Abstract
The mammalian clock regulates major aspects of energy metabolism, including glucose and lipid homoeostasis as well as mitochondrial oxidative metabolism. This study is to identify specific patterns of circadian rhythms for lipid homoeostasis in both female and male mouse livers, and to clarify gender disparity in coupling the peripheral circadian clock to lipid metabolic outputs by nuclear receptors. To achieve this, profiling the diurnal hepatic expression of genes encoding circadian clocks, nuclear receptors and lipid metabolic enzymes was performed. Hepatic lipid levels including cholesterol, triglyceride and non-esterified fatty acids (NEFAs) were monitored over a 24-h period. The cosinor analysis revealed that several genes encoding nuclear receptors and enzymes involved in the lipid metabolic pathway were rhythmically expressed in liver in phase with the peripheral clocks, which were correlated with the diurnal changes of hepatic lipid levels. Gender disparity was observed for circadian characteristics including mesor and amplitude values, accompanied with advances in acrophases in female mouse livers. Accordingly, gender differences were also observed in diurnal lipid homoeostasis. The identification of cycling patterns for lipid metabolic pathways in both female and male mouse livers may shed light on the development of gender-based treatment for human diseases related to the coordination of the cellular clock and control of lipid homoeostasis.
Keywords: Circadian rhythm, gender, lipid homoeostasis, liver, nuclear receptor
Circadian rhythms are daily cycles of multiple molecular, biochemical, physiological and behavioural processes that are driven by an endogenous clock with an approximate 24-h periodicity. Circadian rhythms in mammals are governed substantially by the main time-keeping system, called the central circadian clock, located in the suprachiasmatic nucleus (SCN) of the hypothalamus. Circadian clocks also exist in most peripheral tissues and even in cultured cells. It is believed that the master circadian clock synchronizes the peripheral clocks through both neural and humoral factors, allowing animals to adapt their feeding, activity and metabolism to predictable rhythmic cycles. Environmental light is the predominant Zeitgeber (time-giver) to reset the intrinsic clocks. In addition, other cyclic inputs such as temperature, food, noise or social cues may also influence the phase of clocks (1–3).
The central and peripheral clocks share common molecular circuitry. The circadian clockwork consists of two auto-regulatory feedback loops connected by a central pair of basic-helix–loop–helix (bHLH)-PAS (PER-ARNT-SIM)-containing transcription factors (brain and muscle Arnt-like protein 1-circadian locomotor output cycles kaput heterodimer, Bmal1-Clock) (4, 5). The complex of Bmal1-Clock drives the rhythmic transcriptions of other clock genes including period genes (Per1-3), cryptochrome genes (Cry1-2) and the nuclear receptor Rev-erbα gene through E-box (CACGTG) elements located in their promoters (6, 7). In the first feedback loop, Per proteins and Cry proteins inhibit the transcriptional activity of Bmal1-Clock, while the components of the second loop, retinoid-related orphan receptor alpha (RORα) and Rev-erbα, inversely regulate Bmal1, Cry1 and Rev-erbα transcriptions through the functional ROR response elements (RORE) (1, 3, 7–9).
Lipids encompass molecules such as fatty acids and their derivatives (including tri-, di- and monoglycerides and phospholipids), as well as other sterol-containing metabolites such as cholesterol, which are not only nutritionally important, but also serve as precursors for ligands of many nuclear receptors. Nuclear receptors, as sensors for fat-soluble hormones, vitamins and dietary lipids, direct a wide range of molecular genetic programmes that regulate lipid homoeostasis (3, 10–13). It is well known that general metabolism is subjected to rhythmic fluctuation in accordance with the light–dark cycle (3, 14, 15). In addition, variations in the levels of nuclear receptors per se have been noted in male mouse livers (3, 16).
Although the core molecular clock has been defined, the molecular outputs that ultimately regulate circadian rhythms of cellular physiology, organ function and system-level behaviour are poorly understood. Specifically, despite much evidence supporting the role of nuclear receptors in lipid homoeostasis and the accumulation of data connecting metabolism with circadian biology, few studies have been performed to fully explore the link between circadian transcriptional outputs and lipid homoeostasis. In addition, although anatomical profiling and function of nuclear receptors in various types of tissues have been shown (17–19), little is known about the gender difference in diurnal expression pattern of nuclear receptors in the livers. Furthermore, the gender disparity in coupling the peripheral circadian clock to lipid metabolic outputs by nuclear receptors has not been elucidated.
In the present study, the expression profiles of genes encoding circadian clocks, nuclear receptors and enzymes as well as transporters involved in lipid homoeostasis with focus on fatty acid and cholesterol metabolism were investigated in both female and male mouse livers at an interval of 4 h over a 24-h period (Tables 1 and 2). The present data suggest that lipid homoeostasis requires a member of clock-controlled genes that elicit daily changes in metabolic physiology. Included are previously identified circadian-regulated genes in male-mouse livers, thus validating our findings, along with many other genes previously unknown to be regulated by the circadian clocks. Analysis of expression patterns of these genes further reveals that the dynamic but coordinated changes in nuclear receptor expression play important roles in coupling the peripheral circadian clock to lipid metabolic outputs. Additionally, gender differences observed in the rhythmic profiles of genes examined might provide an explanation for gender disparity in diurnal lipid homoeostasis.
Table 1.
Description of genes encoding circadian clocks and nuclear receptors.
| Official symbol | Gene ID | Official full name |
|---|---|---|
| Circadian clock | ||
| Arntl | 11865 | Aryl hydrocarbon receptor nuclear translocator-like; brain and muscle Arnt-like protein-1 (Bmal1) |
| Clock | 12753 | Circadian locomotor output cycles kaput |
| Cry1 | 12952 | Cryptochrome 1 (photolyase-like) |
| Cry2 | 12953 | Cryptochrome 2 (photolyase-like) |
| Per1 | 18626 | Period homologue 1 (Drosophila) |
| Per2 | 18627 | Period homologue 2 (Drosophila) |
| Per3 | 18628 | Period homologue 3 (Drosophila) |
| Nuclear receptor | ||
| Nr1d1 | 217166 | Nuclear receptor subfamily 1, group D, member 1 (Rev-erbα) |
| Nr1i2 | 18171 | Nuclear receptor subfamily 1, group I, member 2; pregnane X receptor (Pxr) |
| Nr1h3 | 22259 | Nuclear receptor subfamily 1, group H, member 3; liver X receptor (Lxr) |
| Ppara | 19013 | Peroxisome proliferator activated receptor alpha |
| Ppard | 19015 | Peroxisome proliferator activator receptor delta |
| Pparg | 19016 | Peroxisome proliferator activated receptor gamma |
| Ppargc1a | 19017 | Peroxisome proliferative activated receptor, gamma, coactivator 1 alpha (Pgc1α) |
| Ppargc1b | 170826 | Peroxisome proliferative activated receptor, gamma, coactivator 1 beta (Pgc1β) |
| Rora | 19883 | RAR-related orphan receptor alpha |
| Rxra | 20181 | Retinoid X receptor alpha |
Table 2.
Description of genes encoding enzymes and transporters involved in lipid homoeostasis.
| Official symbol | Gene ID | Official full name | Function |
|---|---|---|---|
| Abca1 | 11303 | ATP-binding cassette, subfamily A (ABC1), member 1 | Efflux cholesterol from cell |
| Acaca | 107476 | Acetyl- co-enzyme A carboxylase alpha | Control rate-limiting step in fatty acid synthesis |
| Acadl | 11363 | Acyl-co-enzyme A dehydrogenase, long-chain | Control mitochondrial β-oxidation of long-chain fatty acids |
| Acox1 | 11430 | Acyl-co-enzyme A oxidase 1, palmitoyl | Regulate peroxisomal β-oxidation of fatty acid |
| Acsl1 | 14081 | Acyl-CoA synthetase long-chain family member 1 | Regulate mitochondrial β-oxidation of fatty acid |
| Apob | 238055 | Apolipoprotein B | Carry cholesterol to tissues |
| Apoc3 | 11814 | Apolipoprotein C-III | Inhibit hepatic uptake of triglyceride-rich lipoproteins and/or hydrolysis of triglyceride |
| Cd36 | 12491 | CD36 antigen (fatty acid translocase) | Facilitate long-chain fatty acids uptake |
| Cpt1 | 12894 | Carnitine palmitoyltransferase 1a, liver | Control rate-limiting step in mitochondrial β-oxidation of fatty acid |
| Clps | 109791 | Colipase, pancreatic | Absorb fat in small intestine |
| Cyp4a10 | 13117 | Cytochrome P450, family 4, subfamily a, polypeptide 10 | Regulate ω-oxidation of fatty acid |
| Cyp4a14 | 13119 | Cytochrome P450, family 4, subfamily a, polypeptide 14 | Regulate ω-oxidation of fatty acid |
| Cyp7a1 | 13122 | Cytochrome P450, family 7, subfamily a, polypeptide 1 | Control rate-limiting step in bile acid synthesis from cholesterol |
| Elov16 | 170439 | ELOVL family member 6, elongation of long-chain fatty acids (yeast) (fatty acid elongase, Fae) | Elongate fatty acid chains |
| Fabp1 | 14080 | Fatty-acid-binding protein 1, liver | Bind long-chain fatty acids in hepatocytes |
| Fasn | 14104 | Fatty acid synthase | Synthesize fatty acid |
| Hmgcl | 15356 | 3-hydroxy-3-methylglutaryl co-enzyme A lyase | Control ketosis and break down proteins and fats from the diet |
| Hmgcr | 15357 | 3-hydroxy-3-methylglutaryl co-enzyme A reductase | Control rate-limiting step in cholesterol synthesis |
| Hmgcs1 | 208715 | 3-hydroxy-3-methylglutaryl co-enzyme A synthase 1 | Regulate ketosis |
| Ldlr | 16835 | Low-density lipoprotein receptor | Transport low-density lipoproteins (cholesterol) from bloodstream into cell |
| Lpin1 | 14245 | Lipin 1 | Synthesize triglyceride |
| Mlxipl | 58805 | MLX interacting protein-like (carbohydrate responsive element-binding protein, ChREBP) | Control lipogenesis through the transcriptional regulation of lipogenic genes |
| Mttp | 17777 | Microsomal triglyceride transfer protein | Transport lipid |
| Nt5e | 23959 | 5′ Nucleotidase, ecto (Cd73) | Produce adenosine |
| Pnliprp2 | 18947 | Pancreatic lipase-related protein 2 | Digest intestinal fat |
| Scarb1 | 20778 | Scavenger receptor class B, member 1 (Sr-b1) | Sensor high-density lipoprotein (HDL, cholesterol ester) |
| Scd1 | 20249 | Stearoyl-co-enzyme A desaturase 1 | Synthesize unsaturated fatty acids |
MATERIALS AND METHODS
Animals
C57BL/6 female and male mice (7 weeks old) were maintained at a controlled temperature (24°C) on a 12-h light/dark cycle (lights from 5:00 a.m. to 5:00 p.m.). Female mice, which were separated from male mice, were housed together to ensure a synchronous oestrous cycle (17). Water and standard rodent chow (Harlan Teklad, Madison, WI, USA) were provided ad libitum. Under such conditions, mice tended to feed at night when the light was off (20). All procedures were conducted in accordance with the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals and were approved by University of Kansas Medical Center Institutional Animal Care and Use Committee. After being housed for 2 weeks in the animal facility, the mice were euthanized at 4-h intervals (6:00 a.m., 10:00 a.m., 2:00 p.m., 6:00 p.m., 10:00 p.m. and 2:00 a.m.) over a 24-h period. Five mice were included for each time-point. Livers were dissected immediately and frozen in liquid nitrogen, and stored at –80°C for RNA and lipid extraction.
Quantitative Real-Time Polymerase Chain Reaction (PCR)
Liver samples were lysed using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and total RNA was isolated according to the manufacturer's protocol. RNA purity was confirmed by a 260/280 nm absorbance ratio >1.8. Agarose gel electrophoresis with ethidium bromide staining was utilized to confirm integrity of 18S and 28S ribosomal RNA bands. Reverse transcription was carried out using M-MLV reverse transcriptase (Invitrogen) according to the manufacturer's protocol. Primers and probes for real-time PCR were designed using Primer Express 2.0 according to the manufacturer's instructions (Real-Time PCR Chemistry Guide, Applied Biosystems, Foster City, CA, USA) (Table 3). To eliminate the possibility of contaminating genomic DNA amplification, both primer sequences were designed to cross exon junctions in the corresponding cDNA sequences. In addition, all primer and probe sets were checked for specificity using the NCBI Basic Local Alignment Search Tool (BLAST). Real-time PCR amplification of cDNA was carried out using total RNA (15 ng) in a reaction mixture (12 μl) containing TaqMan® Universal Master Mix (1×), primers (900 nM each) and FAM-BHQ1 dual-labelled probe (250 nM). Amplification and fluorescence detection were carried out using the ABI Prism 7900 HT Real Time PCR System (Applied Biosystems, Foster City, CA, USA). Cycling conditions were initial hold steps (50°C for 2 min, followed by 95°C for 10 min) and 40 cycles of a two-step PCR (92°C for 15 s, then 60°C for 1 min). Relative mRNA levels in comparison with the values obtained from male livers at 6:00 a.m. (as calibrators) were calculated by the comparative delta-CT method, or relative standard curve method according to the manufacturer's instructions (Applied Biosystems, Foster City, CA, USA).
Table 3.
Nucleotide sequences of TagMan probes (with 5′-FAM and 3′-BHQ1 modifications) and primers for the studied genes.
| Official symbol | Probe sequence 5′-3′ | Forward primer 5′-3′ | Reverse primer 5′-3′ |
|---|---|---|---|
| Circadian clock | |||
| Arntl (Bmal1) | 5′-TAGGTCGAATGATCGCGGAGGA | 5′-CAGAGCCGGAGCAGGAA A | 5′-AAGGCGATGACCCTCTTATCC |
| Clock | 5′-CTGCCCTTGAGGCTGGCGTTAAACC | 5′-CCTTGCTCTGGACTGCTTCTGT | 5′-GTCTCTAAGGAAGGAAAGAGGACTTG |
| Cry1 | 5′-CCGGCTCTTCCAACGTGGGCAT | 5′-GTCTATATCCTGGACCCCTGGTT | 5′-AAAAATCGCCACCTGTTGATG |
| Cry2 | 5′-CAGAAACCACCCCTTACCTACAAGCGCT | 5′-CACCCTCTATGACCTAGACAGAATCAT | 5′-GTCCCATGCGGCTGATG |
| Per1 | 5′-CACACTTCGAAACCAGGACACCTTCTCTGT | 5′-CTGGAGGAATTGGAGCATATCAC | 5′-AATATAGACAATCCGGCCTGTCA |
| Per2 | 5′-ATGCCATCTCCACGTCAAGTGACCTG | 5′-AGGCACCTCCAACATGCAA | 5′-GGATGCCCCGCTTCTAGAC |
| Per3 | 5′-ATTAAATGAAGATGTTTTTGCCACCA | 5′-GGTCGACATAAAGTCCGAACGA | 5′-TCGTTACTGGCTGCCTTTTTTATT |
| Nuclear receptor | |||
| Nr1d1 (Rev-erbα) | 5′-CCCATGGACGCAGCGGCC | 5′-GCAAGGCAACACCAAGAATGT | 5′-CTTCCCAGATCTCCTGCACAGT |
| Nr1i2 (Pxr) | 5′-TCGCACCCCTTTGCCACCCC | 5′-CAG TTG CTG CGC ATC CAA | 5′-TGC TGC TAA ATA ACT CTT GCA TGA G |
| Nr1h3 (Lxr) | 5′- CAGGCATGAGGGAGGAGTGTGTGCT | 5′-CCA TGA ATG AGC TGC AAC TCA | 5′-GGC CGG TCT GCA GAG AAG |
| Ppara | 5′-TCTGTCGGGATGTCACACAATGCAATTC | 5′-GAT TCA GAA GAA GAA CCG GAA CA | 5′-TGC TTT TTC AGA TCT TGG CAT TC |
| Pparb | 5′-TGGCACTCGGCATGTCGCACA | 5′-GAA CCG CAA CAA GTG TCA GTA CTG | 5′-CGG CAT CCG TCC AAA GC |
| Pparg | 5′-CTGAAGCTCCAAGAATACCAAAGTGCGATC | 5′-CCC AAT GGT TGC TGA TTA CAA A | 5′-GAG GGA GTT AGA AGG TTC TTC ATG A |
| Ppargc1a (Pgc1α) | 5′-CAAACCCTGCCATTGTTAAGACCGAGAA | 5′-AAC CAC ACC CAC AGG ATC AGA | 5′-TCT TCG CTT TAT TGC TCC ATG A |
| Ppargc1b (Pgc1β) | 5′-CGAGCTCTTCCAGATTGACAGTGAGAATGA | 5′-TGA CTC AGC CAC GTG CTT TG | 5′-TCG TAA GCG CAG CCA AGA G |
| Rora | 5′-GGAATCCATTATGGTGTCATTACG | 5′- GGAATCCATTATGGTGTCATTACG | 5′-GTGGCATTGCTCTGCTGACTT |
| Rxra | 5′-AAACATTTCCTGCCGCTCGACTTCTCTACC | 5′-CATGAGTTAGTCGCAGACATGGA | 5′-CGACCCGTTGGAGAGTTGAG |
| Lipid homoeostasis | |||
| Abca1 | 5′-TGTTCTCAGACGCTCAGAGGCTTCTTCTGTA | 5′-CCC TGC TTC CGT TAT CCA ACT | 5′-GAC CTT GTG CAT GTC CTT AAT GC |
| Acaca | 5′-TCCTCAACTTTGTGCCCACGGTCA | 5′-ATG TCC GCA CTG ACT GTA ACC A | 5′-TGC TCC GCA CAG ATT CTT CA |
| Acadl | 5′-TTCCATGGCAAAATACTGGGCATCTGA | 5′-ACG TCT GGA CTC CGG TTC TG | 5′-CAC ACA TTC ATA AGC TAC ACT GTT TTG |
| Acox1 | 5′-TGGGACCCACAAGCCTCTGCCA | 5′-TTT GTT GTC CCT ATC CGT GAG A | 5′-GCC GAT ATC CCC AAC AGT GA |
| Acsl1 | 5′-CCTACAAAGAGGTGGCAGAACTGGCTGAG | 5′-AGC CAA ACC AGC CCT ATG AGT | 5′-CAG CCC GGA GCC TAT GC |
| Apob | 5′-TGCCA CCCCTGGAGTTGCCAGTT | 5′-TCC TAA CAT CAT GGT GCC TTC AT | 5′-TGA AAT CTG GGA GGA AAA ACT TGA |
| Apoc3 | 5′-TTTACTGACAAGTTCACCGGCTTC | 5′-CCT GAA AGG CTA CTG GAG CAA | 5′-TTG GTC CTC AGG GTT AGA ATC C |
| Cd36 | 5′-TCACCCCTCCAGAATCCAGACAACCA | 5′-TCC AGC CAA TGC CTT TGC | 5′-TGG AGA TTA CTT TTT CAG TGC AGA A |
| Cpt1 | 5′-CTGAAGGTGCTGCTCTCCTACCATTCA | 5′-CGA TCA TCA TGA CTA TGC GCT ACT | 5′-GCC GTG CTC TGC AAA CAT C |
| Clps | 5′-AGAACAGCGAGTGCTCCCCAAAGACC | 5′-GCCCGTTGCACACACAAG | 5′-GGACACCGGTAGTAGATCCCATAG |
| Cyp4a10 | 5′-CCCGACACAGCCACTCATTCCTGC | 5′-TCC AGG TTT GCA CCA GAC TCT | 5′-TCC TGG CTC CTC CTG AGA AG |
| Cyp4a14 | 5′-TGCATGCCTTCCCACTGGCTTTG | 5′-CAA GAC CCT CCA GCA TTT CC | 5′-GAG CTC CTT GTC CTT CAG ATG GT |
| Cyp7a1 | 5′-TGTCATGAGACCTCCGGGCCTTCC | 5′-CCA TGA TGC AAA ACC TCC AAT | 5′-TGA CCC AGA CAG CGC TCT TT |
| Elov16 (Fae) | 5′-CCGTCATGTACTCTTACTACGCCTTGCG | 5′-TGA CTA TGA ACT ATG GCG TGC AT | 5′-CCG GGA GAC TCG GAA ACC |
| Fabp1 | 5′-TCACCATCACCTATGGACCCAAAGTGG | 5′-TGC ATG AAG GGA AGA AAA TCA AA | 5′-CCC CCA GGG TGA ACT CAT T |
| Fasn | 5′-AGCCCATGGCACGGGCACC | 5′-CCC GGA GTC GCT TGA GTA TAT T | 5′-GGA CCG AGT AAT GCC ATT CAG |
| Hmgcl | 5′-AGGTTGCCAAGAAGTTGTACTCGATGGGC | 5′-CTC CCC GGC TAA AGT TGC T | 5′-GGT GTC CCC AAG GGA GAT CT |
| Hmgcr | 5′-CCGACAAGAAGCCTGCTGCCATAAAC | 5′-GGC AGT CAG TGG GAA CTA TTG C | 5′-CAG TCT TTC CTC GTC CTT CGA |
| Hmgcs1 | 5′-TGACCGAGGGCTCCGTGGGAC | 5′-GGG CCA AAC GCT CCT CTA AT | 5′-AGT CAT AGG CAT GCT GCA TGT G |
| Ldlr | 5′-CCTGCATTCACGGTAGCCGCCA | 5′-GCC GAC CTG ATG AAT TCC A | 5′-GCA GTC ATG TTC ACG GTC ACA |
| Lpin1 | 5′-AGACGGAAATGCAGTTTGTGAACGAGG | 5′-ACTGCTCATCCACCAGAGTAAGG | 5′-CCCCCAAGGACTCGAGATC |
| Mlxipl (ChREBP) | 5′-CAAAAAGCGGCTCCGTAAGTCCAGC | 5′-TGA TGC GCG AAT ACC ACA A | 5′-CTG CTT GGG AGC CAG GAA A |
| MTP | 5′-CAAACCAGGTGCTGGGCGTCAGT | 5′-CCGCTGTGCTTGCAGAAGA | 5′-TTTGACACTATTTTTCCTGCTATGGT |
| Nt5e (Cd73) | 5′CAACACCTTTCTCTACACAGGAAATCCACCTTC | 5′-CATCGTGGTGGGAGGACACT | 5′-GCGGTGACTATGAATGGGTACTT |
| Pnliprp2 | 5′-TAGGTTTCGGAATGAGCCAAAAGGTCG | 5′-GTGCCCCCATAATCCCCTAT | 5′-CGTTGGGAAAGAAATCCAGATG |
| Scarb1 | 5′-AGGGAGTTCAGACAAAAGGTCAACATCA | 5′-GGG AGC GTG GAC CCT ATG T | 5′-CAC GGT GTC GTT GTC ATT GAA |
| Scd1 | 5′-CACCGCGCCCACCACAAGTTCT | 5′-CGT TCC AGA ATG ACG TGT ACG A | 5′-AGG GTC GGC GTG TGT TTC |
| Control | |||
| Actb (β-actin) | 5′-CCA CAC CCG CCA CCA GTT CGC C | 5′-CTT CTT TGC AGC TCC TTC GTT G | 5′-CGA CCA GCG CAG CGA TAT C |
Analysis for Transcript Rhythmicity
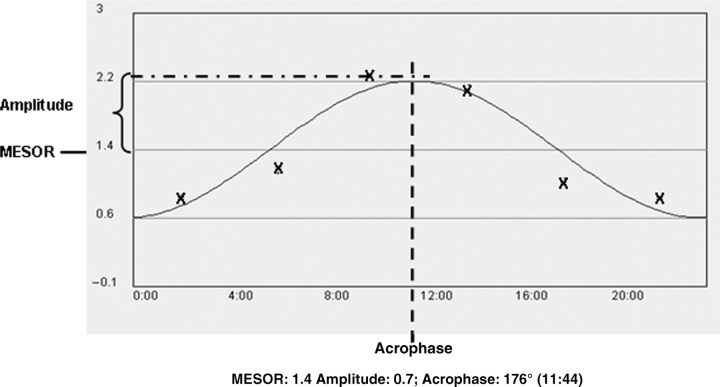
The model curve for diurnal rhythmicity of gene expression was prepared by applying the mean values of the relative mRNA levels (mean ± SD; n = 5 mice/time-point) on the Y-axis and appropriate time-points of tissue collection on the X-axis (Fig. 1). The cosinor program was used for rhythmicity analysis (21–24). In brief, a fitted cosinor curve with a 24-h rhythm from measured values was calculated. The cosinor curve is represented as y = M + Acos (ωt + ϕ), where M = mesor (midline estimating statistic of rhythm, that is the rhythm adjusted mean), A = amplitude (half of the rhythmic variability in a cycle), ω = 2Pi/24 and ϕ = acrophase (peak time of the variable). According to the cosinor analysis, three variables, namely, mesor, amplitude and acrophase characterize the circadian rhythm. Acrophase is given in degrees by this program by converting 24 h to 360°, i.e. 6:00 a.m. to 90° and 12:00 p.m. to 180°. The significance of the circadian rhythm was tested by a zero-amplitude test. Gender differences in mesors and amplitudes were analysed by Student's t-test. A 5% probability was regarded as statistically significant (24). Delay for acrophase between genders is considered to be different when more than 30 min.
Fig. 1.
A typical example of cosinor analysis. The hourly sequential data were fitted to a cosinor curve by the cosinor program. This panel also demonstrates the definition of Mesor (midline estimating statistic of rhythm), amplitude (half of the rhythmic variability in a cycle) and acrophase (peak time of the variable). The calculated values for these circadian parameters are also shown. The symbol ‘X’ indicates experimental values.
Measurement of Hepatic and Serum Lipid Contents
Hepatic and serum triglyceride, cholesterol and non-esterified fatty acids (NEFAs) contents were quantified using colorimetric assays from Wako Chemicals (Richmond, VA) (25). Briefly, 50 mg of liver tissue was homogenized in 1 ml of buffer containing 18 mM Tris (pH 7.5), 300 mM mannitol, 50 mM ethylene glycol tetraacetic acid (EGTA) and 0.1 mM phenylmethysulphonyl fluoride. The homogenate (500 μl) was mixed with chloroform:methanol (2:1, 4 ml) and incubated overnight at room temperature with gentle shaking. After 1 ml of H2O was added, the homogenates were vortexed and centrifuged for 5 min at 3,000 g. The lower lipid phase was collected and concentrated by vacuum. The lipid pellets were dissolved in a mixture of 270 μl of isopropanol and 30 μl of Triton X-100. Triglyceride and cholesterol as well as NEFA levels were quantified by colorimetric assays according to the manufacturer's instructions, using wavelengths of 505 nm for cholesterol, 550 nm for NEFA and 600 nm for triglycerides.
RESULTS
Gender Differences in the Oscillations of Circadian Clock
Using the cosinor program, patterns of gene expression were displayed as indicated in Fig 1, which illustrates a typical result of single cosinor analysis, including hourly sequential data, the fitted cosinor curve and circadian parameters (mesor, amplitude and acrophase). The zero-amplitude test determines the significance of the circadian rhythms.
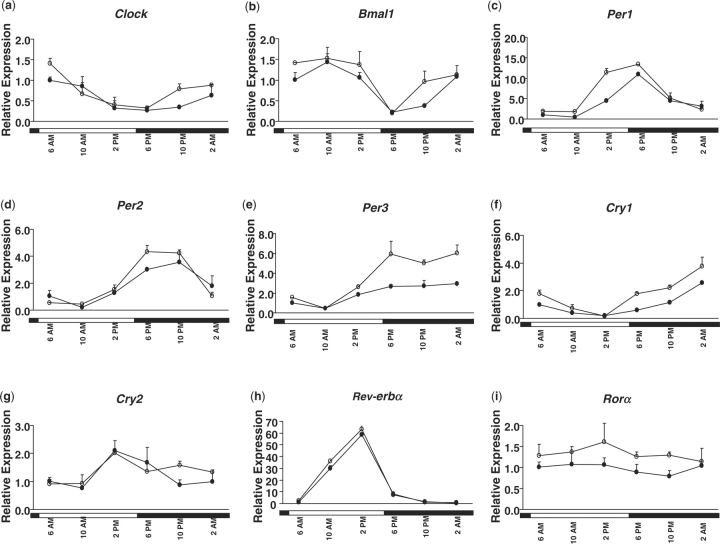
Figure 2 shows the expression profiles of clock genes including Clock, Bmal1, Per 1-3, Cry 1-2 as well as nuclear receptors Rev-erbα and RORα. These genes, except for RORα, exhibited cyclic patterns in both male and female mouse livers, which is consistent with previous reports, in which only male mice were studied (3, 26, 27). An inverse correlation of the expression patterns was observed between the expression of Pers and Bmal1. As shown in Fig 2, Bmal1 reached a nadir around dusk at 6:00 p.m., then gradually increased during the night and decreased after daybreak, while expression of Per 1-3 peaked around 6:00 p.m., then gradually decreased at night, and increased during the daytime. This trend was observed in both genders.
Fig. 2.
Daily expression profiles of the clock genes (A, Clock; B, Bmal1; C, Per1; D, Per2; E, Per3; F, Cry1; and G, Cry2) and nuclear receptors (H, Rev-erbα and I, RORα) in female (open circle) and male (closed circle) mouse livers. Relative mRNA levels of genes were quantified by real-time PCR. Results were normalized by comparison with the amount of β-actin and expressed as relative values to those of male mice at 6:00 a.m. for all genes. Data are means ± SD of five mice at each time-point. Open bar, lights on; closed bar, lights off.
The rhythmic patterns of hepatic clock genes were compared between genders. Table 4 summarizes the characteristics (mesor, amplitude and acrophase) of the circadian patterns in each gender. Most hepatic clock genes had their peak mRNA levels 30 min earlier in female than in male mice. However, the levels of Per3 and Cry2 mRNA peaked earlier in male- than in female-mouse livers, whereas Rev-erbα was coordinately regulated to the same phase in both genders. These findings suggest that multiple gender-specific factors might be involved in regulating these hepatic clock genes. The mesor value of each gene represents a 1-day average mRNA value. The mesors of Per1 and Per3 were significantly higher in female-mouse livers compared to those in male-mouse livers with a 1.46- and 1.49-fold, respectively. Meanwhile, significantly higher amplitudes were also observed for Per3 (5.29 vs. 2.50) and Cry1 (1.45 vs. 0.95) in female- than male-mouse livers. Although no significant difference was observed for mesor or amplitude values of other clock genes in the liver between the genders, the levels of mesors and amplitudes of many circadian genes in the livers tended to be lower in male than in female mice. In addition, RORα expression was also lower in male- compared to that in female-mouse livers over the 24-h period, although no rhythmicity was observed (Fig. 2). The relative higher 1-day average values and earlier peak times in female-mouse livers may result in relatively higher regulation and earlier response of the downstream pathways.
Table 4.
Characteristics [mesor, amplitude and acrophase (ϕ)] of circadian patterns of clock genes as well as nuclear receptor Rev-erbα in male- and female-mouse livers (n = 5).
| Male |
Female |
|||||
|---|---|---|---|---|---|---|
| Mesor | Amplitude | Acrophase (ϕ) | Mesor | Amplitude | Acrophase (ϕ) | |
| Bmal1 | 0.96 ± 0.16 | 0.71 ± 0.13 | 07:42 | 0.95 ± 0.07 | 0.50 ± 0.00 | 06:58a |
| Clock | 0.57 ± 0.10 | 0.38 ± 0.03 | 06:34 | 0.75 ± 0.07 | 0.40 ± 0.00 | 05:10a |
| Per1 | 4.05 ± 0.07 | 4.25 ± 0.49 | 18:44 | 5.90 ± 0.00* | 6.05 ± 0.07 | 16:54a |
| Per2 | 1.82 ± 0.09 | 1.60 ± 0.26 | 21:00 | 2.11 ± 0.01 | 2.19 ± 0.24 | 18:54a |
| Per3 | 2.95 ± 0.07 | 2.50 ± 0.42 | 17:04 | 4.40 ± 0.15* | 5.29 ± 0.13* | 20:58a |
| Cry1 | 0.90 ± 0.00 | 0.95 ± 0.07 | 01:36 | 1.65 ± 0.21 | 1.45 ± 0.07** | 00:50a |
| Cry2 | 1.35 ± 0.07 | 0.65 ± 0.07 | 16:06 | 1.40 ± 0.00 | 0.55 ± 0.07 | 17:56a |
| Rev-erbα | 16.6 ± 0.71 | 25.9 ± 1.56 | 13:04 | 18.5 ± 0.42 | 28.9 ± 0.78 | 12:48 |
Statistically significant differences between female and male mice for mesor and amplitude values are indicated by an asterisk: *P < 0.05; **P < 0.01. aIndicates the time difference between male and female mice for acrophase is longer than 30 min.
Gender Differences in the Diurnal Profiles of Nuclear Receptors
To investigate the potential role of nuclear receptors on gender differences in peripheral circadian circuits, rhythmicity of nuclear receptors expression involved in lipid homoeostasis with focus on fatty acid and cholesterol metabolism was determined (Table 5). The cosinor program revealed that three nuclear receptors (PPARα, β and γ) and two co-regulators [PPARγ coactivator 1 (PGC1α and PGC1β)] exhibited robust diurnal rhythmicity in both male- and female-mouse livers (Figs 3 and 4). Similar rhythmic expression patterns for PPARα, β and γ as well as PGC1α have been reported in previous studies in which only male mice were studied (3, 28). The expression of PXR and RXRα did not demonstrate circadian patterns in either gender using the cosinor program. Consistent with published data, the COSOPT program also revealed a non-rhythmic pattern for PXR in male-mouse livers (3). However, the hepatic expression pattern of RXRα in male-mouse livers was indicated to be cyclic in the previous study (3). Both genders expressed similar levels of hepatic LXRα mRNA, but only cycled in female-mouse livers as analysed by the cosinor program (Fig. 4 and Table 5), which again indicated a gender-difference in hepatic transcriptional rhythmicity.
Table 5.
Characteristics [mesor, amplitude and acrophase (ϕ)] of circadian patterns of nuclear receptors in livers of male and female mice (n = 5).
| Male |
Female |
|||||
|---|---|---|---|---|---|---|
| Mesor | Amplitude | Acrophase (ϕ) | Mesor | Amplitude | Acrophase (ϕ) | |
| Pparα | 1.51 ± 0.06 | 0.82 ± 0.02 | 15:08 | 2.11 ± 0.02* | 1.04 ± 0.01** | 14:16a |
| Pparβ | 0.50 ± 0.14 | 0.35 ± 0.07 | 05:54 | 0.70 ± 0.00 | 0.60 ± 0.14 | 01:08a |
| Pparγ | 1.13 ± 0.25 | 0.43 ± 0.13 | 12:24 | 0.80 ± 0.14 | 0.55 ± 0.07 | 11:56 |
| Lxrα | NC | NC | NC | 1.16 ± 0.01 | 0.22 ± 0.05 | 20:46 |
| Pgc1α | 0.80 ± 0.14 | 0.25 ± 0.07 | 05:02 | 1.72 ± 0.03* | 0.38 ± 0.11 | 04:54 |
| Pgc1β | 1.97 ± 0.18 | 1.64 ± 0.37 | 14:52 | 3.25 ± 0.07* | 2.50 ± 0.28 | 12:30a |
Statistically significant difference between male and female mice for mesor and amplitude values is indicated by an asterisk: *P < 0.05; **P < 0.01. aIndicates the time difference for acrophase is longer than 30 min. NC, non-cyclic.
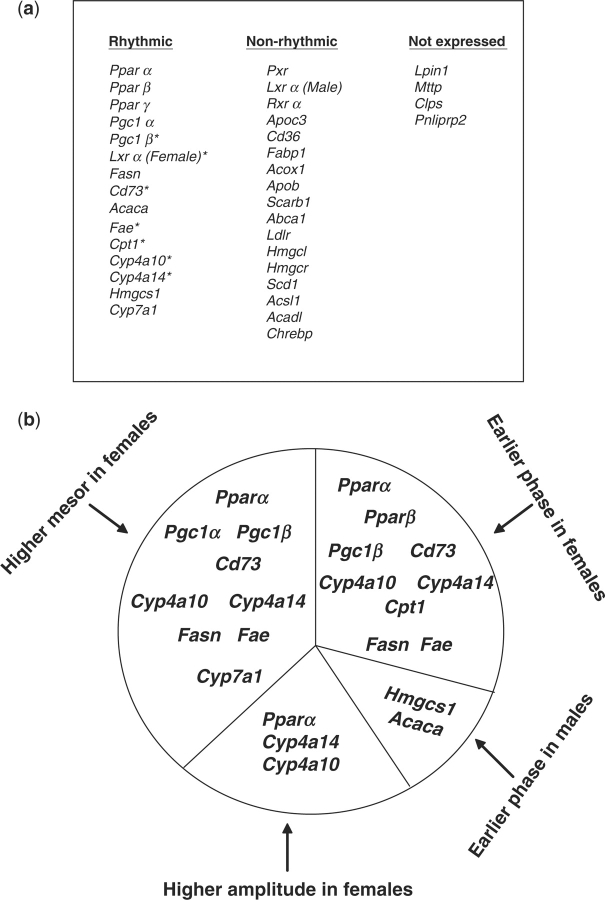
Fig. 3.
Temporal expression profiles of genes encoding nuclear receptors and directing lipid homoeostasis in female- and male-mouse livers. (A) Distribution of genes is categorized according to their expression and rhythmicity in liver tissue. The symbol * indicates genes with rhythmicity identified first in this study. (B) Distribution of genes with rhythmicity is categorized according to their circadian characteristics.
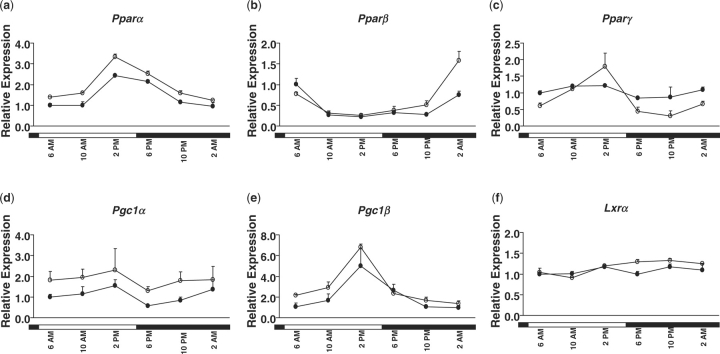
Fig. 4.
Daily expression profiles of genes encoding nuclear receptors and co-factors (A, PPARα; B, PPARβ; C, PPARγ; D, PGC1α; E, PGC1β; and F, LXRα) in female- (open circle) and male- (closed circle)mouse livers. Relative mRNA levels of genes were quantified by real-time PCR. Results were normalized by comparison to the amount of β-actin and expressed as relative values to those of male mice at 6:00 a.m. for all genes. Data are means ± SD of five mice at each time-point. Open bar, lights on; closed bar, lights off.
The peak times for expression of PPARs in male-mouse livers (Table 5) in this study are similar to those reported previously (3). The cycling of each nuclear receptor might be regulated by different Zeitgebers, as indicated by decoupling of peak times, which may result in distinct temporal control of metabolic responses. Generally, the mRNA levels of those nuclear receptors reached their peaks earlier in livers of female than male mice. As indicated in Table 5, the acrophase (time to reach peak level) of PPARα, PPARβ and PGC1β was earlier in female- than in male-mouse livers by at least 30 min.
Similarly, gender differences were also observed for the mesor and amplitude values for the PPARs. The mesor values were consistently higher in livers of female than in male mice for PPARα (2.11 vs. 1.51), PGC1α (1.72 vs. 0.80) and PGC1β (3.25 vs. 1.97). PPARα also had a higher amplitude value in female than in male mouse livers (1.04 vs. 0.82). Such differences might lead to a higher transcriptional activation and greater fluctuation of PPAR target genes in female- than in male-mouse livers.
Gender Differences in the Diurnal Expression of Genes Involved in Lipid Homoeostasis
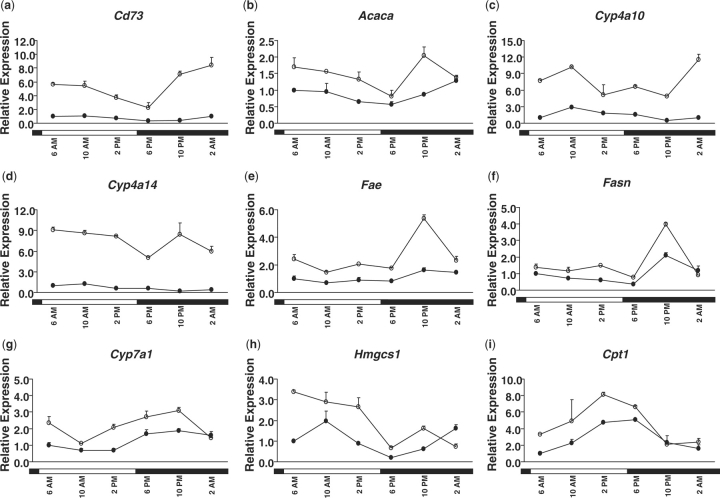
Analyses of the transcripts of genes involved in lipid homoeostasis over a light–dark cycle by the cosinor program revealed that many of these genes exhibited circadian patterns of expression (Figs 3 and 5). Most of the genes encoding lipid metabolism enzymes shared uniform circadian patterns in both female- and male-mouse livers. Genes encoding 5′ nucleotidase, ecto (Nt5e, Cd73); acetyl-coenzyme A carboxylase α (Acaca); cytochrome P450, family 4, subfamily a, polypeptide 10 (Cyp4a10); Cyp4a14; ELOVL family member 6, elongation of long-chain fatty acids (fatty acid elongase, Fae); fatty acid synthase (Fasn); Cyp7a1; 3-hydroxy-3-methylglutaryl co-enzyme A synthase 1 (Hmgcs1); and carnitine palmitoyltransferase 1a, liver (Cpt1) were under circadian control in the liver as analysed by the cosinor program. Rhythmic expressions of Acaca, Fasn, Hmgcs1 and Cyp7a1 have been noted in previous studies (29–32). However, to our knowledge, the rhythmic expression patterns of CD73, Cyp4a10, Cyp4a14, Fae and Cpt1 are novel findings. The peak expressions of these genes were distributed throughout the circadian cycle (Table 6), with the largest clusters of circadianly regulated transcripts occurring in the morning. Notably, a unique rhythmic pattern was observed for Fae and Fasn, in which their transcripts remained at low levels through the daytime yet rapidly increased at night (Fig. 5). The expression levels spiked around 10 p.m. and were followed by a precipitous decline thereafter. Such an unusual rhythmic pattern might be referred to as a single transient pulse (STP) (3). Light might serve as an indirect environmental cue that inhibits the expression of Fae and Fasn in the liver. The hepatic mRNA levels of some genes were extremely low or undetectable in the liver: lipin 1 (Lpin1), microsomal triglyceride transfer protein (Mttp), colipase, pancreatic (Clps) and pancreatic lipase-related protein 2 (Pnliprp2).
Fig. 5.
Daily expression profiles of genes involved in lipid homoeostasis (A, Cd73; B, Acaca; C, Cyp4a10; D, Cyp4a14; E, FAE; F, Fasn; G, Cyp7a1; H, Hmgcs1; and I, Cpt1) in female- (open circle) and male- (closed circle)mouse livers. Relative mRNA levels of genes were quantified by real-time PCR. Results were normalized by comparison with amount of β-actin and expressed as relative values to those of male mice at 6:00 a.m. for all genes. Data are means ± SD of five mice at each time-point. Open bar, lights on; closed bar, lights off.
Table 6.
Characteristics [mesor, amplitude and acrophase (ϕ)] of circadian patterns of genes involved in lipid homoeostasis in livers of male and female mice (n = 5).
| Male |
Female |
|||||
|---|---|---|---|---|---|---|
| Mesor | Amplitude | Acrophase (ϕ) | Mesor | Amplitude | Acrophase (ϕ) | |
| Cd73 | 0.78 ± 0.01 | 0.39 ± 0.08 | 6:46 | 5.90 ± 0.57* | 1.95 ± 0.49 | 1:06a |
| Acaca | 0.87 ± 0.04 | 0.30 ± 0.13 | 4:00 | 1.20 ± 0.14 | 0.40 ± 0.00 | 7:26a |
| Cyp4a10 | 1.40 ± 0.00 | 0.90 ± 0.00 | 11:42 | 7.65 ± 0.35* | 2.30 ± 0.14* | 5:28a |
| Cyp4a14 | 0.73 ± 0.04 | 0.50 ± 0.13 | 10:28 | 6.86 ± 0.37* | 2.48 ± 0.03* | 9:10a |
| Fae | 1.05 ± 0.07 | 0.40 ± 0.00 | 0:02 | 2.65 ± 0.07** | 1.50 ± 0.28 | 23:28a |
| Fasn | 0.95 ± 0.07 | 0.50 ± 0.14 | 0:32 | 2.36 ± 0.04* | 1.61 ± 0.23 | 23:48a |
| Cyp7a1 | 1.27 ± 0.04 | 0.67 ± 0.12 | 22:20 | 2.30 ± 0.14* | 0.85 ± 0.07 | 22:40 |
| Hmgcs1 | 1.25 ± 0.21 | 1.00 ± 0.28 | 6:36 | 1.71 ± 0.01 | 1.75 ± 0.21 | 9:32a |
| Cpt1 | 2.80 ± 0.28 | 2.05 ± 0.21 | 16:18 | 5.90 ± 0.57* | 1.95 ± 0.49 | 1:06a |
Statistically significant differences between male and female mice for mesor and amplitude values is indicated by an asterisk: *P < 0.05; **P < 0.01. aIndicates the time difference for acrophase is longer than 30 min.
Comparison of the temporal patterns for gene expression between livers of male and female mice revealed gender differences. As shown in Fig. 5 and Table 6, average mRNA levels of Cd73, Cyp4a10, Cyp4a14, Fae, Fasn as well as Cyp7a1 were significantly higher in female- than in male-mouse livers. In addition, the peak times of Cd73, Cyp4a10, Cyp4a14, Fae, Fasn and Cpt1 mRNA levels in male-mouse livers were at least half an hour behind female-mouse livers (Table 6). However, the hepatic mRNA levels of Acaca and Hmgcs1 reached their peaks much earlier in male than in female mice.
Quantification of Hepatic and Serum Lipid Contents
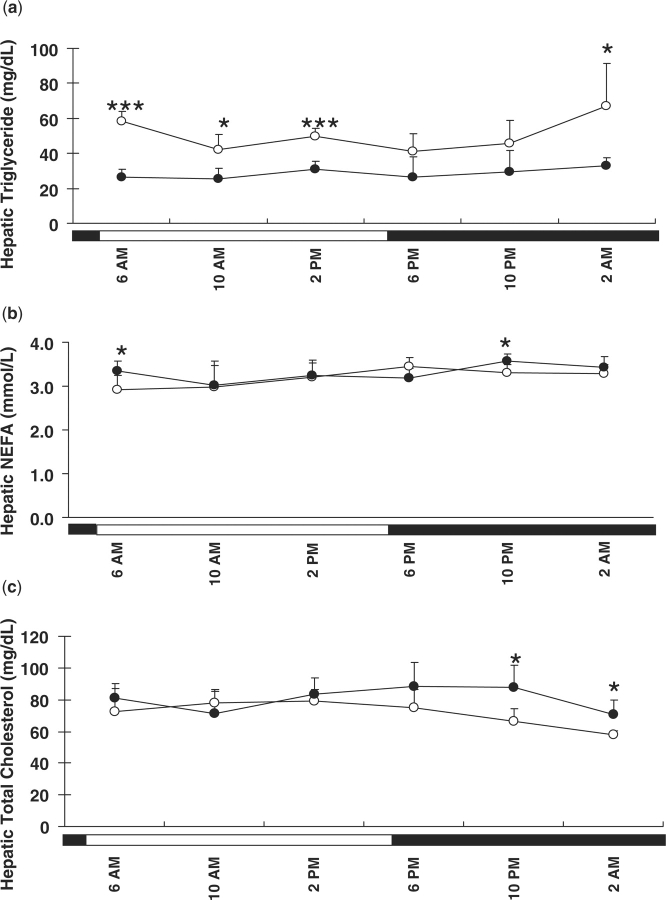
To relate circadian-regulated transcripts with hepatic lipid homoeostasis, hepatic lipid content, including triglyceride, cholesterol and NEFA concentrations were monitored over the 24-h cycle. Although no circadian rhythm was found for hepatic lipid levels (including triglyceride, NEFA and cholesterol) as analysed by the cosinor program, changes were observed throughout the light–dark cycle in both genders.
Gender differences were observed in hepatic triglyceride levels. As shown in Fig. 6A, hepatic triglyceride concentrations in female-mouse livers peaked around 2:00 a.m. From 6:00 to 10:00 a.m., hepatic triglycerides rapidly decreased and were maintained at relatively low levels during the daytime, with a small increase around 2:00 p.m. Hepatic triglyceride levels were significantly higher in female mice at 2:00 a.m. compared to those at 10:00 a.m., 6:00 p.m. and 10:00 p.m. Though a similar pattern was observed for hepatic triglycerides in male mice, lower levels were observed with minor fluctuations over the 24 h. Of note, female mice had higher hepatic triglycerides than male mice. Significant gender differences of hepatic triglycerides were observed at 6:00 a.m., 10:00 a.m., 2:00 p.m. and 2:00 a.m. with a fold change of 2.20, 1.65, 1.59 and 2.05, respectively. Thus, female mice have higher hepatic triglyceride levels than male mice, accompanied with wider fluctuations over the 24-h cycle.
Fig. 6.
Hepatic lipid concentrations in female (open circle) and male (closed circle) mice. Hepatic triglyceride (A), non-esterified fatty acids (NEFAs) (B) and cholesterol (C) were quantified using the livers harvested from each time-point. Data are means ± SD of five mice at each time-point. Open bar, lights on; closed bar, lights off. Gender difference in hepatic lipid contents at different time-points was analysed by Student's t-test. *P < 0.05; ***P < 0.001.
Hepatic NEFA concentrations were also monitored in female and male mice. In female mice, hepatic NEFAs (Fig. 6B) gradually increased during the daytime, peaked around 6:00 p.m., and decreased during the night. However, such a pattern was not apparent in male mice. A small gender difference was observed for hepatic NEFA levels at 6:00 a.m. and 10:00 p.m., with relatively higher levels in male than in female mice. Daily fluctuations and gender differences in hepatic NEFA levels were not as apparent as those of hepatic triglycerides.
Differences were also observed in daily changes in hepatic cholesterol levels between male and female mice. As shown in Fig. 6C, cholesterol in the livers of female mice decreased at night and reached its nadir around 2:00 a.m. Hepatic cholesterol at 2:00 a.m. was significantly lower than those at 10:00 a.m., 2:00 p.m. and 6:00 p.m. In addition, a significant difference was also observed between 2:00 p.m. and 10:00 p.m.; however, in male mice, diurnal fluctuation of hepatic cholesterol was minor. Additionally, hepatic cholesterol concentrations were significantly higher in male than in female mice at 10:00 p.m. and 2:00 a.m. Female mice have relatively lower hepatic cholesterol levels and greater diurnal fluctuations within a day than male mice.
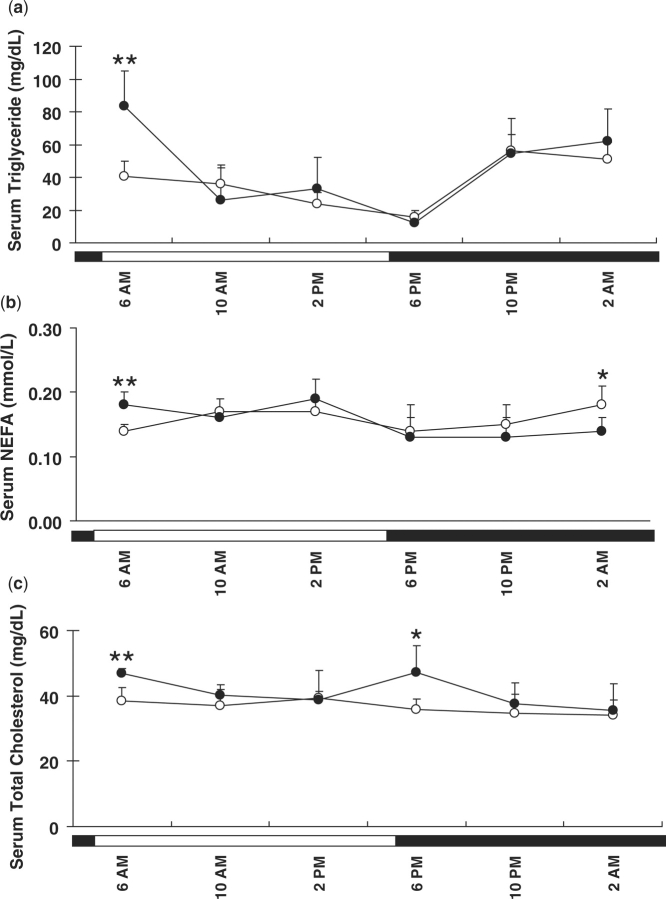
In addition, serum lipid levels were determined. Serum triglyceride levels gradually decreased during daytime and reached nadir around 6:00 p.m., and then increased again at night in both female and male mice (Fig. 7). Significantly higher serum triglyceride levels were observed in male than in female mice at 6:00 a.m. Daily fluctuations in serum NEFA levels were not as apparent as those of serum triglycerides although an increase of serum NEFA was observed in female mice at 2:00 a.m. compared to other time-points. Significant gender differences were observed in serum NEFA levels at 6:00 a.m. and 2:00 a.m. Serum cholesterol levels in male mice decreased from 6:00 a.m. to 2:00 p.m., then increased and reached peak levels at 6:00 p.m. and then decreased again. Such fluctuation was not observed in female mice. In addition, significantly higher cholesterol levels were observed in male mice than in female mice at 6:00 a.m. and 6:00 p.m. As such, serum triglyceride levels increased following feeding at night and decreased during daytime in both genders. Although not as apparent, a similar pattern was found for hepatic triglyceride levels, suggesting the delivery of triglycerides to the liver after absorption. However, unlike triglycerides, serum cholesterol levels did not increase after feeding standard rodent chow, as did hepatic cholesterol levels in our study.
Fig. 7.
Serum lipid concentrations in female (open circle) and male (closed circle) mice. Serum triglyceride (A), non-esterified fatty acids (NEFAs) (B) and cholesterol (C) were quantified using the serum harvested from each time-point. Data are means ± SD of five mice at each time-point. Open bar, lights on; closed bar, lights off. Gender difference in serum lipid contents at different time-points was analysed by Student's t-test. *P < 0.05; **P < 0.01.
DISCUSSION
The current study compiles a dynamic atlas of circadian clocks and nuclear receptors as well as downstream signalling pathways involved in lipid homoeostasis over a light–dark cycle in both male- and female-mouse livers. Gender differences that are regulated by the circadian clock are identified in both lipid levels and lipid metabolic pathway. The results demonstrate the importance of nuclear receptors in coupling peripheral circadian clocks to lipid metabolic outputs. Additional novel findings are the hepatic rhythmic expression patterns of PGC1β, Cd73, Cyp4a10, Cyp4a14, Fae and Cpt1 in both genders, as well as Lxrα in female mice.
The genes examined exhibit similar rhythmicity in both genders, except that Lxrα only cycled in female-mouse livers, which indicates some gender-specific hepatic transcriptional rhythmicity (Fig. 3). As shown in this study, circadian-regulated nuclear receptors are especially noted within the PPAR subfamily, including PPARα, PPARβ/δ and PPARγ. The three PPAR family members are involved in lipid metabolism and energy homoeostasis in adipose tissue, liver and muscle (10). Specifically, PPARα plays a key role in fatty-acid oxidation and ketogenesis in the liver by regulation of many genes involved in lipid homoeostasis (33). Consistently, a subset of fatty-acid-oxidation genes (including Cpt1, Cyp4a10 and Cyp4a14), as well as Hmgcs1, which are involved in ketosis, all exhibited circadian-expression patterns in the present study (Fig. 5 and Table 6), among which Cpt1 fluctuated in phase with Pparα transcripts. Pparα is a positive regulator of these genes upon activation by fatty acids (34). It has also been shown that the daily variation in lipogenic and cholesterogenic gene expression is attenuated or abolished in Pparα-null mice (30). Therefore, Pparα serves not only as a cellular ‘lipostat’, transducing changes in cellular lipid levels to the transcriptional regulation of target genes involved in fatty-acid utilization, but also as a regulator of temporal lipid homoeostasis as the oscillation of Pparα may directly determine the rhythmicity of certain target genes.
Consistent in both genders, Pparα is induced in the daytime with a peak phase of expression around 2:00 p.m. in the livers, which is 9 h after lights come on (5:00 a.m.) (Fig. 4). This is similar to what has been reported previously, in which the acrophase of Pparα was at 08:00, and the lights were on from 00:00 to 12:00 (30, 35). This cycling pattern could be explained by the role of Pparα in meeting energy demands through the regulation of cellular fatty-acid uptake and utilization. Pparα may serve as a metabolic stress-response factor by increasing the expression of enzymes involved in several fatty-acid utilization pathways in response to normal physiological conditions (36). During the daytime, mice are generally in a fasting state that requires ketone bodies. The induction of Pparα is responsible for energy production and ketosis. During the night, when mice utilize hexose as a primary fuel source, the Pparα pathway is decreased. The circadian clock in nocturnal rodents integrates feeding and phases it to the night (26). Recent studies have indicated that circadian-regulated genes exhibit differences in their expression under restricted and ad libitum feeding regimens, suggesting that feeding is a component of rhythmicity in normal conditions (26, 37).
The co-activators of Pparα, Pgc1α and Pgc1β were also rhythmically expressed in phase with Pparα in both genders. Transcripts of Pgc1α and Pgc1β also accumulated in the morning and reached their peaks around 2:00 p.m. The synchronization of the cycling of PPARs and their co-factors is essential for the circadian expression of certain targeted genes. Although Pgc1α is essential for the regulation of cold-induced thermogenesis (3, 38), the induction of Pgc1α in the daytime cannot be explained by cold temperature, as the mice were maintained in a thermally stable environment. On the contrary, it can be explained by regulation of the intrinsic circadian clock.
Although male and female mice share similar rhythmicity of the genes studied, certain gender variations are demonstrated in this study (Fig. 3B and Tables 4, 5 and 6). First, gender differences were observed in the acrophases (time to reach peak level) of the genes studied. Most nuclear receptors (including Pparα, Pparβ and Pgc1β) had their peak mRNA levels at least 30 min earlier in female than in male mice, which correlated well with those of clock genes (except Per3, Cry2 and Rev-erbα). Consistently, the peak times of Pparα-target genes, including Cyp4a10, Cyp4a14 and Cpt1 mRNA, in male-mouse livers were at least half an hour behind those in female-mouse livers. Second, gender differences were noted in the mesors (median levels) for cycling genes in liver. The nuclear receptor cycling genes (Pparα, Pgc1α and Pgc1β) were more highly expressed in livers of female than male mice, which is consistent with the circadian clocks (Per1 and Per3). Such trends were also observed for expression of genes involved in lipid homoeostasis, such as Cyp4a10 and Cyp4a14, which are Pparα target genes. A similar trend was noted for the expression of Cpt1, although not statistically different. Third, female mice exhibited higher amplitudes not only of the circadian clock mRNAs (Per3 and Cry1), but also Pparα and its target genes (Cyp4a10 and Cyp4a14). As a major component of the negative-feedback loop of the circadian clock, it has been shown that the Bmal1-Clock heterodimer governs the circadian transcription of Pparα, by binding to the E-box-rich region located in the second intron (35). Thus, Pparα may play a key role in coupling the peripheral circadian clock to divergent metabolic outputs. However, it has been shown that Bmal1-Clock can transactivate PPAR-target genes, via the PPAR response elements (PPRE) (39). Thus, the direct circadian regulation of these genes by Bmal1-Clock through transcriptional activation via E-box or PPRE is possible.
The temporal fluctuations in nuclear receptors and genes involved in lipid metabolism were paralleled by changes in hepatic lipid levels over the daily light–dark cycle. Hepatic triglyceride levels in female mice basically followed expression of Pparα, but in the opposite direction. With the increased expression of Pparα transcripts in the daytime, more fat may undergo oxidation, thus resulting in decreased hepatic triglyceride levels. At night, the levels of triglycerides increase gradually, with a decrease in Pparα levels. The higher hepatic triglyceride levels in female mice at 2:00 a.m. compared to other times, might be attributed to the lower expression levels of enzymes involved in fat oxidation, including Cpt1 and Cyp4a14, during the night. As the expression levels of these enzymes were relatively stable in male mice, no differences in hepatic triglyceride levels were observed at the various time-points. The higher mesors and amplitudes noted for Pparα and its downstream signalling pathways in female mice may result in greater hepatic capacity for fatty-acid oxidation, thus requiring lower catabolism of fats for energy production. This may finally result in relatively higher triglyceride content in livers of female than male mice, although other pathways including fat absorption, synthesis and transport may also be of importance.
NEFA concentrations in the liver correlate with diurnal changes of Cd73, Fasn and Fae mRNA levels. Cd73 is regarded as the key enzyme in the formation of adenosine from 5′-AMP, which is an inducer of procolipase expression (40, 41). The increased mRNA levels of Cd73 may downregulate 5′-AMP levels, thus subsequently inhibiting pancreatic lipase/procolipase expression, and negatively regulating fat absorption by the small intestine. In livers of female mice, Cd73 mRNA levels reached their nadir at 6:00 p.m., and peaked around 2:00 a.m. (Fig. 5). This might lead to the higher NEFA concentration observed in livers of female mice around dusk (Fig. 6). Such an association was not apparent in livers of male mice. Fatty acids serve as Pparα activating ligands (42). Thus, daily changes in hepatic intra-cellular fatty acids may inversely contribute to the circadian expression of Pparα target genes, although no significant connection between hepatic fatty acids levels with gene expression was observed in this study. Gender differences in the average levels of Cd73 were noted, as indicated by higher mRNA levels of Cd73 in female- than male-mouse livers (5.90 vs. 0.78). This might result in higher fat absorption in male mice, which correlates well with the gender difference observed in hepatic NEFA levels at 6:00 a.m. and 10:00 p.m. (Fig. 6).
With the increased expression levels of Fasn and Fae at night, the time when most feeding occurs in nocturnal rodents, hepatic NEFA content tended to be higher in both genders than during the daytime (Fig. 6). The upregulation of Fasn and Fae at night may facilitate fat synthesis during feeding and the use of fat as a primary energy source for several hours in the post-absorptive period. Additionally, an earlier peak time of Fasn and Fae was also noted in female-mouse livers, which is consistent with the gender difference in phases of the circadian clock. Relatively higher mRNA levels of Fasn and Fae in livers of female mice might lead to higher hepatic synthesis of fatty acids, and subsequently to the increased synthesis of triglycerides, as shown in Fig. 6.
Cellular cholesterol homoeostasis is regulated by the coordinated action of biosynthetic and degradative enzymes, as well as transporters. De novo cholesterol biosynthesis starts with the conversion of acetoacetyl CoA to 3-hydroxy-3-methylglutaryl-CoA (HMGCoA), by the enzyme Hmgcs1. Subsequently, HMGCoA is converted to mevalonate by Hmgcr, which is the rate-limiting enzyme involved in cholesterol synthesis; as a result, cholesterol is converted to bile acids by Cyp7a1. In the present study, rhythmic expression was observed for Hmgcs1 and Cyp7a1 in both female- and male-mouse livers, which is consistent with previous findings (26, 32). Although Hmgcr has been shown to be clock regulated in mammals (43), it exhibited non-rhythmic expression in the present study by cosinor analysis. Hepatic cholesterol levels gradually decreased in female-mouse livers between 10:00a.m. and 2:00 a.m., which might be partially explained by the decreased expression of Hmgcs1 from 10:00 a.m. and thereafter. Increased conversion of cholesterol to bile acids by the increased expression levels of Cyp7a1 during the dark period may also contribute to the decreased hepatic cholesterol levels. Such trends in cholesterol levels were not apparent in male mice. Additionally, Hmgcs1 is the key enzyme involved in the synthesis of ketone bodies. The higher expression of Hmgcs1 will lead to higher synthesis of ketone bodies during the daytime. This is consistent with the fact that during daytime, mice are generally in a fasting state that requires ketone bodies. Coordinately, PPARα was also induced during the daytime, which is responsible for energy production and ketosis. A gender difference was observed for the mesor of Cyp7a1, with a higher mesor value in female than in male mice. Consistent with this data, relatively lower hepatic content of cholesterol was observed in female than in male mice, perhaps due to increased conversion of cholesterol by Cyp7a1 to bile acids. Although Cyp7a1 showed a robust rhythmic pattern in both female- and male-mouse livers, Lxr, the direct regulator of Cyp7a1, exhibited a cyclic pattern only in female mice. Of note, Cyp7a1 cycled in phase with Per2, which was reported previously (26). The peak time of Per2 also preceded that of Cyp7a1 in both male and female mice, suggesting that Per2 might directly regulate the cycling of Cyp7a1.
The coordinated cycling of several enzymes in a single pathway, as observed in the present study, may ensure a tighter circadian regulation of lipid homoeostasis. The consistency in the circadian expression of cycling clocks, nuclear receptors and genes involved in lipid homoeostasis, as well as hepatic lipid levels, suggest that nuclear receptors (including PPAR subfamilies) serve as peripheral signals that regulate circadian patterns of genes involved in lipid homoeostasis by transforming timing information from the clock network into metabolic responses. Significant gender disparity in the circadian expression patterns of these genes is noted in this study. The average expression levels for the circadian clocks, nuclear receptors and genes involved in lipid homoeostasis in livers of female mice were consistently higher and had earlier peak times of expression than in male mice.
One limitation of the present study is that we primarily provided the evidence linking circadian clocks and lipid metabolism as well as gender disparity involved. The mechanism by which the circadian clock is integrated into metabolic system in a gender-specific manner remains to be further explored. Post-menopausal or ovariectomized mice should be used to explore the mechanistic cause and physiological implication of the gender differences in diurnal lipid homoeostasis. In addition, in order to gain a better insight into the regulation and gender disparity in the circadian patterns of lipid homoeostasis, the rhythmicity of expression at the protein level and the sub-cellular localization as well as activity requires more investigation. The cosinor program used in this study does not provide statistical analysis to determine acrophase difference. An arbitrary time-point of 30 min was used to describe the acrophase difference assuming this time difference will result in a difference in protein level as well as biological function.
There are several ways in which the results of this study may find practical application if the findings are confirmed in human. First, gender difference in diurnal lipid levels should be taken into consideration when establishing normal lipid ranges in male and female. When enzyme activity needs to be assessed in clinical setting, the timing of sampling might affect the outcome in a gender specific manner. Second, previous studies have indicated that circadian-rhythm disorders might be associated with diabetes, obesity and cardiovascular disease (44). As nuclear receptors, as well as genes involved in lipid homoeostasis are targets of prescribed drugs, understanding their circadian patterns may provide insights into the development of chronotherapy, and lead to enhanced drug efficacy through delivery timed to match peak levels. For example, as noted in this study, Pparα reached its peak level around 2:00 p.m. in both male and female mice. Thus, administration of a PPARα agonist at 2:00 p.m. may be an effective strategy for the treatment of diseases associated with enzymatic defects in the fatty-acid β-oxidation pathway, such as hypoglycaemia, liver dysfunction and cardiomyopathy. Finally, it has been indicated that tissue-specific circadian regulation of transcription is a distinct feature of the circadian clock in higher organisms (26). Although the core clock resident in the SCN coordinates slave clocks resident in different organs, the peripheral clocks will regulate the specific function relevant to their tissue or organ function separately, as shown in the present study. Thus, to fully understand the circadian organization of lipid homoeostasis in the entire organism, systematic analysis of temporal gene expression involved in lipid homoeostasis in multiple organs is of interest.
CONCLUSIONS
In summary, the present study identified that several genes encoding nuclear receptors and enzymes involved in the lipid metabolic pathway were rhythmically expressed in liver in phase with the peripheral clocks, which were correlated with the diurnal changes of hepatic lipid levels. Furthermore, gender disparity was observed for circadian characteristics, including mesor and amplitude values, accompanied with advances in acrophases in female-mouse livers. Accordingly, gender differences were also observed in diurnal lipid homoeostasis. The identification of cycling patterns for lipid metabolic pathways in both female- and male-mouse livers may shed light on the development of gender-based treatment for human diseases related to the coordination of the cellular clock and control of lipid homoeostasis.
FUNDING
National Institutes of Health (CA53596, AA14147); COBRE (P20 RR021940).
CONFLICT OF INTEREST
None declared.
ACKNOWLEDGEMENTS
The authors would like to acknowledge all the departmental members who joined this project for assistance with tissue collection. We would like to thank Barbara Brede for helpful comments on the manuscript.
Glossary
Abbreviations:
- Bmal1
brain and muscle Arnt-like protein 1
- Clock
circadian locomotor output cycles kaput
- Cry
cryptochrome
- LXR
liver X receptor
- Mesor
midline estimating statistic of rhythm
- NEFA
non-esterified fatty acids
- Per
period
- PGC1
PPARγ coactivator 1
- PPAR
peroxisome proliferator-activated receptor
- ROR
retinoid-related orphan receptor
- RORE
ROR response element
REFERENCES
- 1.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 2.Schibler U, Sassone-Corsi P. A web of circadian pacemakers. Cell. 2002;111:919–922. doi: 10.1016/s0092-8674(02)01225-4. [DOI] [PubMed] [Google Scholar]
- 3.Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, Evans RM. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 4.King DP, Takahashi JS. Molecular genetics of circadian rhythms in mammals. Annu. Rev. Neurosci. 2000;23:713–742. doi: 10.1146/annurev.neuro.23.1.713. [DOI] [PubMed] [Google Scholar]
- 5.Young MW, Kay SA. Time zones: a comparative genetics of circadian clocks. Nat. Rev. Genet. 2001;2:702–715. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- 6.Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 7.Reppert SM, Weaver DR. Molecular analysis of mammalian circadian rhythms. Annu. Rev. Physiol. 2001;63:647–676. doi: 10.1146/annurev.physiol.63.1.647. [DOI] [PubMed] [Google Scholar]
- 8.Preitner N, Brown S, Ripperger J, Le-Minh N, Damiola F, Schibler U. Orphan nuclear receptors, molecular clockwork, and the entrainment of peripheral oscillators. Novartis Found. Symp. 2003;253:89–99. discussion 99–109. [PubMed] [Google Scholar]
- 9.Dumas B, Harding HP, Choi HS, Lehmann KA, Chung M, Lazar MA, Moore DD. A new orphan member of the nuclear hormone receptor superfamily closely related to Rev-Erb. Mol.r Endocrinol. 1994;8:996–1005. doi: 10.1210/mend.8.8.7997240. [DOI] [PubMed] [Google Scholar]
- 10.Li AC, Glass CK. PPAR- and LXR-dependent pathways controlling lipid metabolism and the development of atherosclerosis. J. Lipid Res. 2004;45:2161–2173. doi: 10.1194/jlr.R400010-JLR200. [DOI] [PubMed] [Google Scholar]
- 11.Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ. Nuclear receptors and lipid physiology: opening the X-files. Science. 2001;294:1866–1870. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- 12.Moreau A, Vilarem MJ, Maurel P, Pascussi JM. Xenoreceptors CAR and PXR activation and consequences on lipid metabolism, glucose homeostasis, and inflammatory response. Mol. Pharm. 2008;5:35–41. doi: 10.1021/mp700103m. [DOI] [PubMed] [Google Scholar]
- 13.Tien ES, Negishi M. Nuclear receptors CAR and PXR in the regulation of hepatic metabolism. Xenobiotica. 2006;36:1152–1163. doi: 10.1080/00498250600861827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wijnen H, Young MW. Interplay of circadian clocks and metabolic rhythms. Annu. Rev. Genet. 2006;40:409–448. doi: 10.1146/annurev.genet.40.110405.090603. [DOI] [PubMed] [Google Scholar]
- 15.Zhang J, Kaasik K, Blackburn MR, Lee CC. Constant darkness is a circadian metabolic signal in mammals. Nature. 2006;439:340–343. doi: 10.1038/nature04368. [DOI] [PubMed] [Google Scholar]
- 16.Teboul M, Guillaumond F, Grechez-Cassiau A, Delaunay F. The nuclear hormone receptors family round the clock. Mol. Endocrinol. 2008 doi: 10.1210/me.2007-0521. (doi:10.1210/me.2007-0521) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell. 2006;126:789–799. doi: 10.1016/j.cell.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Umetani M, Domoto H, Gormley AK, Yuhanna IS, Cummins CL, Javitt NB, Korach KS, Shaul PW, Mangelsdorf DJ. 27-Hydroxycholesterol is an endogenous SERM that inhibits the cardiovascular effects of estrogen. Nat. Med. 2007;13:1185–1192. doi: 10.1038/nm1641. [DOI] [PubMed] [Google Scholar]
- 19.Uppal H, Saini SP, Moschetta A, Mu Y, Zhou J, Gong H, Zhai Y, Ren S, Michalopoulos GK, Mangelsdorf DJ, Xie W. Activation of LXRs prevents bile acid toxicity and cholestasis in female mice. Hepatology. 2007;45:422–432. doi: 10.1002/hep.21494. [DOI] [PubMed] [Google Scholar]
- 20.Dudley CA, Erbel-Sieler C, Estill SJ, Reick M, Franken P, Pitts S, McKnight SL. Altered patterns of sleep and behavioral adaptability in NPAS2-deficient mice. Science. 2003;301:379–383. doi: 10.1126/science.1082795. [DOI] [PubMed] [Google Scholar]
- 21.Kolanjiappan K, Manoharan S. Diurnal rhythmicity of thiobarbituric acid reactive substances and antioxidants in experimental mammary carcinogenesis. Exp. Oncol. 2005;27:298–302. [PubMed] [Google Scholar]
- 22.Nelson W, Tong YL, Lee JK, Halberg F. Methods for cosinor-rhythmometry. Chronobiologia. 1979;6:305–323. [PubMed] [Google Scholar]
- 23.Tanaka T, Natsume T, Shibata H, Nozawa K, Kojima S, Tsuchiya M, Ashida T, Ikeda M. Circadian rhythm of blood pressure in primary aldosteronism and renovascular hypertension–analysis by the cosinor method. Jpn. Circ. J. 1983;47:788–794. doi: 10.1253/jcj.47.788. [DOI] [PubMed] [Google Scholar]
- 24.Tabata M, Takeshima T, Burioka N, Nomura T, Ishizaki K, Mori N, Kowa H, Nakashima K. Cosinor analysis of heart rate variability in ambulatory migraineurs. Headache. 2000;40:457–463. doi: 10.1046/j.1526-4610.2000.00068.x. [DOI] [PubMed] [Google Scholar]
- 25.Zhou J, Zhai Y, Mu Y, Gong H, Uppal H, Toma D, Ren S, Evans RM, Xie W. A novel pregnane X receptor-mediated and sterol regulatory element-binding protein-independent lipogenic pathway. J. Biol. Chem. 2006;281:15013–15020. doi: 10.1074/jbc.M511116200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 27.Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 28.Liu C, Li S, Liu T, Borjigin J, Lin JD. Transcriptional coactivator PGC-1alpha integrates the mammalian clock and energy metabolism. Nature. 2007;447:477–481. doi: 10.1038/nature05767. [DOI] [PubMed] [Google Scholar]
- 29.Oishi K, Atsumi G, Sugiyama S, Kodomari I, Kasamatsu M, Machida K, Ishida N. Disrupted fat absorption attenuates obesity induced by a high-fat diet in Clock mutant mice. FEBS Lett. 2006;580:127–130. doi: 10.1016/j.febslet.2005.11.063. [DOI] [PubMed] [Google Scholar]
- 30.Patel DD, Knight BL, Wiggins D, Humphreys SM, Gibbons GF. Disturbances in the normal regulation of SREBP-sensitive genes in PPAR alpha-deficient mice. J. Lipid Res. 2001;42:328–337. [PubMed] [Google Scholar]
- 31.Kudo T, Kawashima M, Tamagawa T, Shibata S. Clock mutation facilitates accumulation of cholesterol in the liver of mice fed a cholesterol and/or cholic acid diet. Am. J. Physiol. Endocrinol. Metab. 2008;294:E120–E130. doi: 10.1152/ajpendo.00061.2007. [DOI] [PubMed] [Google Scholar]
- 32.Oishi K, Amagai N, Shirai H, Kadota K, Ohkura N, Ishida N. Genome-wide expression analysis reveals 100 adrenal gland-dependent circadian genes in the mouse liver. DNA Res. 2005;12:191–202. doi: 10.1093/dnares/dsi003. [DOI] [PubMed] [Google Scholar]
- 33.Desvergne B, Wahli W. Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocr. Rev. 1999;20:649–688. doi: 10.1210/edrv.20.5.0380. [DOI] [PubMed] [Google Scholar]
- 34.Mazzucotelli A, Viguerie N, Tiraby C, Annicotte JS, Mairal A, Klimcakova E, Lepin E, Delmar P, Dejean S, Tavernier G, Lefort C, Hidalgo J, Pineau T, Fajas L, Clement K, Langin D. The transcriptional coactivator peroxisome proliferator activated receptor (PPAR)gamma coactivator-1 alpha and the nuclear receptor PPAR alpha control the expression of glycerol kinase and metabolism genes independently of PPAR gamma activation in human white adipocytes. Diabetes. 2007;56:2467–2475. doi: 10.2337/db06-1465. [DOI] [PubMed] [Google Scholar]
- 35.Oishi K, Shirai H, Ishida N. CLOCK is involved in the circadian transactivation of peroxisome-proliferator-activated receptor alpha (PPARalpha) in mice. Biochem. J. 2005;386:575–581. doi: 10.1042/BJ20041150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Djouadi F, Weinheimer CJ, Saffitz JE, Pitchford C, Bastin J, Gonzalez FJ, Kelly DP. A gender-related defect in lipid metabolism and glucose homeostasis in peroxisome proliferator- activated receptor alpha- deficient mice. J. Clin. Invest. 1998;102:1083–1091. doi: 10.1172/JCI3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kita Y, Shiozawa M, Jin W, Majewski RR, Besharse JC, Greene AS, Jacob HJ. Implications of circadian gene expression in kidney, liver and the effects of fasting on pharmacogenomic studies. Pharmacogenetics. 2002;12:55–65. doi: 10.1097/00008571-200201000-00008. [DOI] [PubMed] [Google Scholar]
- 38.Lowell BB, Spiegelman BM. Towards a molecular understanding of adaptive thermogenesis. Nature. 2000;404:652–660. doi: 10.1038/35007527. [DOI] [PubMed] [Google Scholar]
- 39.Inoue I, Shinoda Y, Ikeda M, Hayashi K, Kanazawa K, Nomura M, Matsunaga T, Xu H, Kawai S, Awata T, Komoda T, Katayama S. CLOCK/BMAL1 is involved in lipid metabolism via transactivation of the peroxisome proliferator-activated receptor (PPAR) response element. J. Atheroscler. Thromb. 2005;12:169–174. doi: 10.5551/jat.12.169. [DOI] [PubMed] [Google Scholar]
- 40.Cunha RA, Brendel P, Zimmermann H, Ribeiro JA. Immunologically distinct isoforms of ecto-5′-nucleotidase in nerve terminals of different areas of the rat hippocampus. J. Neurochem. 2000;74:334–338. doi: 10.1046/j.1471-4159.2000.0740334.x. [DOI] [PubMed] [Google Scholar]
- 41.Lee CC. Constant darkness is a mammalian biological signal. Cold Spring Harb. Symp. Quant. Biol. 2007:287–291. doi: 10.1101/sqb.2007.72.051. [DOI] [PubMed] [Google Scholar]
- 42.Forman BM, Chen J, Evans RM. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors alpha and delta. Proc. Natl. Acad. Sci. USA. 1997;94:4312–4317. doi: 10.1073/pnas.94.9.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shapiro DJ, Rodwell VW. Diurnal variation and cholesterol regulation of hepatic HMG-CoA reductase activity. Biochem. Biophys. Res. Commun. 1969;37:867–872. doi: 10.1016/0006-291x(69)90972-3. [DOI] [PubMed] [Google Scholar]
- 44.Fontaine C, Staels B. The orphan nuclear receptor Rev-erbalpha: a transcriptional link between circadian rhythmicity and cardiometabolic disease. Curr. Opin. Lipidol. 2007;18:141–146. doi: 10.1097/MOL.0b013e3280464ef6. [DOI] [PubMed] [Google Scholar]