | 4 | (1.2%) | 1.95 (0.58–6.84) | N.A‡ | |
| MGMT R178K | AA/AG | 364 | (97.8%) | 359 | (98.7%) | 1.00 (reference) | 1.00 (reference) | |
| rs2308327 | GG | 8 | (2.2%) | 4 | (1.3%) | 1.97 (0.57–6.67) | N.A‡ | |
| DSBR | ||||||||
| RM | XRCC3 T241M | CC/CT | 308 | (83.5%) | 315 | (87.5%) | 1.00 (reference) | 1.00 (reference) |
| rs861539 | TT | 61 | (16.5%) | 45 | (12.5%) | 1.43 (0.93–2.18) | 1.46 (0.92–2.23) | |
| NBS Q185E | GG/GC | 341 | (91.4%) | 318 | (87.1%) | 1.00 (reference) | 1.00 (reference) | |
| rs1805794 | CC | 32 | (8.6%) | 47 | (12.9%) | 0.65 (0.40–1.05) | 0.66 (0.40–1.03) | |
| Others | ||||||||
| DM | LIG1 5’UTR | CC | 285 | (76.6%) | 255 | (69.9%) | 1.00 (reference) | 1.00 (reference) |
| rs20579 | CT/TT | 87 | (23.4%) | 110 | (30.1%) | 0.67 (0.48–0.94) | 0.68 (0.48–0.92) | |
| RM | LIG1 A170A | AA/AC | 278 | (74.7%) | 281 | (77.0%) | 1.00 (reference) | 1.00 (reference) |
| rs20580 | CC | 94 | (25.3%) | 84 | (23.0%) | 1.14 (0.81–1.61) | 1.16 (0.81–1.61) | |
NOTE: The Akaike's information criterion (AIC) was used to determine the genetic model for each SNP.
DM, dominant genetic model; RM, recessive genetic model.
Adjusted for age and gender.
Bootstrapping mean OR and its corresponding 95% CI.
N.A, not available.
Logistic regression analyses revealed that in the dominant-effect model as assessed by the AIC, compared with wild-type homozygote carriers, significant increased risk effects were associated with XRCC1 R399Q (adjusted OR, 1.43; 95% CI, 1.05–1.92), whereas significantly protective effects were associated with PARP1 A762V (adjusted OR, 0.71; 95% CI, 0.52–0.97), MGMT F84L (adjusted OR, 0.66; 95% CI, 0.45–0.95), and LIG1 5’UTR (adjusted OR, 0.67; 95% CI, 0.48–0.94). Meanwhile, in the recessive-effect model (assuming that only the variant homozygotes have an increased risk of glioma), compared with the wild-type homozygotes and heterozygous carriers, a significantly increased risk was associated with the variant homozygotes of ERCC1 3’ UTR (adjusted OR, 1.86; 95% CI, 1.01–3.46), whereas a significant protective effect was associated with the variant homozygotes of APEX1 E148D (adjusted OR, 0.68; 95% CI, 0.48–0.97). All bootstrap ORs were very similar to the presented adjusted Ors (Table 2).
Further stratified analyses by gender, age, IR exposure history, and glioma histological type on the above-mentioned six SNPs, that is, the four protective SNPs (PARP1 A762V, MGMT F84L, LIG1 5’UTR and APEX1 E148D), and the two risk SNPs (ERCC1 3’ UTR and XRCC1 R399Q). As shown in Table 3, the protective effect of PARP1 A762V, MGMT F84L and LIG1 5’UTR were more evident in patients with medium grade tumors (adjusted OR, 0.52, 95% CI, 0.28–0.95; and OR, 0.52, 95% CI, 0.26–0.99, and OR, 0.47, 95% CI, 0.24–0.88, respectively) while the APEX1 E148D variant was more evident in patients with high grade tumors (adjusted OR, 0.50; 95% CI, 0.32–0.78). In addition, the effect of PARP1 A762V variants was more significant in the subjects exposed to IR (adjusted OR, 0.20; 95% CI, 0.08–0.62. P interaction = 0.036). For the risk effect SNPs, the increased risk effect of XRCC1 R399Q was more evident in females (adjusted OR, 2.27; 95% CI, 1.45–3.57. P interaction = 0.0047) whereas, the ERCC1 3’UTR variant was more pronounced in younger cases (adjusted OR, 5.88; 95% CI, 1.64–20.0. P interaction = 0.016). However, both the XRCC1 R399Q and ERCC1 3’UTR variants were more pronounced in patients with high grade gliomas (adjusted OR 1.46, 95% CI, 1.01–2.08; and OR 1.97, 95% CI, 1.00–3.95, respectively) (Table 3)..
Table 3.
Stratified analysis by age, gender, IR exposure history, and histological type of glioma
| Dominant |
Recessive |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| XRCC1 R399Q | PARP1 A762V | MGMT F84L | LIG1 5’UTR | ERCC1 3’ UTR | APEX1 E148D | |||||||
| Variables | OR (95% CI) * | OR (95% CI) * | OR (95% CI) * | OR (95%CI) * | OR (95% CI) * | OR (95% CI) * | ||||||
| No.† | GA/AA vs GG |
No.† | TC/CC vs TT |
No.† | CT/TT vs CC |
No.† | CT/TT vs CC |
No.† | GG vs TT/GT |
No.† | TT vs CC/CT |
|
| Age (years) | ||||||||||||
| ≤45 | 142/143 | 1.47 (0.91–2.38) |
142/143 | 0.62 (0.38–1.06) |
140/143 | 0.57 (0.32–1.00) |
142/143 | 0.46 (0.27–0.80) |
142/143 |
5.88 (1.64–20.0) |
142/143 | 0.96 (0.57–1.64) |
| >45 | 231/221 | 1.39 (0.95–2.03) |
230/221 | 0.76 (0.52–1.14) |
230/220 | 0.76 (0.49–1.20) |
230/221 | 0.85 (0.55–1.29) |
230/220 | 1.08 (0.52–2.27) |
230/220 | 0.54 (0.36–0.85) |
| P for interaction | 0.86 | 0.54 | 0.44 | 0.085 | 0.016 | 0.11 | ||||||
| Gender | ||||||||||||
| Male | 214/159 | 0.94 (0.62–1.44) |
213/159 | 0.58 (0.38–0.91) |
211/159 | 0.53 (0.32–0.88) |
213/159 | 0.79 (0.50–1.24) |
214/157 | 1.92 (0.78–4.76) |
213/158 | 0.70 (0.43–1.14) |
| Female | 158/204 |
2.27 (1.45–3.57) |
158/205 | 0.86 (0.55–1.35) |
156/203 | 0.86 (0.53–1.40) |
158/205 | 0.56 (0.34–0.92) |
156/204 | 1.79 (0.76–4.15) |
156/204 | 0.67 (0.42–1.10) |
| P for interaction | 0.0047 | 0.23 | 0.19 | 0.31 | 0.90 | 0.91 | ||||||
| IR exposure history | ||||||||||||
| No | 333/335 | 1.37 (1.00–1.89) |
332/336 | 0.76 (0.55–1.06) |
329/335 | 0.68 (0.48–1.00) |
332/336 | 0.71 (0.50–1.01) |
331/334 | 1.89 (1.00–3.56) |
331/334 | 0.70 (0.49–1.01) |
| Yes | 35/21 | 3.45 (1.11–11.1) |
35/21 |
0.20 (0.08–0.62) |
35/21 | 0.76 (0.20–2.94) |
35/21 | 0.26 (0.08–0.91) |
35/20 | 1.32 (0.11–16.7) |
34/21 | 0.40 (0.10–1.54) |
| P for interaction | 0.14 | 0.036 | 0.82 | 0.072 | 0.79 | 0.44 | ||||||
| Histological type | ||||||||||||
| High grade | 213/363 |
1.46 (1.01–2.08) |
212/364 | 0.79 (0.54–1.15) |
211/362 | 0.70 (0.46–1.08) |
212/364 | 0.77 (0.51–1.14) |
214/361 |
1.97 (1.00–3.95) |
212/362 |
0.50 (0.32–0.78) |
| Medium grade | 76/363 | 1.15 (0.69–1.90) |
76/364 |
0.52 (0.28–0.95) |
74/362 |
0.52 (0.26–0.99) |
76/364 |
0.47 (0.24–0.88) |
75/361 | 1.77 (0.66–4.71) |
76/362 | 1.32 (0.75–2.30) |
| Low grade | 82/363 | 1.51 (0.91–2.51) |
82/364 | 0.72 (0.41–1.22) |
82/364 | 0.75 (0.42–1.35) |
82/364 | 0.66 (0.38–1.17) |
80/361 | 1.66 (0.63–4.37) |
79/362 | 0.91 (0.51–1.63) |
Adjusted for age and gender, accordingly.
Numbers of cases/controls.
Haplotype Analysis
Strong LD was observed between each pair of the SNPs in the XPC, XPD, XRCC1, and LIG1 genes (data not shown). Table 4 summarizes the associations between the frequency distributions of the haplotypes and the risk of glioma. For XPD, haplotype “TG” showed the greatest protective effect, accounting for a 60% reduction in the risk of glioma (adjusted OR, 0.40; 95% CI, 0.17–0.98). For XRCC1, one risk haplotype “CA” was identified (adjusted OR, 1.28; 95% CI, 1.03–1.59). No haplotypes of the XPC, MGMT, and LIG1 genes were found to be significantly associated with glioma risk or protection. Furthermore, the global score test showed a statistically significant difference in the haplotype distributions between cases and controls for XPD (global P = 0.025) and a borderline association with XRCC1 (global P = 0.071).
Table 4.
Haplotype analyses of glioma risk
| Gene | Haplotype | Frequency |
OR (95%CI)* | Global score test† | ||
|---|---|---|---|---|---|---|
| Total | Cases | Controls | ||||
| XPC | 0.281 | |||||
| C C | 0.394 | 0.387 | 0.401 | 1.00 (reference) | ||
| C A | 0.370 | 0.387 | 0.353 | 1.16 (0.92–1.48) | ||
| T A | 0.235 | 0.225 | 0.246 | 0.95 (0.72–1.23) | ||
| XPD | 0.025 | |||||
| T T | 0.423 | 0.422 | 0.429 | 1.00 (reference) | ||
| G G | 0.340 | 0.370 | 0.307 | 1.20 (0.94–1.54) | ||
| G T | 0.212 | 0.193 | 0.232 | 0.86 (0.65–1.15) | ||
| T G | 0.025 | 0.015 | 0.032 | 0.40 (0.17–0.98) | ||
| XRCC1 | 0.071 | |||||
| C T | 0.563 | 0.542 | 0.584 | 1.00 (reference) | ||
| C A | 0.360 | 0.383 | 0.336 | 1.28 (1.03–1.59) | ||
| T T | 0.077 | 0.074 | 0.079 | 1.05 (0.66–1.69) | ||
| MGMT | 0.280 | |||||
| C A A | 0.774 | 0.788 | 0.759 | 1.00 (reference) | ||
| T A A | 0.113 | 0.097 | 0.128 | 0.77 (0.54–1.08) | ||
| C G G | 0.103 | 0.107 | 0.099 | 1.09 (0.76–1.54) | ||
| T G G | 0.010 | 0.007 | 0.013 | 0.56 (0.11–2.78) | ||
| LIG1 | 0.120 | |||||
| C C | 0.476 | 0.485 | 0.466 | 1.00 (reference) | ||
| C A | 0.382 | 0.391 | 0.372 | 1.03 (0.82–1.31) | ||
| T A | 0.123 | 0.112 | 0.135 | 0.75 (0.53–1.08) | ||
| T C | 0.019 | 0.012 | 0.027 | 0.51 (0.17–1.49) | ||
NOTE: Loci chosen for XPC: W499R, K939Q; Loci chosen for XPD: R156R, Q751K; Loci chosen for XRCC1: W194I, R399Q; Loci chosen for MGMT: F84L, V143I, R178K. Loci chosen for LIG1: 5’UTR, A170A;
Adjusted for age and gender.
Generated by permutation test with 1000 times.
Gene-Gene and Gene-Environment Interactions Analysis
Cumulative Genetic Risk Test of Multiple SNP Association
To test our hypothesis that multiple SNPs in the DNA repair pathways act together to modulate glioma risk, we estimated the global effect of the six adverse SNPs that were significantly associated with glioma risk in the single-locus analysis. Two risk effect SNPs and four protective effect SNPs were included in this analysis; because they exhibited risk in opposite directions, we treated the minor allele of the risk SNPs and the common allele of the protective SNPs as the adverse allele, and regarded individuals with fewer than three adverse alleles as the referent group. As shown in Table 5, the risk of glioma increased progressively as the number of adverse genotypes increased (P trend = 0.0004). That is, the groups with three to four, and five to six adverse genotypes all exhibited a significantly increased glioma risk. Then, we further stratified our analyses by IR exposure. Likewise, a significant dose-response effect on glioma risk was observed (P trend = 0.008). Specifically, compared with the referent group (individuals with fewer than three adverse alleles who had no IR exposure), the non-IR exposure groups with three to six adverse genotypes only showed a marginal significantly increased glioma risk (adjusted OR, 1.45; 95% CI, 0.91–2.25), whereas, the IR exposure groups with three to six adverse genotypes showed an OR of 4.28 (95% CI, 1.65–11.41). These data showed a significant association when interactions among the six SNPs and IR were considered. However, the results from this cumulative genetic risk analyses should be interpreted with caution because of the small size of the subgroup, and the borderline confidence intervals.
Table 5.
Cumulative genetic risk analysis of adverse genotypes in glioma cases and control subjects
| No. of variant or adverse genotypes |
No of Cases/Controls |
OR (95% CI) | P for trend |
|---|---|---|---|
| The total six risk-conferring SNPs | 0.0004 | ||
| 0~2 # | 45/70 | 1.00 (reference) | |
| 3~4 | 223/232 | 1.55 (1.00–2.35) | |
| 5~6 | 93/55 | 2.67 (1.59–4.57) | |
| IR exposure and the total six risk-conferring SNPs | 0.008 | ||
| No IR & 0~2§ | 44/62 | 1.00 (reference) | |
| No IR & 3~6 | 280/269 | 1.45 (0.91–2.25) | |
| Have IR & 0~2 | 9/12 | 1.08 (0.40–2.97) | |
| Have IR & 3~6 | 24/8 | 4.28 (1.65–11.41) | |
Adjusted for age and gender.
We treat the minor allele of the two-risk-effect SNPs and the common allele of the four-protective-effect SNPs as the adverse allele, and set individuals with fewer than three adverse alleles as the reference group. Adverse genotypes: for four protective SNPs, APEX1 E148D (recessive, V V), PARP A762V (dominant, V V + W V), MGMT F84L (dominant, V V + W V), and LIG1 5’UTR (dominant, V V + W V); for the two risk SNPs, ERCC1 3’ UTR (recessive, W W + W V) and XRCC1 R399Q (dominant, WW).
The no IR exposure group and with fewer than three adverse genotypes defined as the reference.
MDR Analysis
Figure 1A summarizes the best interaction models obtained from the MDR analysis. The best one-locus model for predicting glioma risk was MGMT F84L (testing accuracy = 47.25%, CVC = 81, permutation P = 0.982). But the best interaction model was a four-locus model (i.e., IR, PARP1 A762V, MGMT F84L and APEX1 E148D), with an improved testing accuracy of 55.62% (CVC = 100, permutation P = 0.044). The five-locus model (i.e., IR, PARP1 A762V, MGMT F84L, APEX1 E148D and ERCC1 3’ UTR) also had an improved testing accuracy of 52.89% and 100 CVC. However, it only had borderline interactions with an empirical P = 0.096 based on 1000 permutations.
Fig1.
The multifactor dimensionality reduction (MDR) models and interaction dendrogram for gene-gene and gene-environment interactions on glioma risk. (A) Summary of the MDR interaction models. Cross-validation consistency (CVC). P value based on 1000 permutation. (B) Interaction dendrogram. The attributes (SNP or IR), that strongly interact to appear close together at the leaves of the tree, with those not interacting appearing distant from one another.
The MDR analysis indicated that the four-locus model was the best model but did not specify the presence of synergy between the loci. To test this possibility, we applied the interaction dendrogram to determine their relationship. Figure 1B illustrates the interaction dendrogram for these models. Consistent with the stratified analysis, the hierarchical cluster analysis placed IR exposure and PARP1 A762V on the same branch which shows that the strongest interactions exist between them.
CART Analysis
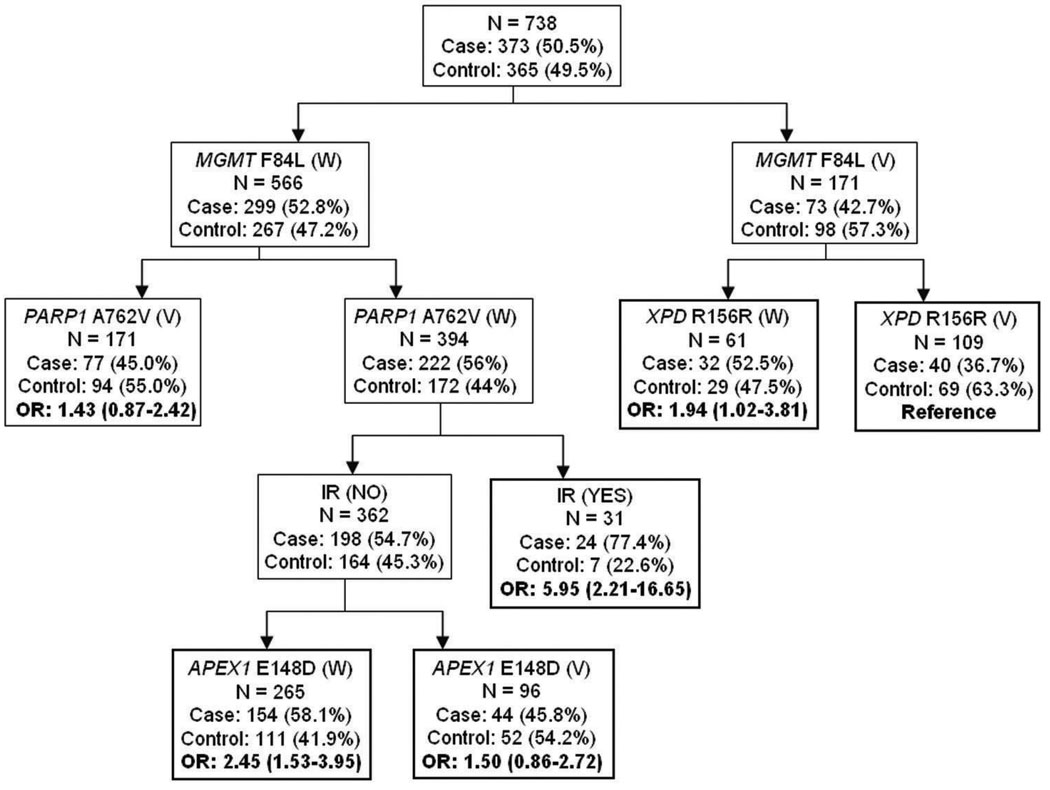
Figure 2 depicts the resulting tree structure generated by the CART analysis. Consistent with the MDR one-locus model, the first split on the decision tree was MGMT F84L, confirming that this SNP was the most important risk factor for glioma among those considered. Further inspection of the CART structure suggested distinct patterns for the wild-type (W) and the variant (V) alleles of MGMT F84L. In particular, individuals with the combined polymorphisms of MGMT F84L (V) and XPD R156R (V) exhibited the lowest glioma risk, with a 36.7% case rate. Using this terminal node as the referent group, the individuals who had IR exposure exhibited the highest glioma risk in the setting of both the MGMT F84L and PARP1 A762V wild-type genotypes (adjusted OR, 5.95; 95% CI, 2.21–16.65), further supporting the strong interaction between PARP A762V and IR exposure shown by the interaction dendrogram. In addition, the subgroups with MGMT F84L (W), PARP1 A762V (W), no IR exposure, and APEX1 E148D (W) also had an increased glioma risk (adjusted OR, 2.45; 95% CI, 1.53–3.95). However, these results are based on small numbers and should be interpreted with care.
Fig2.
Classification and regression tree (CART) analysis of the DNA repair gene polymorphisms and IR exposure history. The number and percentage of cases and controls are shown for each node. W, wild-type genotypes; V, variant genotypes. Referent group is the least percentage of cases. OR and 95% CI were adjusted by age and gender.
DISCUSSION
In this study, we used multi-analytic strategies to systematically examine the associations between a panel of DNA repair genes SNPs and glioma risk. In the single-locus analysis, six SNPs showed a significant association with glioma risk. When evaluating the cumulative genetic risk of these six SNPs, we found a significant trend toward increased risk with an increasing number of adverse genotypes. Moreover, both the MDR and CART analyses identified MGMT F84L as the predominant risk factor for glioma and revealed strong interactions among IR exposure, PARP1 A762V, MGMT F84L and APEX1 E148D. Taken together, these consistent findings imply that gene-gene, gene-environment interactions contribute to glioma susceptibility. However, a definite conclusion can only be reached on external validation by larger studies.
Perhaps the most significant finding in this study was the consistent association of MGMT F84L with glioma risk, which was identified using different analytic approaches. In the single-locus analysis, MGMT F84L had the strongest allelic association with glioma susceptibility, and the genotypic analysis also revealed that it was strongly associated with glioma risk. Consistent with the single-locus association results, the best one-factor model in MDR and the first split in CART both identified MGMT F84L as the predominant risk factor for glioma. Thus, this significant association was consistent across all analyses, which suggests that our results are unlikely due to chance but may reveal a biological link between MGMT F84L and the etiology of glioma.
Although the functional relevance of the MGMT F84L is unknown, several lines of evidence suggest that this finding is biologically plausible. As a pivotal gene involved in repair of DNA lesions induced by alkylating oxidative agents, MGMT is located on chromosome 10q26 and can act alone to reverse alkylation damage (32). Studies have suggested that the region of chromosome 10 (10q24-q26) is one of the commonly deleted regions in glioma and likely to contain the tumor suppressor gene important in glioma tumor formation (33). Moreover, MGMT F84L was previously reported to be associated with risk of endometrial cancer (34) and glioma (7, 35). A recently published paper, Bethke et al found that MGMT F84L was marginally significant in five unique glioma case-control series from four different countries (1013 cases, 1016 controls) (12). Han et al showed that the association between breast cancer and the F84L may be magnified by an increased endogenous estrogen level (36). Another colorectal cancer study reported that significant interactions were found between this SNP and alcohol intake and BMI (37). These studies lend support for a yet unexplained association between environment factors and MGMT F84L for cancer risk. Bugni et al suggested that gene-environment interactions may be particularly important for MGMT because its effect on cancer susceptibility in experimental animals is strongly determined by exposure to alkylating agents (38). Although MGMT is unlikely to be directly related to IR, it may depend on its interaction with other genes in the same pathway, such as p53. It is reported that p53 is involved in regulation of the MGMT by DNA damaging agents such as alkylating agents, IR and UV light (39). Further mechanistic studies are therefore warranted to address the functional relevance of MGMT F84L variant.
It is also important to note that of the six SNPs associated with glioma risk, three (XRCC1 R399Q, PARP1 A762V and APEX1 E148D) were from the BER pathway, suggesting a strong link between BER and glioma. It has been suggested that BER plays a critical role in the maintenance of the central nervous system genomic integrity (40), and recent studies show that BER is active in neuronal cells in culture, in brain cells in experimental animal models and in human postmortem brain tissue (41). Considering that the most important culprit for causing DNA damage in the brain seems to be oxidative stress, and BER is the most important DNA repair pathway to deal with such damage in the brain (42), it is therefore very likely that the SNPs in the BER could play an important role in glioma development.
Several studies have found an increased risk for brain tumors after exposure to high doses of IR such as atomic bomb survivors (2, 43, 44), however, results after exposure to chronic lower doses are not consistent. Although a possible association between low level medical IR exposure and the risk of brain tumors has been suggested (45, 46), occupational exposure to low IR levels and the risk of brain tumors remains, however, not very consistent (45, 47, 48). In the current study, ten study subjects reported medical exposure to IR (for treatment of conditions such as acne, birth mark, and hyperthyroidism) while the remaining 46 were occupationally exposed (occupations such as physicians, radiologists, nurses, pilots and engineers). Our stratified analysis and interaction dendrogram model detected a gene-IR interaction for glioma risk that could potentially be explained by altered protein function resulting from the PARP1 A762V variants thus leading to suboptimal repair of DNA damage. Given that IR is one of the most important risk factors for glioma; our results may have implication on future studies.
Given that our study population is relatively small, several approaches were taken to control for false-positive findings. First, we included only Caucasian cases in the analysis. This ethnic homogeneity of study population reduces the risk of confounding by unmeasured factors, either genetic or environmental. Second, most of the genes and SNPs selected were based on prior biological evidence of their considerable functional importance. Under these circumstances, the frequency of false positive findings would be substantially decreased and the power of association would be improved by accounting for their interactions. Last, we used two different strategies to assess the robustness of associations: an internal validation procedure based on bootstrap re-sampling methods (i.e., using the same data set) and a correction for multiple testing using permutation tests. Two major advantages of bootstrapping are that it makes no assumptions regarding the distribution of the data and that it gives more accurate answers as a result of correction for small sample sizes (24).
Despite the strengths and biological plausibility of the associations observed in our study, there are inherent limitations. A major limitation is the small number of participants with IR exposure in the analyses. Since the IR exposure assessment was based on self-reported information, the possibility of recall bias or misclassification due to subjects not being fully aware of their IR exposures, especially in occupational settings, exists. Quantitative studies on low dose IR exposure in the general population are very difficult to conduct because large samples and long-term complete follow-up are needed in addition to lifetime measures of doses. Several previous studies examined the association between DNA repair gene polymorphisms and cancer risk, but none examined interactions of polymorphisms with IR exposures, except for studies on populations exposed specifically to medical IR (49, 50). Another limitation is the limited power to examine interactions. Given the post hoc data-driven nature of the CART approach, the small sample sizes in some of the terminal nodes and the wide confidence intervals led to the generation of findings that should be cautiously interpreted. However, these findings suggest new directions for the examination of both gene-gene and gene-environment interactions for future studies.
In summary, our study suggested that several polymorphisms in DNA repair genes may act individually or together to contribute to glioma susceptibility. The mechanism of how the variants modify the effect of IR needs further elucidated through experimental studies. In particular, our results support the notion that future risk assessments of complex diseases such as cancer need to move from the analysis of single polymorphisms to the systematic verification of the gene-environment-disease network.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Margaret Spitz for thoughtful comments and assistance; Drs. Charles Conrad, Franco deMonte, Morris Groves, Amy Heimberger, Victor Levin, Sujit Prabhu, Raymond Sawaya, Vinay Puduvalli and Susan Graham for recruiting the subjects, Ms Phyllis Adatto, Georgina N. Armstrong, for data management and laboratory assistance; Dr Beth Notzon (Department of Scientific Publication) for scientific editing.
Grant support: National Cancer Institute grants CA070917 (PI: ML Bondy) and NIEHS center grants P30 ES007784 (PI: John DiGiovanni). This publication was made possible by grant number P30 ES007784 from the National Institute of Environmental Health Sciences, NIH. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIEHS, NIH.
REFERENCE
- 1.Wrensch M, Lee M, Miike R, et al. Familial and personal medical history of cancer and nervous system conditions among adults with glioma and controls. Am J Epidemiol. 1997;145:581–593. doi: 10.1093/oxfordjournals.aje.a009154. [DOI] [PubMed] [Google Scholar]
- 2.Ron E, Modan B, Boice JD, Jr, et al. Tumors of the brain and nervous system after radiotherapy in childhood. N Engl J Med. 1988;319:1033–1039. doi: 10.1056/NEJM198810203191601. [DOI] [PubMed] [Google Scholar]
- 3.Little MP, De Vathaire F, Shamsaldin A, et al. Risks of brain tumour following treatment for cancer in childhood: Modification by genetic factors, radiotherapy and chemotherapy. Int J Cancer. 1998;78:269–275. doi: 10.1002/(SICI)1097-0215(19981029)78:3<269::AID-IJC1>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 4.Bondy ML, Wang LE, El-Zein R, et al. Gamma-radiation sensitivity and risk of glioma. J Natl Cancer Inst. 2001;93:1553–1557. doi: 10.1093/jnci/93.20.1553. [DOI] [PubMed] [Google Scholar]
- 5.Rasheed BK, Wiltshire RN, Bigner SH, Bigner DD. Molecular pathogenesis of malignant gliomas. Curr Opin Oncol. 1999;11:162–167. doi: 10.1097/00001622-199905000-00004. [DOI] [PubMed] [Google Scholar]
- 6.Moore JH, Williams SM. New strategies for identifying gene-gene interactions in hypertension. Ann Med. 2002;34:88–95. doi: 10.1080/07853890252953473. [DOI] [PubMed] [Google Scholar]
- 7.Felini MJ, Olshan AF, Schroeder JC, et al. DNA repair polymorphisms XRCC1 and MGMT and risk of adult gliomas. Neuroepidemiology. 2007;29:55–58. doi: 10.1159/000108919. [DOI] [PubMed] [Google Scholar]
- 8.Kiuru A, Lindholm C, Heinavaara S, et al. XRCC1 and XRCC3 variants and risk of glioma and meningioma. J Neuro-Oncol. 2008;88:135–142. doi: 10.1007/s11060-008-9556-y. [DOI] [PubMed] [Google Scholar]
- 9.Liu Y, Zhou K, Zhang H, et al. Polymorphisms of LIG4 and XRCC4 involved in the NHEJ pathway interact to modify risk of glioma. Hum Mutat. 2007;29:381–389. doi: 10.1002/humu.20645. [DOI] [PubMed] [Google Scholar]
- 10.Liu Y, Zhang H, Zhou K, et al. Tagging SNPs in non-homologous end-joining pathway genes and risk of glioma. Carcinogenesis. 2007;28:1906–1913. doi: 10.1093/carcin/bgm073. [DOI] [PubMed] [Google Scholar]
- 11.Wang LE, Bondy ML, Shen H, et al. Polymorphisms of DNA repair genes and risk of glioma. Cancer Res. 2004;64:5560–5563. doi: 10.1158/0008-5472.CAN-03-2181. [DOI] [PubMed] [Google Scholar]
- 12.Bethke L, Webb E, Murray A, et al. Comprehensive analysis of the role of DNA repair gene polymorphisms on risk of glioma. Hum Mol Genet. 2008;17:800–805. doi: 10.1093/hmg/ddm351. [DOI] [PubMed] [Google Scholar]
- 13.Chang JS, Yeh RF, Wiencke JK, et al. Pathway analysis of single-nucleotide polymorphisms potentially associated with glioblastoma multiforme susceptibility using random forests. Cancer Epidemiol Biomarkers Prev. 2008;17:1368–1373. doi: 10.1158/1055-9965.EPI-07-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartge P, Brinton LA, Rosenthal JF, Cahill JI, Hoover RN, Waksberg J. Random digit dialing in selecting a population-based control group. Am J Epidemiol. 1984;120:825–833. doi: 10.1093/oxfordjournals.aje.a113955. [DOI] [PubMed] [Google Scholar]
- 15.Harlow BL, Davis S. Two one-step methods for household screening and interviewing using random digit dialing. Am J Epidemiol. 1988;127:857–863. doi: 10.1093/oxfordjournals.aje.a114869. [DOI] [PubMed] [Google Scholar]
- 16.Goode EL, Ulrich CM, Potter JD. Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol Biomarkers Prev. 2002;11:1513–1530. [PubMed] [Google Scholar]
- 17.Hung RJ, Hall J, Brennan P, Boffetta P. Genetic polymorphisms in the base excision repair pathway and cancer risk: a HuGE review. Am J Epidemiol. 2005;162:925–942. doi: 10.1093/aje/kwi318. [DOI] [PubMed] [Google Scholar]
- 18.Auranen A, Song H, Waterfall C, et al. Polymorphisms in DNA repair genes and epithelial ovarian cancer risk. Int J Cancer. 2005;117:611–618. doi: 10.1002/ijc.21047. [DOI] [PubMed] [Google Scholar]
- 19.Manuguerra M, Saletta F, Karagas MR, et al. XRCC3 and XPD/ERCC2 single nucleotide polymorphisms and the risk of cancer: a HuGE review. Am J Epidemiol. 2006;164:297–302. doi: 10.1093/aje/kwj189. [DOI] [PubMed] [Google Scholar]
- 20.Fernet M, Hall J. Genetic biomarkers of therapeutic radiation sensitivity. DNA repair. 2004;3:1237–1243. doi: 10.1016/j.dnarep.2004.03.019. [DOI] [PubMed] [Google Scholar]
- 21.Akaike H. A New Look at the Statistical Model Identification. IEEE - Automatic Control, IEEE Transactions on 1974. AC-19:716–723. [Google Scholar]
- 22.Efron B. Bootstrap methods: Another look at the jackknife. Ann Statistics. 1979;7:1–26. [Google Scholar]
- 23.Efron B, Tibshirani RJ. An Introduction to the Bootstrap. London: Chapman and Hall; 1993. [Google Scholar]
- 24.Chernick MR. Bootstrap Methods: A Practioner’s Guide. John Wiley & Sons; 1999. [Google Scholar]
- 25.Lewontin RC. On Measures of Gametic Disequilibrium. Genetics. 1988;120:849–852. doi: 10.1093/genetics/120.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ritchie MD, Hahn LW, Roodi N, et al. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am J Hum Genet. 2001;69:138–147. doi: 10.1086/321276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore JH, Gilbert JC, Tsai CT, et al. A flexible computational framework for detecting, characterizing, and interpreting statistical patterns of epistasis in genetic studies of human disease susceptibility. J Theoret Biol. 2006;241:252–261. doi: 10.1016/j.jtbi.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 29.Jakulin A, Bratko I. Analyzing attribute dependencies. Knowledge Discovery in Databases: Pkdd 2003. Proceedings. 2003:229–240. [Google Scholar]
- 30.Curk T, Demsar J, Xu QK, et al. Microarray data mining with visual programming. Bioinformatics. 2005;21:396–398. doi: 10.1093/bioinformatics/bth474. [DOI] [PubMed] [Google Scholar]
- 31.Zhang HP, Singer B. Recursive Partitioning in the Health Sciences. New York: Springer; 1999. [Google Scholar]
- 32.Glassner BJ, Weeda G, Allan JM, et al. DNA repair methyltransferase (Mgmt) knockout mice are sensitive to the lethal effects of chemotherapeutic alkylating agents. Mutagenesis. 1999;14:339–347. doi: 10.1093/mutage/14.3.339. [DOI] [PubMed] [Google Scholar]
- 33.Rasheed BK, Bigner SH. Genetic alterations in glioma and medulloblastoma. Cancer Metastasis Rev. 1991;10:289–299. doi: 10.1007/BF00554791. [DOI] [PubMed] [Google Scholar]
- 34.Huang J, Ye F, Chen H, Lu W, Xie X. Amino acid substitution polymorphisms of the DNA repair gene MGMT and the susceptibility to cervical carcinoma. Carcinogenesis. 2007;28:1314–1322. doi: 10.1093/carcin/bgm003. [DOI] [PubMed] [Google Scholar]
- 35.Inoue R, Isono M, Abe M, Abe T, Kobayashi H. A genotype of the polymorphic DNA repair gene MGMT is associated with de novo glioblastoma. Neuro Res. 2003;25:875–879. doi: 10.1179/016164103771954005. [DOI] [PubMed] [Google Scholar]
- 36.Han J, Tranah GJ, Hankinson SE, Samson LD, Hunter DJ. Polymorphisms in O6-methylguanine DNA methyltransferase and breast cancer risk. Pharmacogenet Genomics. 2006;16:469–474. doi: 10.1097/01.fpc.0000215065.21718.4c. [DOI] [PubMed] [Google Scholar]
- 37.Tranah GJ, Bugni J, Giovannucci E, et al. O6-methylguanine-DNA methyltransferase Leu84Phe and Ile143Val polymorphisms and risk of colorectal cancer in the Nurses' Health Study and Physicians' Health Study (United States) Cancer Causes Control. 2006;17:721–731. doi: 10.1007/s10552-006-0005-y. [DOI] [PubMed] [Google Scholar]
- 38.Bugni JM, Han J, Tsai MS, Hunter DJ, Samson LD. Genetic association and functional studies of major polymorphic variants of MGMT. DNA repair. 2007;6:1116–1126. doi: 10.1016/j.dnarep.2007.03.023. [DOI] [PubMed] [Google Scholar]
- 39.Grombacher T, Eichhorn U, Kaina B. p53 is involved in regulation of the DNA repair gene O6-methylguanine-DNA methyltransferase (MGMT) by DNA damaging agents. Oncogene. 1998;17:845–851. doi: 10.1038/sj.onc.1202000. [DOI] [PubMed] [Google Scholar]
- 40.Bohr VA, Ottersen OP, Tonjum T. Genome instability and DNA repair in brain, ageing and neurological disease. Neuroscience. 2007;145:1183–1186. doi: 10.1016/j.neuroscience.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 41.Weissman L, de Souza-Pinto NC, Stevnsner T, Bohr VA. DNA repair, mitochondria, and neurodegeneration. Neuroscience. 2007;145:1318–1329. doi: 10.1016/j.neuroscience.2006.08.061. [DOI] [PubMed] [Google Scholar]
- 42.Subba Rao K. Mechanisms of disease: DNA repair defects and neurological disease. Nat Clin Pract Neurol. 2007;3:162–172. doi: 10.1038/ncpneuro0448. [DOI] [PubMed] [Google Scholar]
- 43.Soffer D, Gomori JM, Pomeranz S, Siegal T. Gliomas following low-dose irradiation to the head report of three cases. J Neuro-Oncol. 1990;8:67–72. doi: 10.1007/BF00182089. [DOI] [PubMed] [Google Scholar]
- 44.Schlehofer B, Blettner M, Becker N, Martinsohn C, Wahrendorf J. Medical risk factors and the development of brain tumors. Cancer. 1992;69:2541–2547. doi: 10.1002/1097-0142(19920515)69:10<2541::aid-cncr2820691025>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 45.Neuberger JS, Brownson RC, Morantz RA, Chin TD. Association of brain cancer with dental X-rays and occupation in Missouri. Cancer Detect Prev. 1991;15:31–34. [PubMed] [Google Scholar]
- 46.Ryan P, Lee MW, North B, McMichael AJ. Amalgam fillings, diagnostic dental x-rays and tumours of the brain and meninges. Eur J Cancer B Oral Oncol. 1992;28B:91–95. doi: 10.1016/0964-1955(92)90034-x. [DOI] [PubMed] [Google Scholar]
- 47.Karipidis KK, Benke G, Sim MR, Kauppinen T, Giles G. Occupational exposure to ionizing and non-ionizing radiation and risk of glioma. Occup Med. 2007;57:518–524. doi: 10.1093/occmed/kqm078. [DOI] [PubMed] [Google Scholar]
- 48.Shirangi A, Fritschi L, Holman CD. Maternal Occupational Exposures and Risk of Spontaneous Abortion in Veterinary Practice. Occup Environ Med. 2008 doi: 10.1136/oem.2007.035246. Epub ahead of print, PMID:18388114. [DOI] [PubMed] [Google Scholar]
- 49.Robert C Millikan, Jon S Player, Allan Rene deCotret, Chiu-Kit Tse, Keku T. Polymorphisms in DNA repair genes, medical exposure to ionizing radiation, and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2005;14:2326–2334. doi: 10.1158/1055-9965.EPI-05-0186. [DOI] [PubMed] [Google Scholar]
- 50.Siegal Sadetzki, Pazit Flint-Richter, Sigal Starinsky, et al. Genotyping of patients with sporadic and radiation-associated meningiomas. Cancer Epidemiol Biomarkers Prev. 2005;14:969–976. doi: 10.1158/1055-9965.EPI-04-0366. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.